Identification of candidate piRNAs in the gonads of Paralichthys olivaceus (Japanese flounder)
2016-11-15ChunLeiWANGZhiPengWANGJiaQiWANGMingYouLiXiaoWuCHEN
Chun-Lei WANG, Zhi-Peng WANG, Jia-Qi WANG, Ming-You Li, Xiao-Wu CHEN
Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai 201306, China
Identification of candidate piRNAs in the gonads of Paralichthys olivaceus (Japanese flounder)
Chun-Lei WANG, Zhi-Peng WANG, Jia-Qi WANG, Ming-You Li, Xiao-Wu CHEN*
Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai 201306, China
Piwi-interacting RNA (piRNA) plays an important role in the gonadal development and maintenance of Teleostei. In this study, piRNA libraries derived from the adult gonads of Japanese flounder (Paralichthys olivaceus) were generated using next-generation sequencing technology. Using zebrafish piRNAs as a reference, 5 865 unique candidate piRNAs were identified; 289 candidate piRNA clusters (PRCs)were generated from the above piRNAs. Among the isolated candidate PRCs, a total of 38 ovary-specific,45 ovary-bias, 24 testis-specific, and 131 testis-bias PRCs were found. The relative expression levels of seven PRCs were validated through quantitative reverse transcription-polymerase chain reaction. The results of this study will help facilitate exploration of the development and maintenance of the phenotypic sex mechanism in P. olivaceus.
Paralichthys olivaceus; Ovary; Testis;piRNA
lNTRODUCTlON
In mammals, the corresponding protein of piwi-interacting RNA(piRNA) interacts with a class of germ cell-restricted small RNAs (26-31 nucleotides in length) in the gonads. piRNAs show distinctive localization patterns in the genome and are predominantly grouped into 20-90-kilobase clusters, whereas the long stretches of small RNAs are derived from only one strand. In humans and rats, with major clusters occurring in syntenic locations, both the abundance of piRNAs in germline cells and male sterility of Piwi mutants suggest the possible role of piRNAs in gametogenesis (Aravin et al., 2006; Girard et al.,2006). In zebrafish (Danio rerio), the piwi gene is expressed in both the testis and ovary and is also a component of a germline structure called “nuage”. The loss of piwi function results in a progressive loss of germ cells due to apoptosis during larval development. Many of these small RNAs are derived from transposons; therefore, piRNAs are possibly involved in the silencing of repetitive elements in gonads (Houwing et al.,2007). In general, genome-wide piRNA expression and function have been examined in fruit flies (Drosophilidae) (Aravin et al.,2007), humans (Homo sapiens) (Sigurdsson et al., 2012), mice(Mus musculus) (Zheng et al., 2010), zebrafish (Houwing et al.,2007; Huang et al., 2011), and planaria (Prosorhochmus claparedii) (Palakodeti et al., 2008). Furthermore, piRNAs are found in clusters throughout the genome, and each cluster may contain as few as 10 or up to thousands of piRNAs. The clustering of piRNAs is highly conserved across species,whereas the sequences are not (Malone et al., 2009).1
The gender of Japanese flounder (Paralichthys olivaceus) is easily altered by water temperature during the sex determination stage (Wen et al., 2014). Female P. olivaceus grow faster than males; thus, investigations on the mechanisms of sex differentiation can be potentially useful in increasing production of farmed P. olivaceus. Study on sex manipulation in P. olivaceus has been a topic of research for some time. Sexrelated genes have been studied to elucidate the sexual molecular mechanism in flounder, and include the cyp19a gene,which was first cloned and found to be predominantly expressed in female flounders (Kitano et al., 1999); some transcription factors, such as foxl2 (Yamaguchi et al., 2007),dmrt1 (Wen et al., 2014), and dmrt4 (Wen et al., 2009), and other sex-related genes, such as cyp17 (Ma et al., 2012). However, due to the limited data on these genes, information regarding their expression and regulation mechanisms is also incomplete. As a representative of non-coding RNA,approximately 20.0% and 13.1% of isolated microRNAs(miRNAs) are preferentially expressed in the testis and ovary,respectively, which indicates that miRNAs might play a key role in the regulation of gene expression in P. olivaceus gonads (Guet al., 2014). However, the differential expression and function of piRNAs in fish sex development and maintenance remain poorly investigated. Identifying the expression patterns of piRNAs in P. olivaceus gonads is the first step in clarifying their function in gonadal development. Thus, this study aimed to identify candidate piRNAs in sexually mature gonads of P. olivaceus through next-generation sequencing, which provides not only sequences of low-abundance piRNAs, but also quantitative data on the frequency of sequencing reads that correspond to the abundance of piRNAs in the ovary and testis.
MATERlALS AND METHODS
Fish maintenance
One-year-old P. olivaceus fish with average individual body weights of 450-500 g were obtained from the Beidaihe Central Aquaculture Experiment Station, Chinese Academy of Fishery Sciences. All fish were acclimatized to indoor recirculation systems at the Shanghai Ocean University for two weeks. The gonads were examined through histological sectioning before RNA extraction, and only healthy fish were chosen for RNA isolation. All experiments were performed according to the Experimental Animal Management Law of China and as approved by the Animal Ethics Committee of Shanghai Ocean University.
Histology and RNA isolation
All fish were killed by overdose with tricaine methanesulfonate(MS-222, 200 mg/L). Mature ovaries and testes were collected separately from five females and five males. For histology, the gonads of females and males were fixed with Bouin's solution,then embedded in paraffin (Sigma Aldrich, http://www.sigmaaldrich.com). Cross sections were cut at 5 μm and stained with hematoxylin and eosin. Tissues were isolated,immediately frozen in liquid nitrogen, and stored at -80°C. Total RNA was extracted using a mirVana™ miRNA isolation kit(Ambion; http: //www.thermofisher.com/cn/zh/home/brands/ invitrogen/ambion.html) in accordance with the manufacturer's instructions. Each testis or ovary was separately subjected to RNA extraction. Five RNA samples (1.5 μg per sample) derived from the same type of gonad were pooled for library construction and subsequent sequencing. The quantity and purity of total RNA were monitored using Nanodrop ND-1000(Nanodrop Technologies; http: //www.thermoscientific.com/content/ tfs/en/product/nanodrop-lite-spectrophotometer.html) and denaturing gel electrophoresis. The total RNA was stored at -80°C.
Small RNA library construction and sequencing
Two small complementary DNA (cDNA) libraries were generated from the mixture of total RNAs of mature testes and ovaries, respectively. A combination of RNA samples was used to avoid possible expression bias of piRNAs in different individuals. A small RNA fraction (15-31 nt) was isolated using 15% polyacrylamide/8 mol/L urea/0.5×Tris/borate/ ethylenediaminetetraacetic acid gel electrophoresis and ligated with proprietary 5′ (GTTCAGAGTTCTACAGTCCGACGATC)and 3′ (TCGTATGCCGTCTTCTGCTTGT) adaptors using T4 RNA ligase. The short RNAs were then converted to cDNA through reverse transcription polymerase chain reaction (RTPCR). The results were verified through Pico green staining and fluorospectrophotometry and then quantified using Agilent 2100;afterward, the cDNA libraries were sequenced using HiSeq2000(Illumina; http: //www.illumina.com/) in accordance with the manufacturer's recommended protocols for small RNA sequencing (NEXTflextmSmall RNA-seq kits; BIOO Scientific Corp; http: //www.biooscientific.com/).
Bioinformatics analysis of sequencing data
Raw sequences were filtered to remove low-quality reads(sequences with more than 10% sequencing errors), and those that passed the quality filter were trimmed to remove the adaptor sequences. Sequences with 15-31 nt in the two combined libraries were then selected as raw small RNA sequences. The raw small RNA sequences were mapped to the piRNA database (Lakshmi & Agrawal, 2008) with zebrafish piRNAs used as the templates. The 5 865 candidate piRNAs are listed in Supplemental Table 1. Highly conserved unique piRNAs with different lengths were placed in the same piRNA clusters (PRCs), and total reads of all unique piRNAs were considered as the reads of the PRCs. The expression levels of the PRCs were estimated using RPKM (reads per kilobase of exon model per million mapped reads). Differential expression analyses of the two samples were performed using the DESeq R package (Anders et al., 2013). P-values were adjusted using the Benjamini and Hochberg method. A corrected P-value of 0.05 was set as the threshold for significantly differential expressions. The P-values were also adjusted using Q-values;Q-value <0.01 and log2(fold change) >1 were set as the threshold for significantly differential expressions by default(Storey, 2003).

Table 1 Primers used for piRNA qRT-PCR analysis
Quantitative RT-PCR analysis of piRNAs
The expression profiles of nine PRCs were investigated with real time RT-PCR using a miScript reverse transcription kit and miScript SYBR Green PCR kit (Qiagen: http: //www. transmedchina.com/) in accordance with the manufacturers' instructions. Quantitative RT-PCR (qRT-PCR) was analyzed using iCycler (Bio-Rad; http: //www.bio-rad.com/). The 20 μL reaction mixture consisted of 10 μL of SYBR Premix Taq (2×),0.4 μL of piRNA-specific forward primer (10 μmol/L), 0.4 μL of miScript universal primer (10 μmol/L), and 1 μL of PCR template (cDNA). The amplification protocol was as follows:initial denaturation and enzyme activation for 3 min at 95°C,followed by 40 cycles for 5 sec at 95°C, and for 20 sec at 60°C. Fluorescence was read at the end of each round of amplification. Each assay was performed in duplicate. The relative expression levels of one piRNA in the ovary and testis were determined using themethod, and different piRNAs in the same tissue were determined using themethod (Pfaffl, 2001). The 5S small ribosomal RNA of P. olivaceus was used as the internal control, and piRNA_0002790 was used as the calibrator. The primers of each piRNA are listed in Table 1, with the most conserved sequence of each PRC selected as the primer. The groups were compared via one-way analysis of variance (ANOVA) and Tukey's test to identify statistically distinct groups. Significant differences were accepted using P<0.05.
RESULTS
Histological observation and identification of piRNAs of gonads In the ovary of P. olivaceus, only mature oocytes or those close to maturity were revealed. The previtellogenic oocytes displayed strongly basophilic ooplasms, while mature oocytes were strongly eosinophilic. In the testis, tubules were filled with spermatozoids, while the numbers of spermatogonia and spermatocytes were reduced (Figure 1).
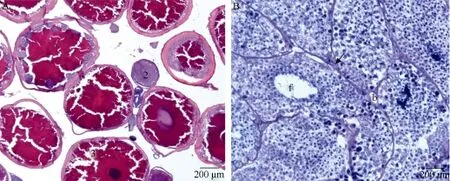
Figure 1 Histological features of the ovary and testis of one-year-old P. olivaceusA: Ovary. 1, Stage III; 2, Stage VI; 3, Stage V. B: Testis. b, blood vessel; f: flagella; arrow indicates interstitial cells.
Combined RNA samples from different ovary and testis samples were used for sequencing. A total of 24 399 395 and 26 417 330 raw reads were obtained from the ovary and testis libraries, respectively. After filtering low-quality sequences,empty adaptors, and single-read sequences, 12 246 743(50.19%) and 13 260 806 (50.19%) clean reads were used for further sequence analyses. After assembly and BLAST searching in the piRNA database, 5 181 and 3 915 unique piRNA sequences showing high homology with zebrafish were obtained from the two libraries. All unique piRNAs were grouped into 289 piRNA clusters (PRCs), where all unique piRNAs in the same PRC shared a conserved sequence with varying lengths. The remaining nucleotides were similar to other types of RNAs, including ncRNA, tRNA, miRNA, rRNA, snRNA,and snoRNA. The size distribution of small RNAs for sequencing was similar between the two libraries. The majority of these small RNAs increased from 18 nt to 30 nt, and lengths ranged from 23 nt to 28 nt (Figure 2A).
Expression patterns of PRCs in P. olivaceus gonads
The relative abundance of various PRCs was determined using the small RNA deep sequencing approach by calculating sequencing frequency. In the ovary and testis libraries, the identified PRCs exhibited a broad range of expression levels. The most abundant PRC in the mature gonads was piR_0000766 (which displayed more than 12 000 reads in the ovary and testis), followed by piR_0034879 (10 766 reads). In contrast, 87 piRNAs displayed less than 10 reads. These results indicate that expression varied significantly among different piRNA families.
Differential expression of PRCs in the ovary and testis
On the basis of changes in the relative piRNA abundance between the two libraries, we found thatof the expressed PRCs varied from -7.65 to 4.89. Approximately 45.32% and 15.57% of isolated PRCs were preferentially expressed in the testis and ovary, respectively. Results showed a total of 289 PRCs, including 38 ovary-specific, 45 ovary-bias,24 testis-specific and 131 testis-bias PRCs (Figure 2B). Seven PRCs (piR_0048450, piR_0006070, piR_0019030,piR_0015958, piR_0055443, piR_0056629, and piR_0050867)with different reads obtained from sequencing results were selected and quantified using real-time RT-PCR to validate the results obtained from the Illumina deep sequencing data. These PRCs were also homologues, according to the piBASE database (http: //www.regulatoryrna.org/database/piRNA/). Three individual experiments using different RNA pools were performed to generate reliable and repeatable results. Results showed that PRCs, including piRNR_0048450, piRNA_0006070,piRNA_0019030, and piRNA_0015958, exhibited high expression levels in both the testis and ovary, though were more highly expressed in the testis (P<0.05). Three other PRCs(piRNA_0055443, piRNA_0056629, and piRNA_0050867)were primarily detected in the ovary (P<0.05). These results areconsistent with the sequencing data (Figure 3).

Figure 2 Length range and differential expression of piRNA in the gonads of P. olivaceusA: Length distribution of sequenced small RNAs from the ovary and testis of P. olivaceus. X-axis denotes the piRNA length of 26-31 nucleotides; Y-axis represents the percentage of piRNA. B: Scatter plot expression level of piRNA in testis versus ovary. Genes are listed in Supplemental Table 2.
DlSCUSSlON
As the largest group of vertebrates, teleosts exhibit remarkably varied sexuality (Godwin, 2010). Genetic, environmental, and social factors can affect sexual development in a large number of fish species at different developmental stages (Heule et al.,2014). Paralichthys olivaceus, which is a common flatfish raised in China, Korea, and Japan, exhibits a temperature-sensitive mechanism of gender determination and differentiation. Artificial breeding and selection is performed to facilitate the production of P. olivaceus, and sex control is regarded as an important technique for fish production (Zhu et al., 2006). Although the sexual development of P. olivaceus needs to be properly understood, information concerning the involved mechanism remains insufficient and unclear. One-year-old Japanese flounder are suitable for supply to the market. The gonad is mature and easy to collect, observe and study in regards to gene expression.

Figure 3 Relationship between relative expression levels of piRNA validated by qRT-PCR and fold changes derived using small RNA sequencingExpression levels of piRNA measured with qRT-PCR are depicted with column charts, and triangles or circles represent the fold changes in sequencing. Error bars represent one standard deviation of three different biological replicates. Statistical significance is reported for each piRNA. F: female; M: male; *: P<0.05.
Three processes take place during sexual development, that is, sex determination, differentiation, and maintenance. The first process determines whether the bipotential primordium will develop into a testis or ovary. The second process involves the actual development of the testes or ovaries from the undifferentiated gonads. The final process ensures the reproductive function of the ovary and testis (Freedberg & Taylor, 2007; Hayes, 1998; Spencer & Janzen, 2014). The protein-coding genes involved in the regulatory network of sexual development described so far originate from mammalian or Teleostei studies and can be divided into different functional groups. The dmy gene has been identified in medaka and plays a key role in gender development (Matsuda et al., 2002); other genes, including dmrt3 (Li et al., 2008), dmrt6 (Zhang et al.,2014), gsdf (Shibata et al., 2010), 11β-hydroxylase cyp11b(Schiffer et al., 2015), and 5α-reductase 1, 2, and 3 (Aquila et al., 2015), are also involved in sexual development. In addition,some genes, such as dmy/dmrt1 in medaka, possess different biological functions in different strains (Matsuda et al., 2002). Plasticity is the main characteristic of sexual development,especially for fish. Studies on noncoding genes, including piRNA, microRNA, and lncRNA, can provide insights into sexual identification in fish (Mishima et al., 2008; Gu et al., 2014;Jing et al., 2014; Xiao et al., 2014). Gender-specific organization of piRNA has been discovered in Drosophila (Li et al., 2009; Malone et al., 2009), as have differences between male and female piRNAs in zebrafish (Houwing et al., 2007). Through microfluidic chip analysis, piRNAs have been found to exhibit quantitative differences, ranging from 1.58 to 64 019.26. A subset comprising more than 20% of piRNA exhibited evident differential expression between the testis and ovary (Houwing et al., 2007). We also observed gender-based differential and specific organization of piRNA in P. olivaceus. Results suggestthat the gender-related organization of piRNA might be conserved in vertebrates and flies, and might possess multiple functions in organisms. Piwi-antisense piRNA complexes mediate cleavage of sense piRNA precursors and transposon(or protein-coding) transcripts, which silence transposon and gene expression at the post-transcriptional level (Weick & Miska, 2014). Therefore, study on piRNA is important for illuminating the function of sex development-related proteincoding genes.
piRNAs are involved in the sexual development of many vertebrates, particularly mammals and teleosts (Aravin et al.,2007; Girard et al., 2006; Houwing et al., 2007). However, the related mechanism remains only partially understood because of the lack of information on piRNA expression patterns in the gonads of non-model organisms. Furthermore, identification of piRNAs is difficult because they lack conservative secondary structure motifs, such as the ones found in miRNA (Zhang et al.,2010). Thus, we examined all potential piRNAs in both male and female gonads through deep sequencing technology,followed by BLAST searching the piBANK database with the zebrafish piRNA library as a reference. With a high frequency of 24-28 nt RNA, the deep sequencing results suggested that piRNAs are present in the male and female gonads of P. olivaceus. These findings are consistent with piRNA length distribution in zebrafish. Mammals, including humans, mice,and rats, have a major distribution interval of 27-31 nt (http://pirnabank.ibab.ac.in/stats.html). The crystal structure of the MID domain is obtained from a Piwi/Argonaute protein, which plays a central role in RNA silencing process as essential catalytic components of the RNA-induced silencing complex(RISC). The RISC complex is responsible for the gene silencing phenomenon known as RNA interference. MID docking experiments showed its capability to specifically recognize the 5′-uridine (1U-bias) of piRNAs (Cora et al., 2014). In this study,the ratios of A, T (U), C, and G in the first nucleotides of the 5 865 unique piRNAs of P. olivaceus were 23.24%, 30.75%,22.95%, and 23.05%, respectively, which also characterized 1U-bias of the molecule. The expression levels of PRC in P. olivaceus presented a broad range. For example, more than several thousand reads of piR_0000766 were found in both male and female piRNA libraries, whereas specific infrequent PRCs showed several reads only. We suggest that the imbalance in the expression levels of different piRNAs might correspond to the different abundance levels of their target genes. Hence, the relationship between the expression levels of PRCs and their target genes should be considered in future studies.
Anders S, McCarthy DJ, Chen YS, Okoniewski M, Smyth GK, Huber W,Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and bioconductor. Nature Protocols, 8(9): 1765-1786.
Aquila S, Montanaro D, Guido C, Santoro M, Perrotta I, Gervasi S, De Amicis F, Lanzino M. 2015. Human sperm molecular anatomy: The enzyme 5α-reductase (SRD5A) is present in the sperm and may be involved in the varicocele-related infertility. Histochemistry and Cell Biology, 144(1): 67-76. Aravin AA, Hannon GJ, Brennecke J. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science,318(5851): 761-764.
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N,Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M,Russo JJ, Ju JY, Sheridan R, Sander C, Zavolan M, Tuschl T. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature,442(7099): 203-207.
Cora E, Pandey RR, Xiol J, Taylor J, Sachidanandam R, McCarthy AA,Pillai RS. 2014. The MID-PIWI module of Piwi proteins specifies nucleotideand strand-biases of piRNAs. RNA, 20(6): 773-781.
Freedberg S, Taylor DR. 2007. Sex ratio variance and the maintenance of environmental sex determination. Journal of Evolutionary Biology, 20(1):213-220.
Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germlinespecific class of small RNAs binds mammalian Piwi proteins. Nature,442(7099): 199-202.
Godwin J. 2010. Neuroendocrinology of sexual plasticity in teleost fishes. Front in Neuroendocrinology, 31(2): 203-216.
Gu YF, Zhang L, Chen XW. 2014. Differential expression analysis of Paralichthys olivaceus microRNAs in adult ovary and testis by deep sequencing. General and Comparative Endocrinology, 204: 181-184.
Hayes TB. 1998. Sex determination and primary sex differentiation in amphibians: Genetic and developmental mechanisms. The Journal of Experimental Zoology, 281(5): 373-399.
Heule C, Salzburger W, Bohne A. 2014. Genetics of sexual development:an evolutionary playground for fish. Genetics, 196(3): 579-591.
Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RHA, Hannon GJ,Draper BW, Ketting RF. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell, 129(1): 69-82.
Huang HY, Houwing S, Kaaij LJT, Meppelink A, Redl S, Gauci S, Vos H,Draper BW, Moens CB, Burgering BM, Ladurner P, Krijgsveld J, Berezikov E, Ketting RF. 2011. Tdrd1 acts as a molecular scaffold for Piwi proteins and piRNA targets in zebrafish. The EMBO Journal, 30(16): 3298-3308.
Jing J, Wu JJ, Liu W, Xiong ST, Ma WG, Zhang J, Wang WM, Gui JF, Mei J. 2014. Sex-biased miRNAs in gonad and their potential roles for testis development in yellow catfish. PLoS One, 9(9): e107946.
Kitano T, Takamune K, Kobayashi T, Nagahama Y, Abe SI. 1999. Suppression of P450 aromatase gene expression in sex-reversed males produced by rearing genetically female larvae at a high water temperature during a period of sex differentiation in the Japanese flounder (Paralichthys olivaceus). Journal of Molecular Endocrinology, 23(2): 167-176.
Lakshmi SS, Agrawal S. 2008. PiRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Research, 36(S1):D173-D177.
Li CJ, Vagin VV, Lee S, Xu J, Ma SM, Xi HL, Seitz H, Horwich MD,Syrzycka M, Honda BM, Kittler ELW, Zapp ML, Klattenhoff C, Schulz N,Theurkauf WE, Weng ZP, Zamore PD. 2009. Collapse of germline piRNAs in the absence of argonaute3 reveals somatic piRNAs in flies. Cell, 137(3):509-521.
Li Q, Zhou X, Guo YQ, Shang X, Chen H, Lu H, Cheng HH, Zhou RJ. 2008. Nuclear localization, DNA binding and restricted expression in neural andgerm cells of zebrafish Dmrt3. Biology of the Cell, 100(8): 453-463.
Ma RQ, He F, Wen HS, Li JF, Mu WJ, Liu M, Zhang YQ, Hu J, Qun L. 2012. Polymorphysims of CYP17-I gene in the exons were associated with the reproductive endocrine of Japanese flounder (Paralichthys olivaceus). Asian-Australasian Journal of Animal Sciences, 25(6): 794-799.
Malone CD, Brennecke J, Dus M, Stark A, McCombie WR,Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell, 137(3): 522-535. Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T,Morrey CE, Shibata N, Asakawa S, Shimizu N, Hori H, Hamaguchi S,Sakaizumi M. 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature, 417(6888): 559-563.
Mishima T, Takizawa T, Luo SS, Ishibashi O, Kawahigashi Y, Mizuguchi Y,Ishikawa T, Mori M, Kanda T, Goto T, Takizawa T. 2008. MicroRNA(miRNA) cloning analysis reveals sex differences in miRNA Expression profiles between adult mouse testis and ovary. Reproduction, 136(6): 811-822.
Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. 2008. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA, 14(6): 1174-1186.
Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research, 29(9): e45.
Schiffer L, Anderko S, Hannemann F, Eiden-Plach A, Bernhardt R. 2015. The CYP11B subfamily. The Journal of Steroid Biochemistry and Molecular Biology, 151: 38-51.
Shibata Y, Paul-Prasanth B, Suzuki A, Usami T, Nakamoto M, Matsuda M,Nagahama Y. 2010. Expression of gonadal soma derived factor (GSDF) is spatially and temporally correlated with early testicular differentiation in medaka. Gene Expression Patterns, 10(6): 283-289.
Sigurdsson MI, Smith AV, Bjornsson HT, Jonsson JJ. 2012. The distribution of a germline methylation marker suggests a regional mechanism of LINE-1 silencing by the piRNA-PIWI system. BMC Genetics,13: 31.
Spencer RJ, Janzen FJ. 2014. A novel hypothesis for the adaptive maintenance of environmental sex determination in a turtle. Proceedings of the Royal Society B: Biological Sciences, 281(1789): 20140831.
Storey JD. 2003. The positive false discovery rate: a Bayesian interpretation and the q-Value. The Annals of Statistics, 31(6): 2013-2035. Weick EM, Miska EA. 2014. piRNAs: from biogenesis to function. Development, 141(18): 3458-3471.
Wen AY, You F, Sun P, Li J, Xu DD, Wu ZH, Ma DY, Zhang PJ. 2014. CpG methylation of dmrt1 and cyp19a promoters in relation to their sexual dimorphic expression in the Japanese flounder Paralichthys olivaceus. Journal of Fish Biology, 84(1): 193-205.
Wen AY, You F, Tan XG, Sun P, Ni J, Zhang YQ, Xu DD, Wu ZH, Xu YL,Zhang PJ. 2009. Expression pattern of dmrt4 from olive flounder(Paralichthys olivaceus) in adult gonads and during embryogenesis. Fish Physiology and Biochemistry, 35(3): 421-433.
Xiao J, Zhong H, Zhou Y, Yu F, Gao Y, Luo YJ, Tang ZY, Guo ZB, Guo EY,Gan X, Zhang M, Zhang YP. 2014. Identification and characterization of microRNAs in ovary and testis of Nile tilapia (Oreochromis Niloticus) by using solexa sequencing technology. PLoS One, 9(1): e86821.
Yamaguchi T, Yamaguchi S, Hirai T, Kitano T. 2007. Follicle-stimulating hormone signaling and Foxl2 are involved in transcriptional regulation of aromatase gene during gonadal sex differentiation in Japanese Flounder,Paralichthys olivaceus. Biochemical and Biophysical Research Communications, 359(4): 935-940.
Zhang DP, Duarte-Guterman P, Langlois VS, Trudeau VL. 2010. Temporal expression and steroidal regulation of piRNA pathway genes (mael, piwi,vasa) during Silurana (Xenopus) tropicalis embryogenesis and early larval development. Comparative Biochemistry and Physiology Part C:Toxicology & Pharmacology, 152(2): 202-206.
Zhang T, Murphy MW, Gearhart MD, Bardwell VJ, Zarkower D. 2014. The mammalian Doublesex homolog DMRT6 coordinates the transition between mitotic and meiotic developmental programs during spermatogenesis. Development, 141(19): 3662-3671.
Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A,Sachidanandam R, Pillai RS, Wang PJ. 2010. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proceedings of the National Academy of Sciences of the United States of America, 107(26): 11841-11846.
Zhu XP, You F, Zhang PJ, Xu YL, Xu JH. 2006. Effects of cold shock on microtubule organization and cell cycle in gynogenetically activated eggs of olive flounder (Paralichthys olivaceus). Marine Biotechnology, 8(3): 312-318.
08 April 2016; Accepted: 05 September 2016
s: This research was supported by grants from the China Agriculture Research System (CARS-49), National Natural Science Foundation of China (31372520) and Shanghai Collaborate Innovation Center for Aquatic Animal Genetics and Breeding (ZF1206)
, E-mail: xwchen@shou.edu.cn
10.13918/j.issn.2095-8137.2016.5.301
杂志排行
Zoological Research的其它文章
- Lamprey: a model for vertebrate evolutionary research
- What is the destiny of a threatened fish, Ptychobarbus chungtienensis, now that non-native weatherfishes have been introduced into Bita Lake, Shangri-La?
- Evolution and phylogenetic application of the MC1R gene in the Cobitoidea (Teleostei: Cypriniformes)
- A new species of the genus Triplophysa (Cypriniformes:Nemacheilidae), Triplophysa daochengensis, from Sichuan Province, China
- A new cave species of the Genus Triplophysa from Yunnan, China
- Identification of Sphaeroma terebrans via morphology and the mitochondrial cytochrome c oxidase subunit I(COI) gene
