两个相关基因表达量和SNP与玉米雄穗大小相关
2016-06-23任小丹李晚忱付凤玲
任小丹, 陈 玲, 杨 琳, 李晚忱, 付凤玲
( 四川农业大学 玉米研究所, 成都 611130 )
两个相关基因表达量和SNP与玉米雄穗大小相关
任小丹, 陈玲, 杨琳, 李晚忱, 付凤玲*
( 四川农业大学 玉米研究所, 成都 611130 )
摘要:玉米雄穗通常较发达,散粉量大于授粉需要,过量消耗能量会影响光合产物向果穗的分配,过于发达的雄穗还会影响群体透光性、降低光合效率。生产实践和育种研究证明,由于雄穗大小与玉米籽粒产量负相关,因此成为品种选育的间接选择指标。该研究根据前人的报道,从11个雄穗大小不同的玉米自交系中扩增角蛋白相关蛋白基因KAP5-4和受体样蛋白激酶基因CLV1的基因组序列,多重比较后用以分析其开放阅读框、保守结构和单核苷酸多态性,用荧光实时定量PCR检测其在雄穗原基中的差异表达,并与雄穗分枝数和雄穗干重两个度量雄穗小的指标进行了相关分析。结果表明:KAP5-4基因的相对表达量与雄穗分枝数(r=0.77,P<0.01)和雄穗干重正相关(r=0.83,P<0.01)。11个自交系的CLV1 基因开放框在2 104 bp存在单核苷酸多态性,其中5个自交系的2 014~2 016 bp核苷酸组成密码子GAC,编码受体样蛋白第702位酸性的天冬氨酸,另6个自交系的2 014~2 016 bp核苷酸组成密码子AAC,编码受体样蛋白第702位极性天冬氨酰胺。在前5个自交系中,CLV1 基因的相对表达量与雄穗分枝数(r=-0.92, P<0.01)和雄穗干重(r=-0.91, P<0.05)负相关;而在后6个自交系中,仅与雄穗干重负相关(r=-0.91, P<0.05)。综上所述,KAP5-4和CLV1基因的表达和单核苷酸多态性与玉米雄穗大小关系密切,可开发功能性的DNA标记用于玉米育种的分子标记辅助选择。
关键词:差异表达, 角蛋白伴侣基因, 玉米, 受体类蛋白激酶, 雄穗大小
玉米雄穗通常比较发达,散粉量大于授粉需要,过量消耗能量会影响光合产物向果穗的分配,过于发达的雄穗还会影响群体透光性、降低光合效率(Gue et al,1996)。前人研究表明,雄穗大小与玉米籽粒产量负相关(Lambert & Johnson,1978; Geraldi et al等,1985; Fischer et al,1987; Bodi et al,2008)。玉米生产和育种实践证明,在授粉前部分去除雄穗或选择小雄穗品种,均可显著提高籽粒产量(Subedi,1996; Upadyayula et al,2006; Duvick et al,1999; Neto & Filho,2000)。在美国等玉米生产较为发达的国家和地区,推广玉米杂交种的雄穗一般较小(Duvick & Cassman,1999; Neto & Filho,2000)。而在发展中国家,逆境胁迫是玉米生产的主要限制因素,适当大小的雄穗可保证授粉所需的花粉量,具有一定的稳产作用(Monneveux et al,2008; Tiwari et al,2004)。因此,虽然雄穗大小并不是玉米籽粒产量的构成因素,但在育种中常被用作间接选择指标,一般以雄穗分枝数和雄穗干重来度量。然而,雄穗大小的遗传性质也很复杂,是一个受众多基因调控的数量性状。现已发现ba2(barren stalk 2)、bif2(barren inflorescence 2)、bd1(branched silkless 1)、fe2(fasciated ear 2)、ra1(ramosal 1)、ra2(ramosa 2)、ra3(ramosa 3)、Ts6(Tassel seed 6)、td1(thick tassel dwarf 1)等多个染色体位点,均与玉米雄穗分生组织发育和形态建成有关(Vollbrecht et al,2005; Bommert et al,2005; Bortiri et al,2006; Chuck et al,2002; Irish,1997; Skirpan et al,2009; Taguchi-Shibara et al,2001)。而且,与雄穗分枝数和雄穗重相关的QTL位点也不少(Upadyayula et al,2006; Mickelson et al,2002)。然而,这些染色体或QTL位点对雄穗大小的具体调控关系,却不甚了解。
在水稻上,与穗密度、穗粒数和穗直立性相关的DEP1(Dense and Erect Panicle 1)基因与编码角蛋白相关蛋白5-4(KAP 5-4)的QTL位点qPE9-1等位。进化分析还发现,其他温带谷物也具有相应功能的等位基因,而且早在小麦与大麦分化前就已存在(Huang et al,2009; Kong et al,2007; Yan et al,2007; Zhou et al,2009)。FON1(Floral Organ Number 1)基因。FON1基因与水稻花序分生组织的发育, 以及后续的分枝、小穗和小花增殖相关。该基因与拟南芥编码富含亮氨酸重复受体蛋白激酶的CLV1(CLAVATA1)基因高度同源(Chu et al,2006; Suzaki et al,2004)。在拟南芥上,CLV/WUS信号转导途径被证实参与细胞分裂的调节,进而调控茎端分生组织的增殖和花芽的分化(Dievarta et al,2003; Godiard et al,2003; Schoof et al,2000; Suzaki et al,2006; Torri,2004)。
本研究根据同源比对结果,克隆与水稻DEP1和FON1基因同源的玉米基因KAP5-4和CLV1,检测其在雄穗发育过程中的差异表达,以及在雄穗大小差异明显自交系间的单核苷酸多态性(single nucleotide polymorphism, SNP),分析其与雄穗分枝数和雄穗干重的相关性。
1材料与方法
1.1 田间鉴定与样品制备
2010-2011年,按随机区组设计、3次重复种植雄穗大小及其他农艺性状差异明显的11个玉米自交系。取六叶期脚叶,用DNA quick Plant System试剂盒(Tiangen,北京)提取基因组DNA。在顶端分生组织伸长期,每小区随机取样3株,解剖雄穗原基,用Eeasy Spin Plant RNA Mini Kit试剂盒(Aidlab,北京)提取总RNA,并用PrimeScript RT Reagent Kit试剂盒(TakaRa,大连)立即反转录成cDNA。授粉后2 d,每小区随机抽样5株收获雄穗,调查雄穗分枝数后105 ℃杀青1 h,95 ℃烘干至恒重,调查雄穗干重。
1.2 多重比较和保守结构域预测
从NCBI数据库(http://www.ncbi.nlm.nih.gov/guide/)下载水稻DEP1和FON1基因的推导氨基酸序列GI: 208293844和GI: 75112604),以此为探针从UniProt数据库(http://www.uniprot.org)搜索玉米推导氨基酸序列。先用MEGA 5.05软件(http: www.megasoftware.net/index.php)与模式植物的同源序列进行多重比较, 然后用Sanger Pfam 25.0软件(http://pfam.sanger.ac.uk)预测保守结构域。
1.3 KAP5-4和CLV1基因组序列的SNP分析
根据与水稻DEP1和FON1基因推导氨基酸序列的同源比对结果,分别设计3对(KF1:5′-TCCTCGCTTCCTCATCCAGAC-3′/KR1:5′-GGTAGCCACAGGTTGCCATAA-3′; KF2:5′-GGTTGAAGGGAG-
AAGGCTGAG-3′/KR2:5′-GGTCATACATCGGTATTAGTGGG-3′; KF3:5′-TACCGATGTATGACCTATAC-
TTGAT-3′/KR3:5′-CTTCTGGTGATATGGGATTTCTT-3′)和2对(CF1:5′-TCGGCTCCACTTCCACTGGTCTA-3′/CR1:5′-CGACAACACGATCAATCCCAAAC-3′/CF2:5′-CGTGCTCTTTCACCCTGTCA-3′/CR2:5′-TCATGGTTCCAGAGTCCCG-3′)特异PCR引物,以11个玉米自交系的基因组DNA为模板,用PrimeSTAR HS DNA聚合酶(TakaRa,大连),分段扩增玉米KAP5-4和CLV1基因。PCR温度循环为94 ℃变性1 min;94 ℃变性10 s,按不同引物的退火温度退火30 s,72 ℃延伸2 min,循环35次;72 ℃延伸5 min。扩增产物用7%琼脂糖凝胶分离,Universal DNA纯化试剂盒(Tiangen,北京)回收,用Taq酶(TaKaRa,大连)在3′-端加(A)尾后,插入pMD19-T质粒(TakaRa,大连),分别在Majorbio(上海)和SinoGenoMax(北京)重复测序2次。用NCBI网站(http://www.ncbi.nlm.nih.gov/gorf/gorf.html)的开放阅读框分析软件ORF finder查找开放阅读框,再用DNAMAN软件(http://www.lynnon.com)进行多重比较,分析两个基因在11个自交系间的SNP。
1.4 荧光实时定量PCR
根据测序结果,分别设计特异PCR引物(KF4:5′-CCCACTAATACCGATAACGAAG-3′/KR4: 5′-CA-
ATGGCAGGAACAGCAGA-3′; CF3:5′-TCAGATTGACAACAACAGC-3′/CR3:5′-GGAGAAACGGTACATGGT-3′),用SsoFast EvaGreen Supermix反应体系(Bio-Rad,美国)和iQTM5荧光定量PCR仪(Bio-Rad,美国),分别扩增KAP5-4和CLV1基因的103 bp和75 bp片段,分析雄穗发育中两个基因在11个自交系间的表达差异,并用引物GR(5′-CCATCACTGCCACACAGAAAC-3′)/GR(5′-AGGAACACGGAA-
GGACATACCAG-3′)扩增GAPDH基因片段作内参。10 μL反应体系含SsoFast EvaGreen Supermix 5 μL、稀释cDNA 1 μL和10 pmol/μL 引物各0.5 μL。 温度循环为95 ℃变性30 s;95 ℃变性5 s, 60 ℃(KAP5-4)/ 59 ℃(CLV1)延伸10 s,循环40次。最后一次循环后,将温度以0.5 ℃ / 10 s的速度升至95 ℃,计算标准曲线,检测扩增产物的特异性。iQ 5系统(Bio-Rad,美国)自带软件以绝对表达量最低的样品为标准),计算其他样品的相对表达量(Vandesompele et al,2002)。
2结果与分析
2.1 玉米KAP 5-4和CLV1基因的同源性比较
KAP5-4基因的开放阅读框长1 227 bp,编码蛋白长408个氨基酸。多重比较表明,KAP5-4基因的编码蛋白与水稻DEP1基因(gi: 208293844)的推导氨基酸序列具有51%的同源性。进化树分析表明,植物KAP5-4蛋白质家族可分为单子叶和双子叶两个明显的亚家族(图1)。第26至第98位氨基酸的多肽序列包含一个G-蛋白γ-样结构域,在物种间高度保守(图2)。
CLV1基因的开放阅读框长2 619 bp,编码蛋白长872个氨基酸,包含1个富含亮氨酸重复的胞外结构域、1个小的跨膜结构域和1个细胞质丝氨酸/苏氨酸蛋白激酶结构域(图3)。多重比较表明,其推导氨基酸序列与高粱(gi: 242073424)和大麦(gi: 326531976)受体样蛋白激酶1(CLV1)高度同源,但与水稻FON1基因(gi: 75112604)推导蛋白的氨基酸序列却只有31%的同源性。 进化树分析表明,12个植物物种的CLV1基因间没有明显的亚家族划分(图4)。
2.2 CLV1基因基因组序列SNP
通过多重比较,在11个自交系间未发现KAP5-4基因的基因组序列内存在一致性的SNP。但在11个自交系CLV1基因的基因组序列第2 104 bp存在SNP。自交系“郑58”、“Mo17”、“178”、“478”和“自330”第2 104 bp碱基为鸟嘌呤(G),组成密码子GAC,编码的702位氨基酸为天冬氨酸。自交系“R08”、“昌7-2”、“丹340”、“苏37”、“ES40”和“RP125”第2 104 bp碱基为腺嘌呤(A),组成密码子AAC,编码的702位氨基酸为天冬酰胺(图5)。根据图3所示的结构预测,702位氨基酸正是CLV1蛋白丝氨酸/苏氨酸蛋白激酶结构域的质子受体。
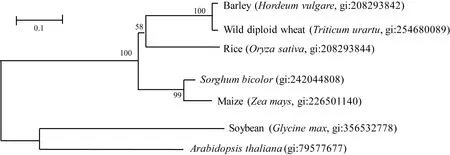
图 1 玉米与其他6种植物角质蛋白相关蛋白5-4(KAP5-4)进化树Fig. 1 Phylogenetic tree of keratin-associated protein 5-4 (KAP 5-4) among maize and six other plant species
2.3 KAP 5-4和CLV1基因的差异表达及其与雄穗大小的相关性
KAP5-4和CLV1基因在雄穗原基中的相对表达量在11个自交系间差异明显(图6,图7)。值得注意的是,CLV1基因在第2 104 bp碱基为鸟嘌呤(G)的5个自交系中的相对表达量,均低于其他6个第2 104 bp碱基为腺嘌呤(A)的自交系。可能是该位点的碱基替代引起了基因表达的下调,或是酸性天冬氨酸与极性天冬酰胺的差异对基因表达的反馈调节。
相关分析表明,KAP5-4基因在伸长期雄穗原基中的相对表达量,与雄穗分枝数(r=0.77,P<0.01)和雄穗干重(r=0.83,P<0.01)正相关。然而,CLV1基因相对表达量与雄穗分枝数(r=-0.17,P>0.05)和雄穗干重(r=-0.25,P>0.05)的相关性不显著。这一基因序列SNP造成氨基酸序列第702位酸性天冬氨酸与极性天冬酰胺的差异,可能影响其质子受体的活性(图5)。因此,将第702位氨基酸为天冬氨酸的5个自交系和第702位氨基酸为天冬酰胺的6个自交系,分别对CLV1基因相对表达量与雄穗大小进行相关分析。在自330、478、郑58、Mo17和178五个自交系中,CLV1基因相对表达量与雄穗分枝数(r=-0.92,P<0.01)和雄穗干重(r=-0.91,P<0.05)负相关。在丹340、昌7-2、RP125、苏37、ES40和R08六个自交系中,CLV1基因相对表达量与雄穗干重(r=-0.91,P<0.05)负相关,而与雄穗分枝数的相关性仍不显著(r=-0.62,P>0.05)。
3讨论
KAP5-4基因首先发现于哺乳动物,是KAPty家族5的成员,编码角质蛋白相关蛋白,是发基质的组成成分(Wu et al,2008)。在水稻上发现,与KAP5-4同源的DEP1基因与穗大小密切相关(Huang et al,2009; Kong et al,2007; Yan et al,2007; Zhou et al,2009)。The feedback regulation of another ortholog 与CLV1同源的CLV3基因发现有反馈调节现象,而且与拟南芥顶端分生组织的增殖和分化有关(Brand et al,2000)。在本研究中,KAP5-4基因的相对表达量与雄穗大小正相关(图6)。根据结构域预测,玉米及其他6种植物的KAP5-4蛋白,均包含一个高度保守的G-蛋白γ-样结构域。这些结果说明,KAP5-4蛋白在植物中可能是一种调节蛋白,而不是发基质的组成成分。
本研究克隆的CLV1基因,位于第10染色体第125009858至125013205 bp。在第5染色体第121675658至121681684 bp,还有另外一个同源基因(GRMZM2G300133),通过重穗型突变鉴定为td1(thick tassel dwarf1)。这一突变体的雄穗顶端分生组织发育不良,造成穗轴缩短、小穗增密、颖片和雄蕊增生等畸形(Bommert et al,2005)。在本研究中,11个雄穗大小不同自交系的CLV1基因相对表达量与其雄穗大小相关性不显著,但根据其2 104 bp的SNP将11个自交系分为两组后再作相关分析,却发现存在显著的负相关关系(图7),说明CLV1基因可能与玉米雄穗发育存在密切关系。拟南芥与CLV1同源的基因,编码富含亮氨酸重复的受体样蛋白激酶,参与CLV/WUS信号转导,通过对细胞分裂的负调控,对茎顶端分生组织的细胞增殖和分化起重要调控作用(Dievarta et al,2003; Godiard et al,2003; Schoof et al,2000; Suzaki et al,2006; Torri ,2004; Trotochaud et al,1999)。水稻与CLV1同源的FON1基因,也发现参与花序分生组织的发育,以及后续的分枝、小穗和小花分生组织的增殖(Chu et al,2006; Suzaki et al,2004)。
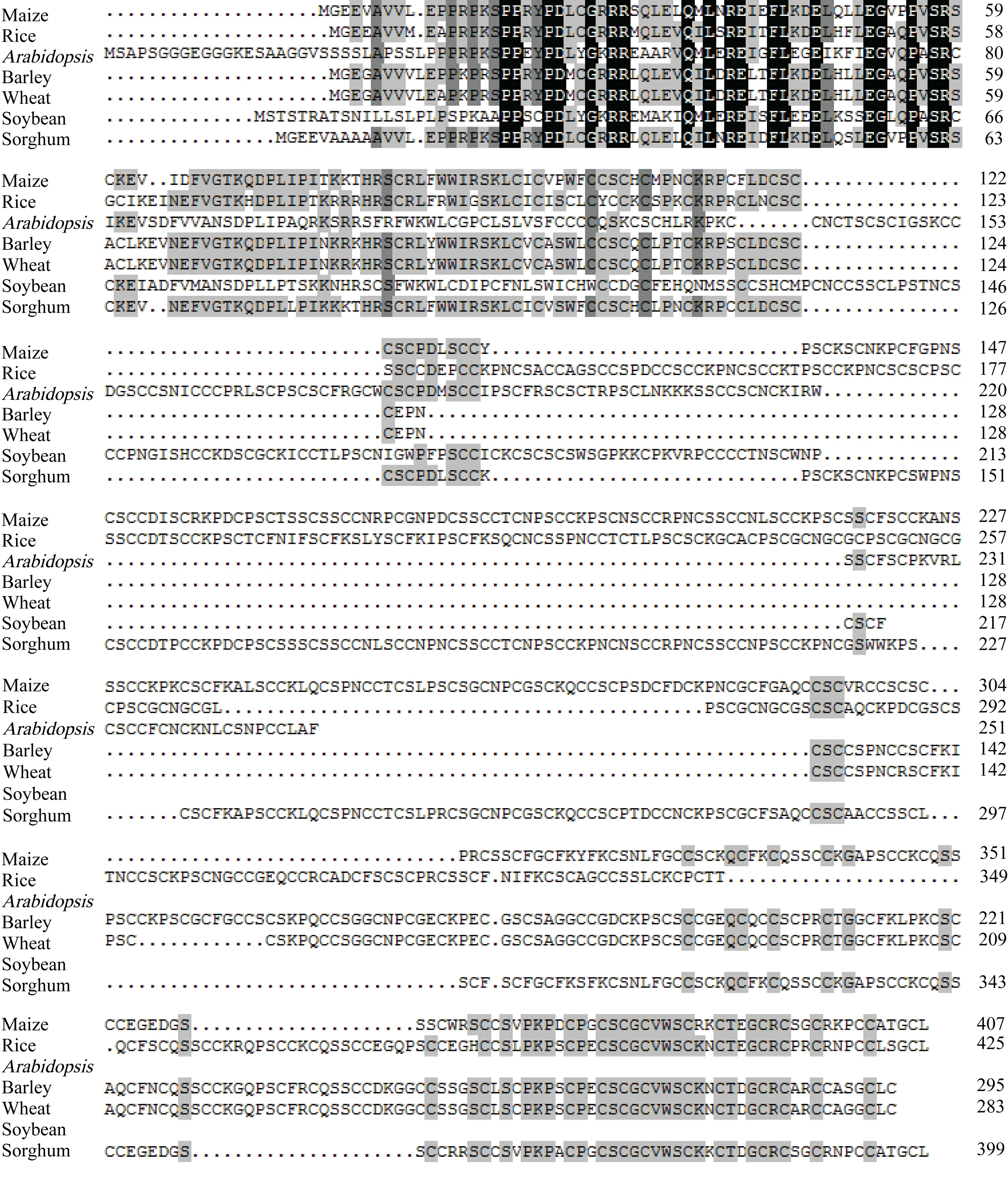
图 2 玉米与其他6种植物角质蛋白相关蛋白5-4(KAP5-4)的氨基酸序列多重比较 黑色背景表示完全(100%)保守; 深灰背景表示高度(75%)保守; 浅灰背景表示中度(50%)保守; 白色背景表示低度(25%)保守。Fig. 2 Muliple alignment of amino acid sequence of KAP 5-4 protein in maize and other six plant species Black background indicates completely conservative (100%); Dark grey background indicates highly conservative (75%); Light grey background indicates moderate conservative (50%); White background indicates low conservative (25%).
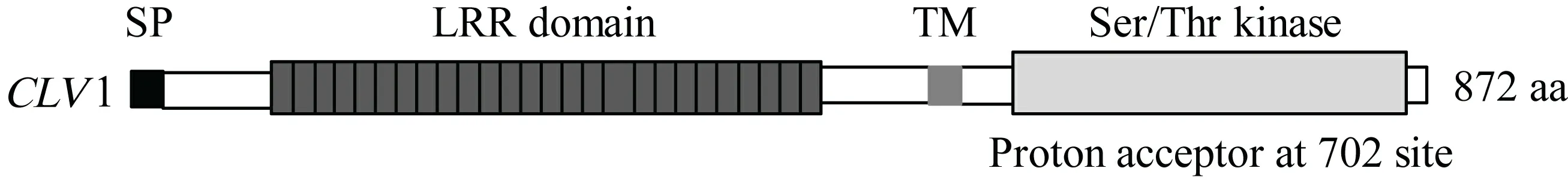
图 3 玉米CLV1蛋白的域结构 SP: 信号肽; LRR domain: 富含亮氨酸重复结构域; TM: 跨膜结构域; Ser/Thr kinase: 丝氨酸/苏氨酸蛋白激酶。Fig. 3 Domain structure of maize CLV1 protein SP. Signal peptide; LRR domain. Leucine-rich repeat domain; TM. Transmembrane domain; Ser/Thr kinase. Serine/threonine protein kinase.
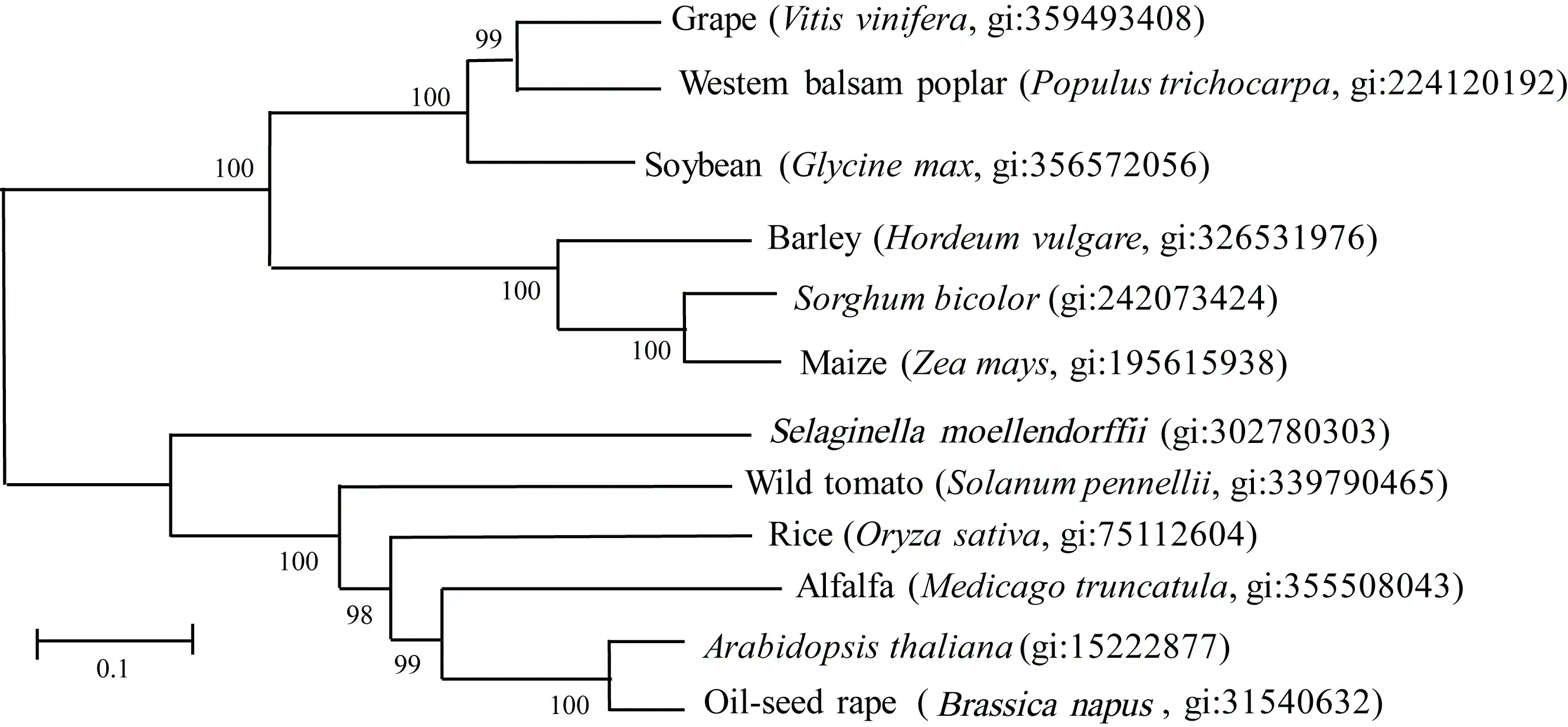
图 4 玉米与其他7种植物受体样蛋白激酶1(CLV1)进化树Fig. 4 Phylogenetic tree of receptor-like protein kinase gene CLV1 among maize and seven other plant species
参考文献:
BODI Z, PEPO P, KOVACS A, 2008. Morphology of tassel components and their relationship to some quantitative features in maize [J]. Cereal Res Comm, 36(2): 353-360.
BOMMERT P, LUNDE C, NARDMANN J, et al, 2005. Thick tassel dwarf1 encodes a putative maize ortholog of theArabidopsisCLAVATA1 leucine-rich repeat receptor-like kinase [J]. Development, 132(6): 1 235-1 245.
BORTIRI E, CHUCK G, VOLLBRECHT E, et al, 2006. Ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize [J]. Plant Cell, 18(3): 574-585.
BRAND U, FLETCHER JC, HOBE M, et al, 2000. Dependence of stem cell fate inArabidopsison a feedback loop regulated by CLV3 activity [J]. Science, 289(5479): 617-619.
CHUCK G, MUSZYNSKI M, KELLOGG E, et al, 2002. The control of spikelet meristem identity by the branched silkless1 gene in maize [J]. Science, 298(5 596): 1 238-1 241.
CHU HW, QIAN Q, LIANG WQ, et al, 2006. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice [J]. Plant Physiol, 142(3): 1 039-1 052.
DUVICK DN, CASSMAN KG, 1999. Post-green revolution trends in yield potential of temperate maize in the north-central United States [J]. Crop Sci, 39(6): 1 622-1 630.
DIEVART A, DALAL M, TAX FE, et al, 2003. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development [J]. Plant Cell, 15(5): 1 198-1 211.
FISCHER KS, EDMEADES GO, JOHNSON EC, 1987. Recurrent selection for reduced tassel branch number and reduced leaf area density above the ear in tropical maize populations [J]. Crop Sci, 27(6): 1 150-1 156.GUEI RG, WASSOM CE, 1996. Genetic analysis of tassel size and leaf senescence and their relationships with yield in two tropical lowland maize populations [J]. Afr Crop Sci J, 4(3): 275-281.
GERALDI IO, FILHO JBM, VENCOVSKY R, 1985. Estimates of genetic parameters of tassel characters in maize (Zea mays L) and breeding perspectives [J]. Maydica, 30(1): 1-14.
GODIARD L, SAUBIAC L, TORRI KU, et al, 2003. ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt [J]. Plant J, 36(3): 353-365.
HUANG X, QIAN Q, LIU Z, et al, 2009. Natural variation at the DEP1 locus enhances grain yield in rice [J]. Nat Gen, 41(4): 494-497.
IRISH EE, 1997. Experimental analysis of tassel development in the maize mutant Tassel Seed 6 [J]. Plant Physiol, 114(3): 817-825.
KONG FN, WANG JY, ZOU JC, et al, 2007. Molecular tagging and mapping of the erect panicle gene in rice [J]. Mol Breed, 19(4): 297-304.
LAMBERT RJ, JOHNSON RR, 1978. Leaf angle, tassel morphology, and the performance of maize hybrids [J]. Crop Sci, 18(3): 499-502.
MONNEVEUX P, SANCHEZ C, TIESSEN A, 2008. Future progress in drought tolerance in maize needs new secondary traits and cross combinations [J]. Agric Sci, 146(3): 287-300.
MICKELSON SM, STUBER CS, SENIOR L, et al, 2002. Quantitative trait loci controlling leaf and tassel traits in a B73×Mo17 population of maize [J]. Crop Sci, 42(6): 1 902-1 909.
NETO ALF, FILHO JBM, 2000. Inbreeding in two maize subpopulations selected for tassel size [J]. Sci Agric, 57(3): 487-490.
SUBEDI KD, 1996. Effect of leaf stripping, de-tasselling and topping of maize on the yield of maize and relay intercropped finger millet [J]. Exp Agric, 32(1): 57-61.
SKIRPAN A, CULLER AH, GALLAVOTTI A, et al, 2009. BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development [J]. Plant Cell Physiol, 50(3): 652-657.SUZAKI T, SATO M, ASHIKARI M, et al, 2004. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous toArabidopsisCLAVATA1 [J]. Development, 131(22): 5 649-5 657.
SCHOOF H, LENHARD M, HAECKER A, et al, 2000. The stem cell population ofArabidopsisshoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes [J]. Cell, 100(6): 635-644.
SUZAKI T, TORIBA T, FUJIMOTO M, et al, 2006. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene [J]. Plant Cell Physiol, 47(12): 1 591-1 602.
TIWARI TP, BROOK RM, SINCLAIR FL, 2004. Implications of hill farmers′ agronomic practices in Nepal for crop improvement in maize [J]. Exp Agric, 40(4): 397-417.
TAGUCHI-SHIBARA F, YUAN Z, HAKE S, et al, 2001. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize [J]. Gen Dev, 15(20): 2 755-2 766.
TORRI KU, 2004. Leucine-rich repeat receptor kinases in plants: structure, function and signal transduction pathways [J]. Int Rev Cytol, 234: 1-46.
TROTOCHAUD AE, HAO T, WU G, et al, 1999. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein [J]. Plant Cell, 11(3): 393-405.
UPADYAYULA N, DA SILVA HS, BOHN MO, et al, 2006. Genetic and QTL analysis of maize tassel and ear inflorescence architecture [J]. Theor Appl Genet, 112(4): 592-606.
VOLLBRECHT E, SPRINGER PS, GOH L, et al, 2005. Architecture of floral branch systems in maize and related grasses [J]. Nature, 436(7 054): 1 119-1 126.
VANDESOMPELE J, PRETER KD, PATTYN F, et al, 2002. Accurate normalization of real time quantitative RT-PCR data by geometric averaging of multiple internal control genes [J]. Gen Biol, 3(7): Research0034- Research0034.11.WU DD, IRWIN DM, ZHANG YP, 2008. Molecular evolution of the keratin associated protein gene family in mammals, role in the evolution of mammalian hair [J]. BMC Evol Biol, 8(1): 241-256.
YAN CJ, ZHOU JH, YAN S, et al, 2007. Identification and characterization of a major QTL responsible for erect panicle trait in japonica rice (OryzasativaL.) [J]. Theor Appl Genet, 115(8): 1 093-1 100.ZHOU Y, ZHU J, LI Z, et al, 2009. Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication [J]. Genetics, 183(1): 315-324.
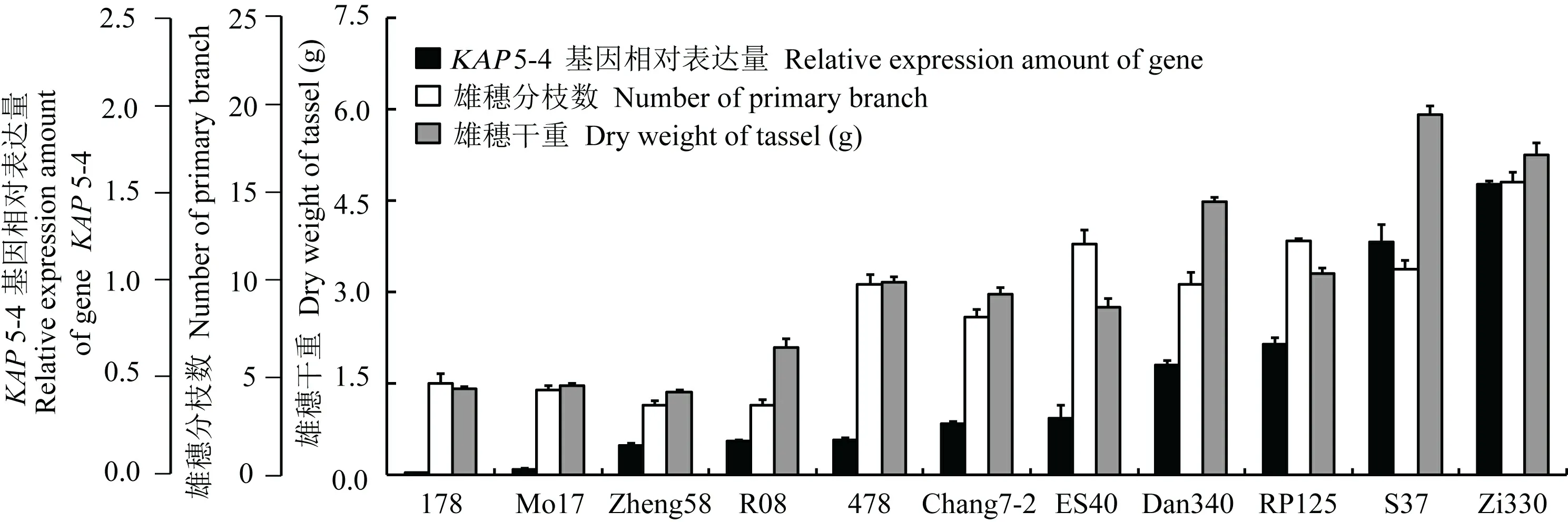
图 6 KAP5-4基因在雄穗原基中的相对表达量与雄穗性状的相关性Fig. 6 Relative expression of gene KAP5-4 in tassel primordial and its correlation with tassel character
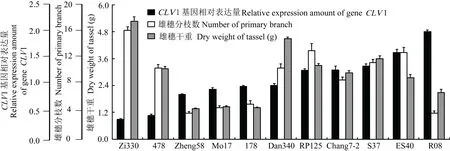
图 7 CLV1基因在雄穗原基中的相对表达量与雄穗性状的相关性Fig. 7 Relative expression of gene CLV1 in tassel primordial and its correlation with tassel character
Association of expression amount and SNP of two candidate genes to tassel size in maize
REN Xiao-Dan, CHEN Ling, YANG Lin, LI Wan-Chen, FU Feng-Ling*
(MaizeResearchInstitute,SichuanAgriculturalUniversity, Chengdu 611130, China )
Abstract:Maize tassel usually sheds powders much more than the requirement of pollination. The overdeveloped tassel not only consumes excessive energy, preventing the transferring of photosynthate to the developing ear, but also decreases the sunlight transmittance of the population, resulting in photosynthetic rate decrease. Therefore, tassel size becomes an indirect selection criterion in maize variety selection because its negative correlation with grain yield was found in maize production practice and breeding study. According to previous reports, the genomic sequences of the keratin-associated protein gene KAP 5-4 and the receptor-like protein kinase gene CLV1 were amplified piece by piece from eleven maize inbred lines with different tassel size, and used for the analysis of multiple alignment, open reading frames, domain structures, and single nucleotide polymorphism. Their differential expressions in tassel primordial among these inbred lines were detected by fluorescence real-time quantitative PCR, and used for correlation analysis with tassel size measured by number of primary branch and dry weight of tassel. The results showed that the relative expression amount of the KAP 5-4 gene was positively correlated with number of primary branch(r=0.77, P<0.01) and dry weight of tassel (r=0.83, P<0.01). A single nucleotide polymorphism at the 2 104 bp base of the open reading frame of the CLV1 gene from eleven maize inbred lines consisted codon GAC encoding acidic aspartic acid at the 702nd site of the receptor-like protein kinase at the 2 104 to 2 016 bp base in five of the inbred lines, and consisted codon AAC encoding polar asparagine at the same site in the other six inbred lines. The relative expression amount of the CLV1 gene was negatively correlated with number of primary branch (r=-0.92, P<0.01) and dry weight of tassel (r=-0.91, P<0.05) within the former five inbred lines, and negatively correlated only with dry weight of tassel (r=-0.91, P<0.05) within the later six inbred lines. It was concluded that the expression and single nucleotide polymorphism of the KAP 5-4 and CLV1 genes were closely associated with tassel size of maize inbred lines, and functional DNA markers can be developed for DNA marker-assisted selection in maize breeding.
Key words:differential expression, keratin-associated protein, maize, receptor-like protein kinase, tassel size
DOI:10.11931/guihaia.gxzw201410030
收稿日期:2014-12-20修回日期: 2015-03-26基金项目: 国家自然科学基金(31071433)[Supported by the National Natural Science Foundation of China(31071433)]。
作者简介:任小丹(1989-),女,新疆石河子人, 硕士研究生,研究方向为植物分子生物学,(E-mail)1035775907@qq.com。 *通讯作者: 付凤玲,教授,博士生导师,研究方向为玉米遗传育种与生物技术,(E-mail)ffl@sicau.edu.cn。
中图分类号:Q945.4, Q949.9
文献标识码:A
文章编号:1000-3142(2016)03-0253-08
任小丹,陈玲,杨琳,等. 两个相关基因表达量和SNP与玉米雄穗大小相关 [J]. 广西植物, 2016, 36(3):253-260
REN XD,CHEN L,YANG L,et al.Association of expression amount and SNP of two candidate genes to tassel size in maize [J]. Guihaia, 2016, 36(3):253-260
