Genetic variation for phytic acid content in mungbean(Vigna radiata L.Wilczek)
2015-11-12VinodJanardanDholeKandaliSrinivasaluReddy
Vinod Janardan Dhole*,Kandali Srinivasalu Reddy
Nuclear Agriculture and Biotechnology Division,Bhabha Atomic Research Centre,Mumbai 400 085,India
Genetic variation for phytic acid content in mungbean(Vigna radiata L.Wilczek)
Vinod Janardan Dhole*,Kandali Srinivasalu Reddy
Nuclear Agriculture and Biotechnology Division,Bhabha Atomic Research Centre,Mumbai 400 085,India
A R T I C L E I N F O
Article history:
form 4 December 2014
Accepted 16 February 2015
Available online 23 February 2015
Mungbean
Phytic acid
Inorganic phosphorus
Cluster analysis
Mungbean(VignaradiataL.Wilczek)isashort-durationlegumecropcultivatedforseedsthatare rich in protein and carbohydrates.Mungbeans contain phytic acid(PA),an anti-nutritional factor that is the main storage form of organic phosphorus in seeds.It is a strong inhibitor againsttheabsorptionofnutrientsincludingiron,zinc,calciumandmagnesiuminmonogastric animals.Genotypes with low phytic acid(lpa)in seed may show increased assimilation of nutrients and be useful in breeding lpa cultivars.The present study was conducted to identify lpa sources,genetic variation,heritability,and association with seed coat color,inorganic phosphorus(IP),and seed size in 102 mungbean genotypes including released varieties,land races,mutants,andwildspeciesgrownintwoseasons:summer2011andrabi2012.PAandIPin dry seeds were estimated by modified colorimetric method and Chen's modified method,respectively.PA,IP,and 100-seed weightdiffered significantly inthe two seasons.PA content in 102 genotypes ranged from 5.74 to 18.98 mg g-1and 5.85 to 20.02 mg g-1in summer 2011 and rabi 2012,respectively.High heritability was found for PA(0.87 and 0.86)and seed size(0.82 and 0.83)but low heritability for IP(0.61 and 0.60).A negative correlation was found between PA and seed size(r=-0.183 and-0.267).Yellow and green seed coat genotypes contained significantly less PA than black seed coat genotypes.Cluster analysis revealed the distinctness of wild species,land races and cultivated varieties on the basis of PA content.The genotypes YBSM(6.001 mg g-1)and JL-781(6.179 mg g-1)showed lowest PA.These lpa sources can be used to develop high-yielding mungbean cultivars with low phytic acid.
©2015 Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.This is an open access article under the CC BY-NC-ND license
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
1.Introduction
Mungbean(Vigna radiata L.Wilczek),an important shortduration grain legume crop,is cultivated for its dry seeds,which are a rich source of easily digestible protein,carbohydrates,vitamin C,folic acid,thiamin,iron,zinc,potassium,magnesium,copper,manganese,and phosphorus[1-3].However,mungbean alsocontainsphytic acid(PA,myo-inositol hexakisphosphate),an anti-nutritional factor that is the main storage form of organic phosphorus(P).As an effective chelator of positively charged cations,PA binds to nutritionally important mineral cations such as calcium,iron,and zinc.Phytate also inhibits trypsin[4]. Humans as well as other non-ruminants such as poultry,swine and fish lacking the enzyme phytase are enable to digest PA and excrete a large fraction of these salts.This phenomenon can contributetohumanmineraldeficiency,particularlywithrespectto iron and zinc,and also causes eutrophication of waterways[5-7].Vegetarian populations in developing countries such as India are at greatest risk of mineral deficiencies caused by dietary PA,particularly children and child-bearing women in rural communities that depend on cereals,and legumes as staple foods[8].Recently there has been increasing interest in the development of crops with low phytic acid(lpa)content to enhance the bioavailability of minerals and other nutrients. Earlier breeding efforts have identified several lpa mutants,resulting in reduction of seed phytic acid phosphorus(PAP)by 50 to>95%in crops such as barley,wheat,maize,soybean,and common bean[9-13].But no lpa mutant was identified in mungbean.PAP typically represents from 65%to 85%of seed total P[14].Mungbean seeds contain 6.17-9.90 mg g-1of PA[3,15].PAPandinorganicphosphorus(IP)contentsinmungbean seeds ranged between 1.77 and 5.79 mg g-1and 0.25 to 0.73 mg g-1respectively[16].PA content was reported to be lowerinyellowthaningreenseedcoatmungbeancultivars[15]. If a source is available,genes for lpa can be transferred into improvedhigh-yieldingvarieties,astheheritabilityofPAishigh(0.80)[16].For the improvement of any trait,genetic variation is a prerequisite.Very few reports on thePAcontent inmungbean germplasm are available.Since mungbean is native to India,natural genetic variation is much higher in Indian germplasm than the rest of the world collection hence need to be evaluated foridentificationoflpasources.Onceasourceisavailable,genes for lpacan be transferred tohigh-yieldingvarieties.For effective transfer of genes,information on heritability as well as on the correlation of PA with important traits such as IP and 100-seed weight is needed to avoid negative correlated response to selection.Differences in PA content among yellow,green and black seed coat mungbean genotypes should be evaluated to identify possible associations of PA with seed coat color.
The present study was conducted to identify genetic variation for phytic acid in 102 mungbean genotypes and its correlation with seed coat color,IP,and 100-seed weight.
2.Material and methods
2.1.Plant materials and field experiments
Material consisted of 102 diverse mungbean germplasm lines,including released varieties,mutants,newly developed genotypes,land races,and wild species(Table S1 of supplementary information).These lines were selected on the basis of their origin;morphological traits including plant type,seed size,and seed coat color;resistant/susceptible reaction to major diseases and pests;and molecular diversity studies performed earlier using SSR and ISSR markers[17-19].These genotypes were grown in a randomized complete block design with two replications at the experimental field facility section,Bhabha Atomic Research Centre,Mumbai during summer 2011 and rabi 2012.Single plants were harvested at maturity and seeds were dried in an oven for 72 h at 50°C.The 100-seed weight was recorded in grams(g).Seeds from the two seasons were used to estimate PA and IP contents.Samples of 20 to 30 randomly selected seedsfromeach genotypeand replicationwere ground tofinepowderandsievedthrough40 meshtoremoveseedcoat particles.
2.2.Measurement of PAP and PA
In a 2 mL microcentrifuge tube,50 mg of the fine powder was thoroughly mixed with 1 mL of 2.4%HCl.The tubes were shaken overnight in a Lab-Line Incubator Shaker(Lab-Line Instruments Inc.,Melrose Park,IL,USA)and centrifuged at 10,000 r min-1in a tabletop centrifuge(Eppendorf,Hamburg,Germany)at 25°C for 20 min.Crude acid extracts were transferred to 1.5 mL microcentrifuge tubes containing 100 mg NaCl.The contents were vortexed to dissolve the salt and incubated at-20°C for 20 min to precipitate remaining matrix components that could interfere with the colorimetric reaction. The mixtures were then centrifuged at 10,000 r min-1for 20 min at 25°C to yield clear supernatant.The supernatant was diluted 25 times by addition of deionized distilled water and 750 μL of the diluted supernatant was mixed with 250 μL of modified Wade reagent(0.03%FeCl3,and 0.3%sulfosalicylic acid).Thesupernatantwasthencollectedforthedetermination of PAP using the colorimetric method[20,21].A series of calibration standards containing 0,0.5,1.0,1.5,2.0,3.0,4.0,5.0,7.5,10.0,and 12.0 mg PAP mL-1were also prepared from the sodium salt of phytic acid(Sigma,St Louis,MO,USA)and treated in the same way as described above.The phosphorus content of sodium phytate was 18.38%.The absorbance of color reaction products for both samples and standards was measuredat500 nmonaUV/Vspectrophotometer(Jasco,Cambridge,UK).The pink color of the Wade reagent is due to the reaction between ferric ion and sulfosalicylic acid with absorbance maxima at 500 nm.PA content was calculated as PA=3.552 PAP[16].
2.3.Measurement of IP
In a 2 mL microcentrifuge tube,400 μL of 12.5%trichloroacetic acid with 25 mmol L-1MgCl2was added to 50 mg of the powdered seed sample and vortexed.The suspension was shaken overnight at room temperature(25°C)for complete extraction,and then centrifuged at 10,000 r min-1for 20 min. The supernatant was diluted with deionized distilled water(1:2).Then,100 μL of the diluted supernatant was mixed with 900 μL of Chen's reagent(6 mol L-1H2SO4,2.5%ammonium molybdate,10%ascorbic acid,and water,in 1:1:1:2 proportions)and incubated in a water bath at 50°C for 1 h.A series of standards containing 0.15,0.31,0.46,0.62,0.77,0.93,1.08,1.24,1.39,and 1.55 mgIP mL-1of sodium dihydrogen phosphate were prepared and processed in the same way as described above.The absorbance of color reaction products for both samples and standards was measured at 660 nm. Total IP was estimated by Chen's modified method[22].
2.4.Data analysis
The data were subjected to analysis of variance for each year and combined over both years,where replication and years were fitted as random effects and genotypes as fixed effects were tested for significance using PROC GLM of SAS 9.3.1(SAS Institute Inc.,Cary,NC).Pearson's correlation coefficients and cluster analysis were estimated with SAS 9.3.1 separately for each season.The mixed model was used to identify significant differences between seasons among the three seed coatcolor genotypes for PA,IP,and 100-seed weight.Genotypic and phenotypic variances were estimated using means of two replications of PA,IP,and 100-seed weight.Broad-sense heritability(h2BS)was estimated for PA,IP,and 100-seed weight according to the standard method[23].
3.Results
3.1.Variation between genotypes and seasons
The 102 mungbean genotypes were evaluated in two seasons with different environmental conditions.Genotype×season interaction was nonsignificant for PA and 100-seed weight but significantforIP.PA,IP and100-seedweightdiffered significantly between the two seasons(Table 1).The PA accumulation in seed was greater in rabi 2012(8.26 mg g-1)than in summer 2011(8.01 mg g-1).In contrast,IP(0.99 mg g-1)and 100-seed weight(5.18 g)were higher in summer 2011 than IP(0.92 mg g-1)and 100-seed weight(4.78 g)in rabi 2012.This observation indicated the influence of environment on PA,IP,and 100-seed weight. Among the102genotypes,42and 31genotypesshowedlessthan 7.5 mg g-1,28 and 39 genotypes showed between 7.5 and 8.5 mg g-1,and 32 genotypes showed more than 8.5 mg g-1of PA in summer 2011 and rabi 2012,respectively.Seasonwise analysis of variance indicated that considerable genetic variation was present between genotypes for all three traits(Table 2).The PA content varied significantly among genotypes and ranged from5.74 mg g-1inVC-6379(58-97)to18.98 mg g-1inThokalwadi wild in summer 2011,but 5.85 mg g-1in YBSM to 20.02 mg g-1in Thokalwadi wild in rabi 2012.The lowest IP content was found in ML-131-03(0.69 mg g-1)and YBSM(0.62 mg g-1)and the highest was found in TARM-2(1.39 mg g-1)and LRM(1.36 mg g-1)in summer 2011 and rabi 2012 respectively.Sonamung had the smallest seed size(2.38 and 2.77 g per 100 seeds)in both seasons and TMB-26(8.16 g per 100 seeds)had the largest seed size in summer 2011 and VC-6379(83-8-1)(7.47 g per 100 seeds)in rabi 2012(Table S1 of supplementary information).
3.2.Heritability,and correlations among PA,IP,and 100-seed weight
Broad-sense heritability was high for PA(0.87 and 0.86)and 100-seed weight(0.82 and 0.83)but low for IP(0.61 and 0.60)in both summer 2011 and rabi 2012,respectively(Table 3). Significant negative correlations were observed between PA and 100-seed weight(r=-0.183 and-0.267)in both summer 2011 and rabi 2012(Table 4).Some of the largest seed size genotypes(100-seed weight greater than 6.5 g)showed lowest PA in both summer 2011 and rabi 2012:JL-781(5.886 and 6.472 mg g-1),VC-6379(58-97)(5.740and6.289 mg g-1),VC-6357(44-55-2)(5.997 and 6.780 mg g-1),and SML-668(6.602 and 7.440 mg g-1).These genotypes can be used as donors for lpa genes in mungbean.
3.3.Variation between yellow,green,and black seed coat genotypes
Among 102 germplasm lines,4,93,and 5 genotypes had yellow,green,and black seed coats,respectively(Table 5).Lowest mean PA(7.648 mg g-1)was found in yellow seed coat genotypes,followed by green(8.016 mg g-1)and black(10.715 mg g-1)over the seasons(summer 2011 and rabi 2012).The yellow and green seed coat genotypes showed significantly less PA than the black seedcoatgenotypes.However,differencesinPAcontentwerenot significant between yellow and green seed coat genotypes.The lowest PA contents were shown by the green seed coat genotype VC-6379(58-97)(5.74 mg g-1)in summer 2011 and the yellow seedcoatgenotypeYBSM(5.85 mg g-1)inrabi2012.ThemeanPA ofblackseedcoatgenotypeswashighest,owingtoonegenotype,Thokalwadi wild(18.98 and 20.02 mg g-1)which showed almost two times higher PA than the rest of the genotypes.The black seed coat color genotype Alibag local had 6.83 and 7.66 mg g-1of PA in summer 2011 and rabi 2012 respectively,similar to the means of yellow and green seed coat genotypes.
3.4.Cluster analysis
On the basis of two seasons of data for PA,IP,and 100-seed weight,102 mungbean genotypes were grouped into two major clusters.Cluster I contained 101 genotypes and cluster II only one(Fig.S1 of supplementary information).The first major cluster showed four subclusters containing cultivated varieties belonging to V.radiata var.radiata.Thokalwadi wild,which belongs to V.radiata var.sublobata,fell into the second cluster.Genotypes containing the highest PA were grouped in subclusters II(9.994 mg g-1)and IV(8.215 mg g-1),whereas subclusters I(7.534 mg g-1)and III(7.214 mg g-1)contained genotypes with low PA(Table 6).The mean IP was highest in subclusterIV(0.984 mg g-1)followedbysubclustersII(0.981 mg g-1),I(0.935 mg g-1),and III(0.892 mg g-1).Genotypes with large seed size were grouped in subcluster I(6.27 g per 100 seeds),those with medium size in subcluster III(4.56 g per 100 seeds),and those with small size in subcluster II(4.11 g per 100 seed)and subcluster IV(4.26 g per 100 seed).
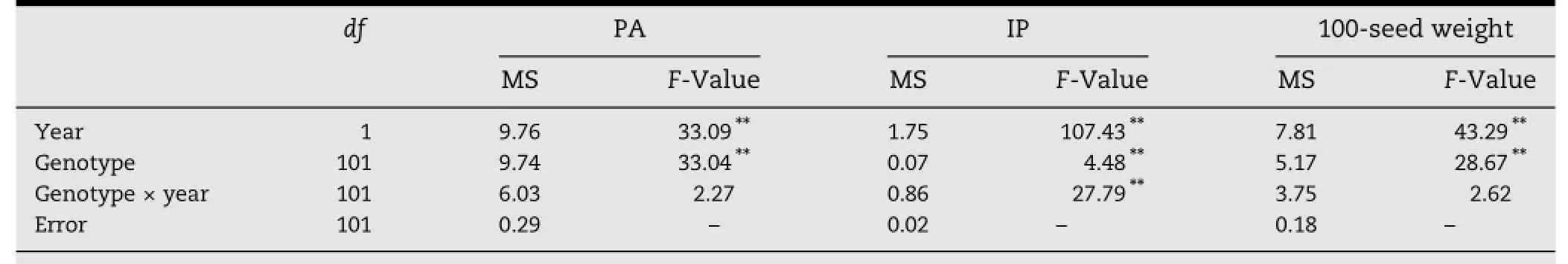
Table 1-Mean sums of squares for PA,IP,and 100-seed weight over two seasons.
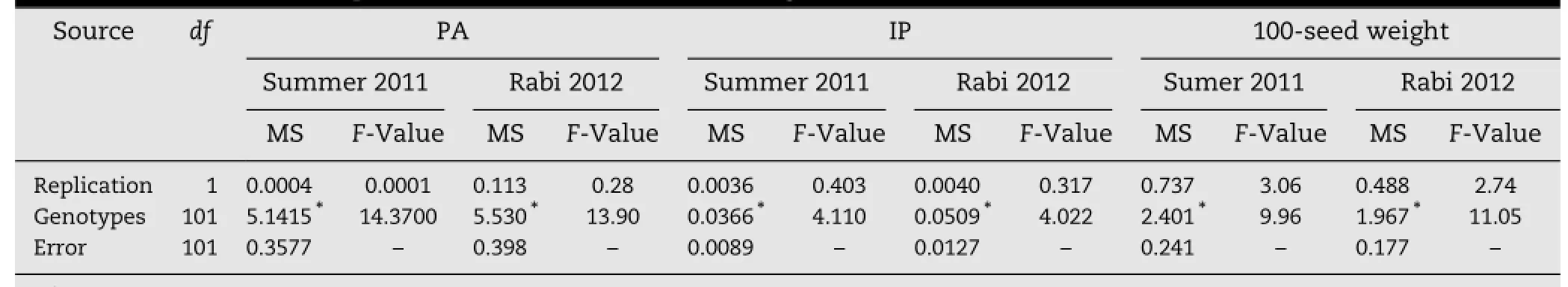
Table 2-Mean sums of squares for PA,IP,and 100-seed weight in summer 2011 and rabi 2012.
On the basis of two seasons of data for PA,IP,and 100-seed weight,102 mungbean genotypes were grouped into two major clusters.Cluster I contained 101 genotypes and cluster II only one(Fig.S1 of supplementary information).The first major cluster showed four subclusters containing cultivated varieties belonging to V.radiata var.radiata.Thokalwadi wild,which belongs to V.radiata var.sublobata,fell into the second cluster.Genotypes containing the highest PA were grouped in subclusters II(9.994 mg g-1)and IV(8.215 mg g-1),whereas subclusters I(7.534 mg g-1)and III(7.214 mg g-1)contained genotypes with low PA(Table 6).The mean IP was highest in subclusterIV(0.984 mg g-1)followedbysubclustersII(0.981 mg g-1),I(0.935 mg g-1),and III(0.892 mg g-1).Genotypes with large seed size were grouped in subcluster I(6.27 g per 100 seeds),those with medium size in subcluster III(4.56 g per 100 seeds),and those with small size in subcluster II(4.11 g per 100 seed)and subcluster IV(4.26 g per 100 seed).
4.Discussion
The present study identified significant differences due to environment in PA,IP,and 100-seed weight.Temperature and humidity were high in the summer but low in the rabi season. In the summer season,plant growth was fast and mungbean matured early with good seed development,whereas in the rabi season,plant growth was slow with late maturity and seed was not developed to its maximum potential.The seasonal difference was also reflected in accumulation of PA and IP in seeds.PA content was greater in the rabi than in the summer season.The PA content reported in the present study was comparable to those in earlier reports,whereas the IP content was slightly higher[3,15,16].The main objective of the present study,to identify lpa sources,was achieved with the identification of two lpa genotypes:YBSM and JL-781.These genotypes showed almost 25%lower PA than the mean PA of the 102 genotypes over the two seasons,40%lower PA than the mean of subcluster II with the highest mean PA,which contained the cultivated mungbean genotypes,and 70%lower than Thokalwadi wild,which contained the highest PA.
The high heritability found in the present study was also reported earlier for PA and 100-seed weight,but heritability for IP content was slightly lower than those reported earlier[16,24].This slight difference may be due to the type of population used in the present study(germplasm or pure lines)in contrast to the F2population used in the previous study[16].The high heritability values also suggested that there is low environmental effect on PA content and that the trait is controlled either by major genes or by polygenes with additive gene action.PA content in mungbean has been reported to be controlled by dominant genes with duplicate recessive epistasis,and three quantitative trait loci were identified using molecular markers[16,25].High heritability with stable values across environments(summer 2011 and rabi 2012)for PA and 100-seed weight further indicated that selection in the desired direction can be made efficiently.No significant correlation of IP with PA and 100-seed weight was observed in either season,whereas a significant negative correlation was observed between PA and 100-seed weight.The possibility of negative correlation between seed size and PA will correspond to the proportion of seed coat in the sample.Smaller seeds have a lower proportion of seed coat.In earlier reports of mungbean,no correlation was found between PA and 100-seed weight[24,25].This finding was probably due to the size and type(F2)of the population used in the previous studies,which may have shown lower variation for seed size.In the present study,102 diverse genotypes were used in which 100 seed weight ranged from 2.38 to 8.16 g and 2.27 to 7.47 g in summer 2011 and rabi 2012,respectively.The large sample size along with variation for seed size yielded a reliable estimate of negative correlation between PA and 100-seed weight.We infer that simultaneous improvement is possible for large seed size and lpa.At the same time,we can improve IP content without affecting PA content,given that no correlation was found between them.
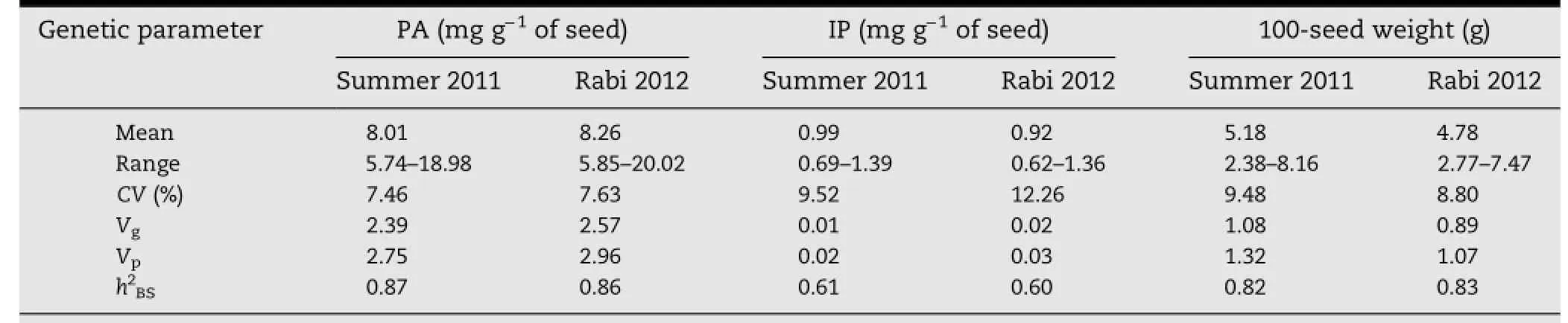
Table 3-Estimates of genetic components of variance and broad-sense heritability(h2BS)for PA,IP,and 100-seed weight in summer 2011 and rabi 2012.
Seed coat color is one of the important parameters for consumer preference.Green seed coat mungbean cultivars are most preferred,while black seed coat cultivars are least preferred by consumers.Three seed coat colors,yellow,green,and black,were found among the 102 genotypes in the present study.The range of PA within each of the three seed coat color genotypes suggested that PA content is not always associated with seed coat color,although a lower mean PA contentwasobservedinyellowthaninblackandgreenseedcoat genotypes in both seasons.Lower PA content in yellow seed coat color mungbean has been reported earlier[15].Mean IP content was highest in green followed by yellow and black seed coatgenotypes,with no significant differences.During domestication ofmungbean,yellowandgreenseedcoatcolorwerepreferredfor consumption and cultivation over black,which was assumed to be associated with some antinutritional factors[15].
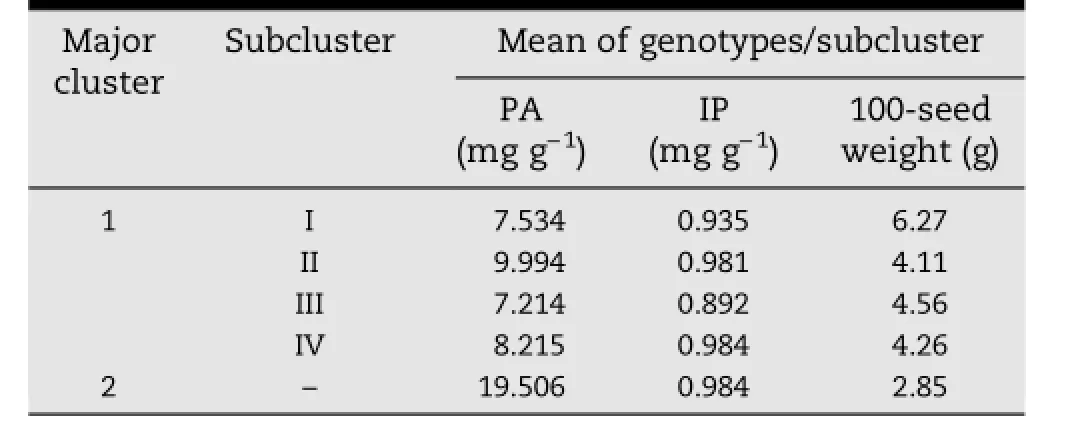
Table 6-Mean PA,IP,and 100-seed weight per cluster and subcluster.
Theclusteranalysisrevealedgeneticvariationpresentinthe mungbean genotypes used in the present study.The two major clusters separated cultivated and wild genotypes.The wild species Thokalwadi wild is separated from the cultivated mungbean genotypes mainly by its PA content,which was almosttwicethemeanPA.ThesubclustersofthemajorclusterI differed from one another for PA,IP and seed size.Low PA with high 100-seed weight in one subcluster and high PA with low 100-seed weight in another subcluster are the result of negative correlation between these traits.This information is useful for mungbean breeders to select lpa genotypes for hybridization with high yielding genotypes from different clusters so as to develop lpa mungbean cultivars with broad genetic bases.The lpa genotype YBSM and JL-781 identified in the present study can be used as a source for lpa genes.
5.Conclusions
In mungbean,considerable genetic variation and high heritability were observed for PA content.Negative correlation was found between seed size and PA,indicating that simultaneous improvement in different directions is possible for these traits.No significant differences in PA content were observedbetween yellow and green seed coat genotypes,but black contained significantly greater PA than yellow and green seed coat genotypes.The sources for lpa genes identified in the present study can be utilized for breeding high-yielding lpa mungbean cultivars with broad genetic bases by use of cluster analysis information.Consumption of mungbean grains with lpa will increase the assimilation of nutrients in monogastric animals.
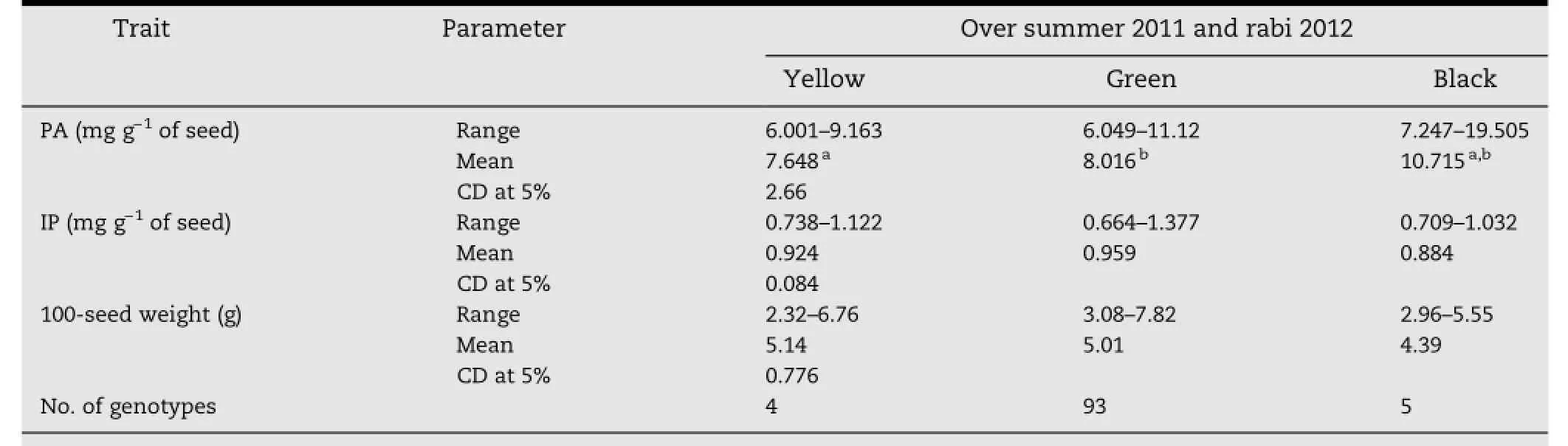
Table 5-Range,mean and CD of PA,IP,and 100-seed weight in yellow,green,and black seed coat mungbean over two seasons,summer 2011 and rabi 2012.
Acknowledgments
The authors are grateful to Dr.S.P.Kale,Head,NABTD,BARC,Mumbai,forhisguidanceandsupportthroughoutthisresearch. We are thankful to Dr.S.Mondal,Dr.Ashok Badigannavar,and Mr.P.Dhanasekar for help in the laboratory experiments.We are especially thankful to Mr.S.M.Bhatkar,and P.N.Thokal for help in conducting the field experiments.
Supplementary material
Supplementary material to this article can be found online at http://dx.doi.org/10.1016/j.cj.2014.12.002.
R E F E R E N C E S
[1]A.E.Mubarak,Nutritional composition and anti-nutritional factors of mungbean seeds 40(Phaseolus aureus)as affected by some home traditional processes,Food Chem.89(2005)489-495.
[2]J.Taunk,N.R.Yadav,R.C.Yadav,R.Kumar,Genetic diversity among green gram[Vigna radiata(L.)Wilczek]genotypes varying in micronutrients(Fe and Zn)content using RAPD markers,Indian J.Biotechnol.11(2012)48-53.
[3]P.K.Dahiya,A.R.Linnemann,M.J.R.Nout,M.A.J.S.Boekel,R.B. Grewal,Nutrient composition of selected newly bred and established mungbean varieties,LWT-Food Sci.Technol.54(2013)249-256.
[4]M.Singh,A.D.Krikorian,Inhibition of trypsin activity in vitro by phytate,J.Agric.Food Chem.30(1982)799-800.
[5]J.W.Erdman,Bioavailability of trace minerals from cereals and legumes,Cereal Chem.58(1981)21-26.
[6]G.L.Cromwell,R.D.Coffey,Phosphorus:a key essential nutrient,yet a possible major pollutant-its central role in animal nutrition,in:T.P.Lyons(Ed.),Biotechnology in the Feed Industry,Alltech Tech Publishers,Nicholaville,KY,1991,pp.133-145.
[7]K.H.Brown,N.W.Solomons,Nutritional problems of developing countries,Infect.Dis.Clin.N.Am.5(1991)297-317.
[8]V.Raboy,Progress in breeding low phytate crops,J.Nutr.132(2002)S503-S505.
[9]S.R.Larson,K.A.Young,A.Cook,T.K.Blake,V.Raboy,Linkage mapping of two mutations that reduce phytic acid content of barley grain,Theor.Appl.Genet.97(1998)141-146.
[10]M.Guttieri,D.Bowen,J.A.Dorsch,V.Raboy,E.Souza,Identification and characterization of low phytic acid wheat,Crop Sci.44(2004)418-424.
[11]V.Raboy,P.Gerbasi,Genetics of myo-inositol phosphate synthesis and accumulation,in:B.Biswas,S.Biswas(Eds.),Myo-inositol Phosphates,Phosphoinositides and Signal Transduction,Plenum Press,New York,1996,pp.257-285.
[12]S.Yuan,F.J.Zhao,H.J.Ren,X.L.Zhu,S.L.Fu,X.J.Shu,Generation and characterization of two novel low phytate mutations in soybean(Glycine max L.Merr.),Theor.Appl. Genet.115(2007)945-957.
[13]B.Campion,F.Sparvoli,E.Doria,G.Tagliabue,I.M.Galasso Fileppi,R.Bollini,E.Nielsen,Isolation and characterization of an lpa(low phytic acid)mutant in common bean(Phaseolus vulgaris L.),Theor.Appl.Genet.118(2009)1211-1221.
[14]V.Raboy,Accumulation and storage of phosphate and minerals,in:B.A.Larkins,I.K.Vasil(Eds.),Cellular and Molecular Biology of Plant Seed Development,Kluwer Academic Publishers,Dordrecht,The Netherlands,1997,pp.441-477.
[15]M.D.Tajoddin,M.Shinde,J.Lalitha,In vivo reduction the phytic acid content of mungbean(Phaseolus aureus L.)cultivars during germination,Am-Euras,J.Agric.Environ.Sci. 10(2011)127-132.
[16]U.Sompong,C.Kaewprasit,S.Nakasathien,P.Srinives,Inheritance of seed phytate in mungbean(Vigna radiate L. Wilczek),Euphytica 171(2010)389-396.
[17]K.S.Reddy,J.Souframanien,K.S.Reddy,P.Dhanasekar,Genetic diversity in mungbean as revealed by SSR and ISSR markers,J.Food Legum.21(2008)15-21.
[18]S.K.Gupta,R.Bansal,U.J.Vaidya,T.Gopalakrishna,Assessment of genetic diversity at molecular level in mungbean(Vigna radiata L.Wilczek),J.Food Legum.26(2013)19-24.
[19]S.K.Gupta,R.Bansal,T.Gopalakrishna,Development and characterization of genic SSR markers for mungbean(Vigna radiata L.Wilczek),Euphytica 195(2014)245-258.
[20]M.Latta,M.Eskin,A simple and rapid colorimetric method for phytate determination,J.Agric.Food Chem.28(1980)1315-1317.
[21]Y.Gao,C.Shang,M.A.Saghai Maroof,R.M.Biyashev,E.A. Grabau,P.Kwanyuen,J.W.Burton,G.R.Buss,A modified colorimetric method for phytic acid analysis in soybean,Crop Sci.47(2007)1797-1803.
[22]P.S.Chen,T.Y.Toribara,H.Warner,Micro determination of phosphorus,Anal.Chem.28(1956)1756-1758.
[23]D.S.Falconer,T.F.C.Mackey,Introduction to Quantitative Genetics,fourth ed.Pearson Education Limited,Essex,England,1996.
[24]U.Sompong,S.Nakasathien,C.Kaewprasit,P.Srinives,Variation in phytic acid content in seed and correlation with agronomic traits of mungbean(Vigna radiata L.Wilczek),Agric.Sci.J.38(2007)243-250.
[25]U.Sompong,P.Somta,V.Raboy,P.Srinives,Mapping quantitativetraitlociforphyticacid andphosphoruscontentsin seed and seedling of mungbean(Vigna radiata L.Wilczek),Breed. Sci.62(2012)87-92.
21 April 2014
in revised
.
E-mail address:vjdhole@yahoo.co.in(V.J.Dhole).
Peer review under responsibility of Crop Science Society of China and Institute of Crop Science,CAAS.
http://dx.doi.org/10.1016/j.cj.2014.12.002
2214-5141/©2015 Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.This is an open access article under the CC BY-NC-ND license(http://creativecommons.org/licenses/by-nc-nd/4.0/).
杂志排行
The Crop Journal的其它文章
- Inhibition of the spread of endophytic Sporisorium reilianum renders maize resistance to head smut
- Maize response to elevated plant density combined with lowered N-fertilizer rate is genotype-dependent
- Organic amendments increase corn yield by enhancing soil resilience to climate change
- Bed planting of wheat(Triticum aestivum L.)improves nitrogen use efficiency and grain yield compared to flat planting
- Reduced grain chalkiness and its possible physiological mechanism in transgenic rice overexpressingL-GalLDH
- Characterization and mapping of QTLs on chromosome 2D for grain size and yield traits using a mutant line induced by EMS in wheat
