基于文献挖掘的前列腺癌蛋白组学与基因组学差异基因的生物信息学分析
2015-09-14陈晨曹笑歌张立国么安亮刘健康绍叁高伟兴韩会曹凤宏李治国
陈晨,曹笑歌,张立国,么安亮,刘健,康绍叁,高伟兴,韩会,曹凤宏,李治国
基于文献挖掘的前列腺癌蛋白组学与基因组学差异基因的生物信息学分析
陈晨,曹笑歌,张立国,么安亮,刘健,康绍叁,高伟兴,韩会,曹凤宏,李治国
目的通过前列腺癌(PCa)蛋白组学与基因组学文献,挖掘与PCa存在相关性并缺乏具体研究的差异基因,并分析其参与的生物过程与通路。方法计算机检索PubMed数据库,检索策略:“(prostate cancer[Title]) AND Proteomics”“(prostate cancer[Title])AND Genomics”,时间限定为建库至2015年1月。按照PCa与前列腺增生(BPH)组织(A组)、PCa与邻近良性组织(B组)、PCa高Gleason评分与低Gleason评分(C组)蛋白谱和基因谱的比较提取差异蛋白或基因,输入“The Protein Information Resource(PIR,Georgetown University Medical Center,Washington,DC 20007,USA)”,按照“official gene symbol”统一名称。采用“DAVID Bioinformatics Resources 6.7 (National Institute of Allergy and Infectious Diseases,NIH,USA)”在线工具对差异基因进行GO(Gene Ontology)、KEGG等生物信息学分析。结果共纳入35篇文献,提取差异基因764个,其中A组差异基因162个,B组差异基因423个,C组差异基因209个,3组共同差异基因21个。A组差异基因中DES报道最多,为6次;B组差异基因ACPP报道最多,为4次;C组差异基因ACTN1、HSPB1、LMNA报道最多,均为3次。GO分析结果显示,差异基因涉及的生物过程主要有细胞死亡调控、细胞增殖调控、创伤反应、蛋白质转运、稳态过程(差异基因频数≥72,频率≥9.4%),细胞成分主要涉及胞外区、膜封闭腔、细胞骨架、囊泡以及线粒体(差异基因频数≥85,频率≥11.1%),分子功能主要涉及核苷酸结合、钙离子结合、相同蛋白结合以及酶结合(差异基因频数≥60,频率≥7.9%)。KEGG通路分析发现,差异基因主要参与癌症通路、黏着斑、肌动蛋白细胞骨架调控、MAPK信号等生物学通路(差异基因频数≥29,频率≥3.8%)。对各组差异基因进行KEGG通路分析结果显示,各组差异基因共同参与的KEGG通路主要有黏着斑、补体及凝血级联、ECM受体相互作用等生物学通路。结论差异基因DES、ACTN1、ATP5B、TLN1、COL6A2、MYH9、OGN、PGAM1报道次数较多,而其参与PCa发生发展的具体机制未见报道,值得进一步实验验证。黏着斑、肌动蛋白细胞骨架的调控以及MAPK信号通路可能在PCa发生发展过程中发挥重要作用,对其进一步分析将为临床治疗PCa提供新的靶点。
前列腺肿瘤;基因组学;蛋白组学;生物信息学
陈晨,曹笑歌,张立国,等.基于文献挖掘的前列腺癌蛋白组学与基因组学差异基因的生物信息学分析[J].中国全科医学,2015,18(32):4011-4016.[www.chinagp.net]
Chen C,Cao XG,Zhang LG,etal.Bioinformatics analysis of differentially expressed genes of prostate cancer proteomics and genomics based on literaturemining[J].Chinese General Practice,2015,18(32):4011-4016.
前列腺癌(PCa)是男性最常见的癌症之一。2014年,美国新增PCa患者23万例,死亡患者近3万例[1]。目前,针对PCa的蛋白组学和基因组学研究非常广泛,发现表达差异具有统计学意义的基因数以千计,但其中仅少数可能与PCa的发生发展相关[2]。通过文献挖掘对癌症相关基因进行提取和整理,是癌症分子机制研究的良好方法。但若文献挖掘只局限于摘要,且利用自动文献挖掘工具未免机械,在文章题目和摘要中未出现的差异基因可能被漏掉。本研究通过检索PCa蛋白组学及基因组学相关文献,阅读全文人工提取差异基因,并对其进行GO(Gene Ontology)、KEGG通路等生物信息学分析,进一步挖掘可能与PCa相关的基因和通路,从而为PCa发生的分子机制研究提供依据。
1 资料与方法
1.1文献纳入与排除标准纳入标准:(1)研究对象为人PCa组织或人PCa细胞株;(2)提供差异蛋白或基因名称。排除标准:(1)某种干预措施对PCa蛋白谱变化的影响研究; (2)未提供差异蛋白或基因名称;(3)文献综述。
1.2文献检索计算机检索PubMed数据库,检索策略:“(prostate cancer[Title])AND Proteomics”“(prostate cancer[Title])AND Genomics”,时间限定建库至2015年1月。
1.3资料提取文献中涉及PCa与前列腺增生(BPH)组织或细胞(A组)、PCa与邻近良性组织(B组)、PCa高Gleason评分与低Gleason评分(C组)蛋白谱和基因谱的比较,提取差异基因或蛋白输入“The Protein Information Resource(PIR,Georgetown University Medical Center,Washington,DC 20007,USA)”,按照“official gene symbol”统一名称。
1.4生物信息学分析
1.4.1差异基因的GO分析采用“DAVID Bioinformatics Resources 6.7(National Institute of Allergy and Infectious Diseases,NIH,USA)”在线工具分别对差异基因及各组的差异基因进行GO分析,探索共同出现的生物过程、细胞成分以及分子功能。
1.4.2差异基因的KEGG通路分析通过“DAVID Bioinformatics Resources 6.7(National Institute of Allergy and Infectious Diseases,NIH,USA)”在线工具分别对差异基因及各组的差异基因进行KEGG通路分析,探索共同出现的生物学通路。
2 结果
2.1文献筛选结果检索PubMed数据库获得文献564篇,排除重复文献179篇,阅读文题和摘要排除282篇,阅读全文排除文献68篇,共纳入35篇文献[3-37],文献筛选流程见图1。
2.2差异基因纳入文献提取差异基因764个,其中A组差异基因162个,B组差异基因423个,C组差异基因209个,3组共同差异基因21个(见表1)。报道大于4次的差异基因共14个,其中DES报道8次,HSPB1、VCL各报道7次,ACPP、ACTN1、AZGP1、ENO1、HSPA5和HSPD1各报道6次,ANXA1、EZR、KRT8、LMNA和SERPINF1各报道5次。A组差异基因DES报道最多,为6次;B组差异基因ACPP报道最多,为4次;C组差异基因ACTN1、HSPB1、LMNA报道最多,均为3次。
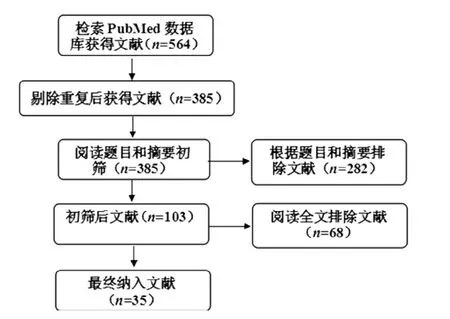
图1 文献筛选流程图Figure 1 Flow chart of literature screening
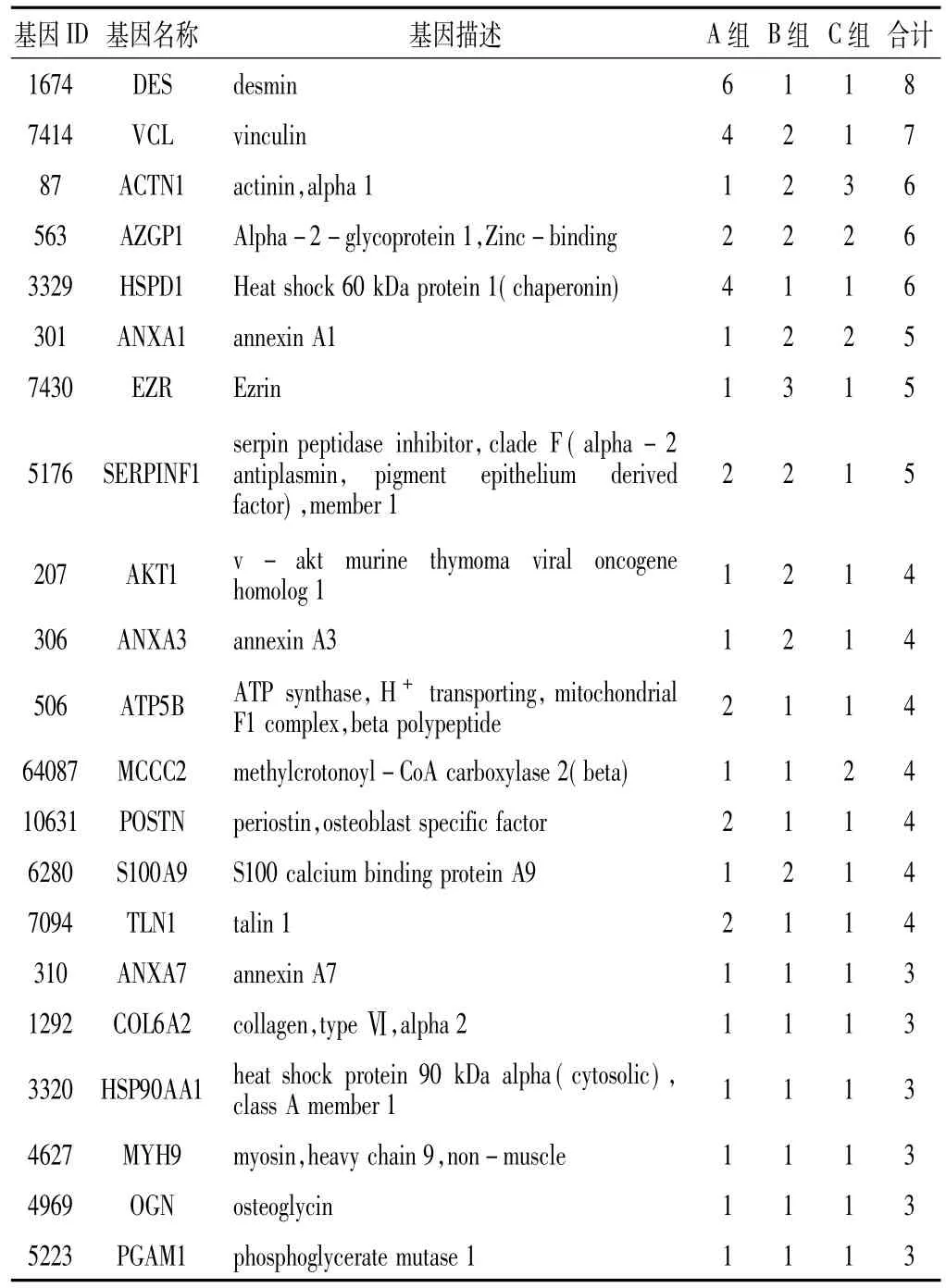
表1 各组共同差异基因及其被报道次数(次)Table 1 Co-occurrence of the differentially expressed genes in different groups and the reported frequency
2.3差异基因的GO分析GO分析结果显示,差异基因涉及的生物过程主要有细胞死亡调控、细胞增殖调控、创伤反应、蛋白质转运、稳态过程(差异基因频数≥72,频率≥9.4%),细胞成分主要涉及胞外区、膜封闭腔、细胞骨架、囊泡以及线粒体(差异基因频数≥85,频率≥11.1%),分子功能主要涉及核苷酸结合、钙离子结合、相同蛋白结合以及酶结合(差异基因频数≥60,频率≥7.9%,见表2)。
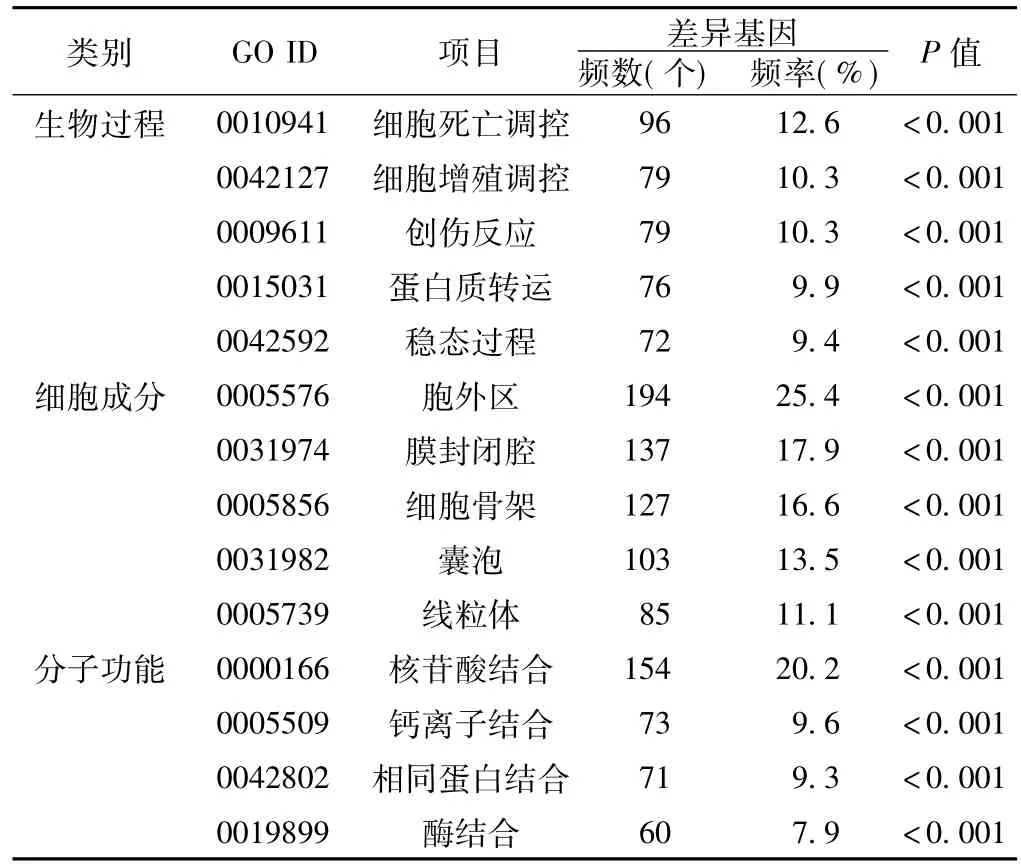
表2 差异基因的GO分析Table 2 GO analysis of differentially expressed genes in PCa
对各组差异基因进行GO分析结果显示,各组差异基因共同涉及的生物过程主要有细胞死亡调控、细胞增殖调控、创伤反应以及稳态过程;共同涉及的细胞成分主要有胞外区、膜封闭腔、细胞骨架、囊泡以及线粒体;共同涉及的分子功能主要有钙离子结合、相同蛋白结合、结构分子活性以及肌动蛋白结合。
2.4差异基因的KEGG通路分析KEGG通路分析发现,差异基因主要参与癌症通路、黏着斑、肌动蛋白细胞骨架调控、MAPK信号等生物学通路(差异基因频数≥29,频率≥3.8%,见表3)。
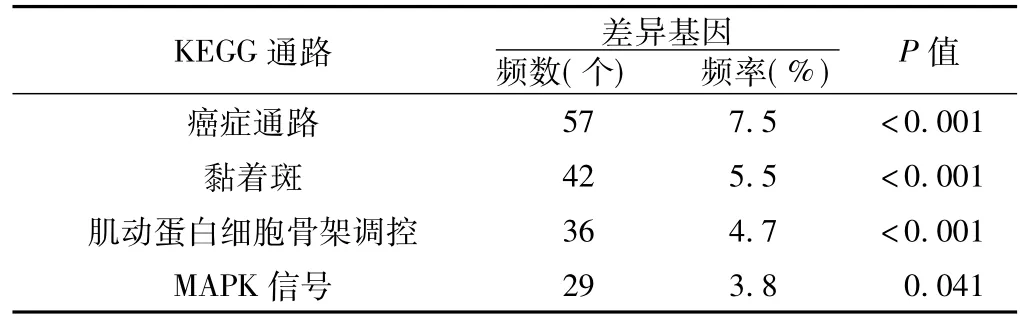
表3 所有差异基因的KEGG通路分析Table 3 KEGG pathway analysis of all differentially expressed genes in PCa
对各组差异基因进行KEGG通路分析结果显示,各组差异基因共同参与的KEGG通路主要有黏着斑、补体及凝血级联、ECM受体相互作用等生物学通路。
3 讨论
目前,尚缺少通过文献挖掘对PCa蛋白组学及基因组学差异基因数据进行整理并开展生物信息学分析的研究。Hu等[2]采用MedGene文献挖掘工具对乳腺癌及正常组织蛋白组学和基因组学数据进行分析,确定了一组研究相对充分、在雌激素受体阴性肿瘤高表达的基因。李铁求等[38]通过文献挖掘对雄激素非依赖型PCa特异表达基因进行生物信息学分析,发现MMP9、EGFR等基因在雄激素依赖型转变为雄激素非依赖型PCa过程中可能发挥重要作用。本研究首次将PCa与蛋白组学及基因组学数据整合,对更新至2015年1月的相关文献进行差异基因挖掘,并观察其在PCa与BPH、PCa与邻近良性组织、PCa高低Gleason评分不同分类比较中差异基因出现的频率,更直观地了解差异基因与癌症发生、发展、转移过程中的关联性。
本研究发现,所有差异基因中DES报道次数最多,同时也是PCa与BPH比较报道最多的差异基因,提示其可能与PCa存在一定联系。目前表明,高表达的DES是结肠癌、胃肠道间质瘤、子宫内膜癌等内皮细胞分化和肿瘤侵袭的高度敏感标志物[39]。也有研究发现,DES在肿瘤早期毛细血管形成过程中持续高表达[40]。然而,PCa组学实验结果发现,与BPH组织比较,DES在PCa组织中表达下调;同时,与局限性PCa相比,DES在淋巴结转移PCa中表达同样下调[5]。这似乎与上文所述DES参与肿瘤血管内皮细胞形成互相矛盾,具体机制值得后续进一步研究。另外,HSPB1、VCL、ACPP、ACTN1、AZGP1、ENO1、HSPA5、HSPD1、ANXA1、EZR、KRT8、LMNA以及SERPINF1等差异基因被报道5次及以上,提示上述差异基因可能参与PCa的发生发展过程。ACPP在PCa与邻近良性组织比较中被报道次数最多,其编码前列腺酸性磷酸酶,由前列腺上皮细胞分泌并受雄激素调节。ACTN1、HSPB1、LMNA在PCa高Gleason评分与低Gleason评分比较中被报道次数最多,提示其可能在PCa进展转移的过程中发挥重要作用。多篇研究报道了HSPB1与PCa的相关性,其参与PCa发展过程的机制也已被广泛探讨[41]。LMNA也已发现在PCa组织中高表达,并通过PI3K/AKT/PTEN通路促进肿瘤细胞生长、迁移和入侵[42]。HSPA5、AZGP1、EZR参与PCa的机制研究比较深入[43-46],ENO1、HSPD1、ANXA1、SERPINF1、VCL以及KRT8参与PCa的具体机制有待进一步探讨和多角度分析。
本研究发现,21个差异基因在各组均有报道,提示其可能在PCa发生、进展以及转移的过程中均发挥重要作用。其中,HSP90AA1、S100A9、POSTN、ANXA3、ANXA7、AKT1与PCa的相关性研究较为深入[47-49],而MCCC2仅初步证实其高表达可提升PCa细胞的迁移能力[50],但其确切机制缺乏具体阐述。ATP5B、TLN1、COL6A2、MYH9、OGN、PGAM1参与PCa的机制尚未见报道。部分被报道次数较高且在各组中频繁出现的差异基因,如DES、ACTN1、ATP5B、TLN1、COL6A2、MYH9、OGN、PGAM1,提示其可能与PCa存在潜在的联系,同时又缺乏具体的相关性报道,有进一步开展研究的价值,以验证其参与PCa差异表达的真实性,以及详细阐明其参与PCa发生、发展、转移的具体机制。
本研究KEGG通路分析发现,差异基因主要参与癌症通路、黏着斑、肌动蛋白细胞骨架的调控、MAPK信号通路,提示这些通路可能涉及PCa的发生发展过程。黏着斑是细胞外基质黏附在细胞膜某个特殊的区域[38],是黏着斑激酶(FAK)介导的重要细胞过程。FAK在PCa中常高表达和过度活跃,通过主要致癌途径的激活,FAK促进雄激素非依赖PCa的生长、存活、迁移及转移[51]。肌动蛋白细胞骨架的调控提供了PCa分子机制研究的新靶点,Wang等[52]通过研究Wnt/Ca2+信号在PCa中的机制,发现癌细胞特异性抑制CaMKⅡ(Wnt/ Ca2+信号的一个主要传感器)可引起肌动蛋白细胞骨架的重构,CaMKⅡ可能通过肌动蛋白结合蛋白的中间信号传导,导致癌细胞中细胞骨架的重构。Zhang等[53]认为,肿瘤抑制蛋白ZNF185通过调节PCa中的肌动蛋白细胞骨架动力学发挥其功能。可以预见,未来对肌动蛋白细胞骨架的研究,将为临床肿瘤靶向治疗带来巨大的导向作用。MAPK信号通路近年来也是PCa分子研究的热点,PCa的发生与发展、癌细胞的增殖、癌症的复发与信号转导与MAPK信号通路密切相关[54]。在未经雄激素阻断治疗的PCa最初阶段,雄激素的刺激通过激活PCa细胞MAPK信号通路,诱导细胞增殖[55],激活的MAPK水平随PCa的进展而升高。此外,研究证实,在PCa进展到后期阶段,生长因子信号、神经多肽信号、致癌基因HER2/Neu等均可作用于MAPK信号通路,使PCa不依赖雄激素继续发展,即进展到雄激素非依赖型PCa[54]。尽管目前MAPK信号通路的作用及具体机制有待进一步的研究和探索,但可以确定,MAPK信号转导调控机制的阐明将为临床提供治疗PCa的新思路。此外,本研究通过对各组差异基因的KEGG通路分析结果发现,黏着斑、补体及凝血级联、ECM受体相互作用通路在各组均有出现,提示其可能与PCa存在一定的相关性,其中补体及凝血级联、ECM受体相互作用参与PCa的具体机制尚未见报道。Spans等[56]也发现在LNCaP和C4-2B PCa细胞株中,黏着斑与ECM受体相互作用通路发生变化,具体机制值得持续关注和深入调查研究。
综上所述,本研究通过对PCa蛋白组学及基因组学文献进行挖掘,筛选出可能与PCa有较大联系且缺乏具体相关报道的差异基因,如DES、ACTN1、ATP5B、TLN1、COL6A2、MYH9、OGN、PGAM1,为下一步研究提供了方向。黏着斑、肌动蛋白细胞骨架的调控、MAPK信号通路可能在PCa分子机制中发挥重要作用,对其进一步分析有利于揭示PCa的发病机制,为临床治疗PCa提供新的靶点。
[1]Nandana S,Chung LW.Prostate cancer progression and metastasis: potential regulatory pathways for therapeutic targeting[J].Am JClin Exp Urol,2014,2(2):92-101.
[2]Hu Y,Hines LM,Weng H,et al.Analysis of genomic and proteomic data using advanced literature mining[J].J Proteome Res,2003,2(4):405-412.
[3]Saraon P,Musrap N,Cretu D,etal.Proteomic profiling of androgen-independent prostate cancer cell lines reveals a role for protein S during the development of high grade and castration-resistant prostate cancer[J].JBiol Chem,2012,287(41):34019-34031.
[4]Sun C,Song C,Ma Z,et al.Periostin identified as a potentialbiomarker of prostate cancer by iTRAQ-proteomics analysis of prostate biopsy[J].Proteome Sci,2011,9(22):22.
[5]Pang J,Liu WP,Liu XP,et al.Profiling proteinmarkers associated with lymph node metastasis in prostate cancer by DIGE-based proteomics analysis[J].J Proteome Res,2010,9(1):216-226.
[6]Ummanni R,Junker H,Zimmermann U,et al.Prohibitin identified by proteomic analysis of prostate biopsies distinguishes hyperplasia and cancer[J].Cancer Lett,2008,266(2):171-185.
[7]Burgess EF,Ham AJ,Tabb DL,et al.Prostate cancer serum biomarker discovery through proteomic analysis of alpha-2 macroglobulin protein complexes[J].Proteomics Clin Appl,2008,2(9):1223.
[8]Glen A,Gan CS,Hamdy FC,et al.iTRAQ-facilitated proteomic analysis of human prostate cancer cells identifies proteins associated with progression[J].JProteome Res,2008,7(3):897-907.
[9]Han ZD,Zhang YQ,He HC,et al.Identification of novel serological tumormarkers for human prostate cancer using integrative transcriptome and proteome analysis[J].Med Oncol,2012,29 (4):2877-2888.
[10]Kim Y,Ignatchenko V,Yao CQ,et al.Identification of differentially expressed proteins in direct expressed prostatic secretions ofmen with organ-confined versus extracapsular prostate cancer[J].Mol Cell Proteomics,2012,11(12):1870-1884.
[11]Chen P,Wang L,Li N,et al.Comparative proteomics analysis of sodium selenite-induced apoptosis in human prostate cancer cells[J].Metallomics,2013,5(5):541-550.
[12]Chen J,Huang P,Kaku H,et al.A comparison of proteomic profiles changes during17beta-estradiol treatment in human prostate cancer PC-3 cell line[J].Cancer Genomics Proteomics,2010,6(6):331-335.
[13]Song DX,Chen AM,Guo FJ,etal.Differential proteomic analysis and function study of human prostate carcinoma cells with different osseousmetastatic tendency[J].NationalMedical JournalofChina,2008,88(17):1197-1201.(in Chinese)宋登新,陈安民,郭风劲,等.人前列腺癌细胞骨转移潜能差异表达蛋白的研究[J].中华医学杂志,2008,88(17): 1197-1201.
[14]Zhang XM,Shen Y,Xianyu ZQ.Serum proteomic study of prostate cancer with bone metastasis[J].National Journal of Andrology,2010,16(8):721-725.(in Chinese)张雪梅,沈影,鲜于志群.前列腺癌骨转移血清蛋白质组学研究[J].中华男科学杂志,2010,16(8):721-725.
[15]Zhang XB,Tang ZY,Qi L,et al.Modified serum-guided immunoblotting for differential proteomic study of prostate cancer[J].National Journal of Andrology,2010,16(5):438-444.(in Chinese)张晓波,唐正严,齐琳,等.改进的血清免疫印迹引导的前列腺癌差异蛋白质组学研究[J].中华男科学杂志,2010,16 (5):438-444.
[16]Pin E,Fredolini C,Petricoin EF.The role of proteomics in prostate cancer research:biomarker discovery and validation[J].Clin Biochem,2013,46(6):524-538.
[17]Bigot P,Mouzat K,Lebdai S,et al.Quantitative proteomic determination of diethylstilbestrol action on prostate cancer[J].Asian JAndrol,2013,15(3):413-420.
[18]Everley PA,Krijgsveld J,Zetter BR,et al.Quantitative cancer proteomics:stable isotope labeling with amino acids in cell culture (SILAC)as a tool for prostate cancer research[J].Mol Cell Proteomics,2004,3(7):729-735.
[19]Alaiya AA,Al-Mohanna M,Aslam M,et al.Proteomics-based signature for human benign prostate hyperplasia and prostate adenocarcinoma[J].Int JOncol,2011,38(4):1047-1057.
[20]Skvortsov S,Schäfer G,Stasyk T,et al.Proteomics profiling of microdissected low-and high-grade prostate tumors identifies Lamin A as a discriminatory biomarker[J].J Proteome Res,2011,10(1):259-268.
[21]Wood SL,Knowles MA,Thompson D,et al.Proteomic studies of urinary biomarkers for prostate,bladder and kidney cancers[J].Nat Rev Urol,2013,10(4):206-218.
[22]Sandvig K,Llorente A.Proteomic analysis of microvesicles released by the human prostate cancer cell line PC-3[J].Mol Cell Proteomics,2012,11(7):M111.012914.
[23]Sardana G,Jung K,Stephan C,et al.Proteomic analysis of conditioned media from the PC3,LNCaP,and 22Rv1 prostate cancer cell lines:discovery and validation of candidate prostate cancer biomarkers[J].J Proteome Res,2008,7(8):3329-3338.
[24]Lee EK,Cho H,Kim CW.Proteomic analysis of cancer stem cells in human prostate cancer cells[J].Biochem Biophys Res Commun,2011,412(2):279-285.
[25]Sardana G,Marshall J,Diamandis EP.Discovery of candidate tumor markers for prostate cancer via proteomic analysis of cell cultureconditionedmedium[J].Clin Chem,2007,53(3):429-437.
[26]Kallioniemi O.Functional genomics and transcriptomics of prostate cancer:promises and limitations[J].BJU Int,2005,96(S2): 10-15.
[27]Rowehl RA,Crawford H,Dufour A,et al.Genomic analysis of prostate cancer stem cells isolated from a highly metastatic cell line[J].Cancer Genomics Proteomics,2009,5(6):301-310.
[28]Wiklund F.Prostate cancer genomics:can we distinguish between indolent and fatal disease using genetic markers?[J].Genome Med,2010,2(7):45.
[29]Nakagawa H,Akamatsu S,Takata R,et al.Prostate cancer genomics,biology,and risk assessment through genome-wide association studies[J].Cancer Sci,2012,103(4):607-613.
[30]Roychowdhury S,Chinnaiyan AM.Advancing precision medicine for prostate cancer through genomics[J].J Clin Oncol,2013,31(15):1866-1873.
[31]Chaudhary J,Schmidt M.The impact of genomic alterations on the transcriptome:a prostate cancer cell line case study[J].Chromosome Res,2006,14(5):567-586.
[32]Chow A,Amemiya Y,Sugar L,et al.Whole-transcriptome analysis reveals established and novel associations with TMPRSS2: ERG fusion in prostate cancer[J].Anticancer Res,2012,32 (9):3629-3641.
[33]Shipitsin M,Small C,Choudhury S,et al.Identification of proteomic biomarkers predicting prostate cancer aggressiveness and lethality despite biopsy-sampling error[J].Br JCancer,2014,111(6):1201-1212.
[34]Kiprijanovska S,Stavridis S,Stankov O,et al.Mapping and identification of the urine proteome of prostate cancer patients by 2D PAGE/MS[J].Int JProteomics,2014,2014:594761.
[35]Amaro A,Esposito AI,Gallina A,et al.Validation of proposed prostate cancer biomarkerswith gene expression data:a long road to travel[J].Cancer Metastasis Rev,2014,33(2/3):657-671.
[36]Bergamini S,Bellei E,Reggiani Bonetti L,et al.Inflammation: an important parameter in the search of prostate cancer biomarkers[J].Proteome Sci,2014,12:32.
[37]Williams KA,Lee M,Hu Y,et al.A systems genetics approach identifies CXCL14,ITGAX,and LPCAT2 as novel aggressive prostate cancer susceptibility genes[J].PLoS Genet,2014,10 (11):e1004809.
[38]Li TQ,Feng CQ,Zou YG,et al.Literature-mining and bioinformatic analysis of androgen-independent prostate cancerspecific genes[J].National Journal of Andrology,2009,15 (12):1102-1107.(in Chinese)李铁求,冯春琼,邹亚光,等.基于文献挖掘的雄激素非依赖型前列腺癌特异表达基因的生物信息学分析[J].中华男科学杂志,2009,15(12):1102-1107.
[39]Ma Y,Peng J,LiuW,etal.Proteomics identification of desmin as a potential oncofetal diagnostic and prognostic biomarker in colorectal cancer[J].Mol Cell Proteomics,2009,8(8):1878-1890.
[40]Arentz G,Chataway T,Price TJ,et al.Desmin expression in colorectal cancer stroma correlates with advanced stage disease and marks angiogenic microvessels[J].Clin Proteomics,2011,8 (1):16.
[41]Shiota M,Bishop JL,Nip KM,et al.Hsp27 regulates epithelial mesenchymal transition,metastasis,and circulating tumor cells in prostate cancer[J].Cancer Res,2013,73(10):3109-3119.
[42]Kong L,Schäfer G,Bu H,et al.Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth,migration and invasion through the PI3K/AKT/PTEN pathway[J].Carcinogenesis,2012,33(4): 751-759.
[43]Ruiz C,Holz DR,Oeggerli M,et al.Amplification and overexpression of vinculin are associated with increased tumour cell proliferation and progression in advanced prostate cancer[J].J Pathol,2011,223(4):543-552.
[44]Yu L,Shi J,Cheng S,et al.Estrogen promotes prostate cancer cellmigration via paracrine release of ENO1 from stromal cells[J].Mol Endocrinol,2012,26(9):1521-1530.
[45]Mu D,Gao Z,Guo G,et al.Sodium butyrate induces growth inhibition and apoptosis in human prostate cancer DU145 cells by up-regulation of the expression of annexin A1[J].PLoS One,2013,8(9):e74922.
[46]Guan M,Jiang H,Xu C,et al.Adenovirus-mediated PEDF expression inhibits prostate cancer cell growth and results in augmented expression of PAI-2[J].Cancer Biol Ther,2007,6 (3):419-425.
[47]Altieri DC.Mitochondrial Hsp90 chaperones as novelmolecular targets in prostate cancer[J].Future Oncol,2010,6(4):487-489.
[48]Srivastava M,Leighton X,Starr J,etal.Diverse effects of ANXA7 and p53 on LNCaP prostate cancer cells are associated with regulation of SGK1 transcription and phosphorylation of the SGK1 target FOXO3A[J].Biomed Res Int,2014,2014:193635.
[49]Goc A,Liu J,Byzova TV,et al.Akt1 mediates prostate cancer cellmicroinvasion and chemotaxis tometastatic stimuli via integrinβ3 affinity modulation[J].Br J Cancer,2012,107(4):713-723.
[50]李科,陈怡,黄文涛,等.MCCC2在前列腺癌细胞系中的表达及其对前列腺癌转移影响的初步研究[J].中华腔镜泌尿外科杂志:电子版,2014,8(2):140-146.
[51]Figel S,Gelman IH.Focal adhesion kinase controls prostate cancer progression via intrinsic kinase and scaffolding functions[J].Anticancer Agents Med Chem,2011,11(7):607-616.
[52]Wang Q,Symes AJ,Kane CA,et al.A novel role for Wnt/Ca2+signaling in actin cytoskeleton remodeling and cellmotility in prostate cancer[J].PLoSOne,2010,5(5):e10456.
[53]Zhang JS,Gong A,Young CY.ZNF185,an actin-cytoskeletonassociated growth inhibitory LIM protein in prostate cancer[J].Oncogene,2007,26(1):111-122.
[54]查树伟,查佶,王兴海.MAPK信号转导通路与前列腺癌的发病及治疗关系的研究进展[J].中国男科学杂志,2008,22 (6):57-60.
[55]Gioeli D,Mandell JW,Petroni GR,et al.Activation ofmitogenactivated protein kinase associated with prostate cancer progression[J].Cancer Res,1999,59(2):279-284.
[56]Spans L,Helsen C,Clinckemalie L,et al.Comparative genomic and transcriptomic analyses of LNCaP and C4-2B prostate cancer cell lines[J].PLoSOne,2014,9(2):e90002.
(本文编辑:吴立波)
Bioinform atics Analysis of Differentially Expressed Genes of Prostate Cancer Proteom ics and Genom ics Based on Literature M ining
CHEN Chen,CAO Xiao-ge,ZHANG Li-guo,et al.School of Graduate,North China University of Science and Technology,Tangshan 063000,China
Objective To mine differentially expressed genes which have strong correlation with prostate cancer (PCa)but have not been specifically covered through literatures about proteomics and genomics of PCa and analyze the biological processes and pathways in which these genes are involved.M ethods With PubMed public database,we used the advanced search by inputting"(prostate cancer[Title])AND Proteomics""(prostate cancer[Title])AND Genomics"for literaturesbefore January 2015.We extracted differentially expressed proteins or genes according to the comparison of protein and gene expression profiles between PCa and benign prostatic hyperplasia(BPH)(Group A),between PCa and adjacent benign tissues (Group B)and between high and low Gleason scores of Pca(Group C).We input all the differentially expressed genes or proteins into"The Protein Information Resource(PIR,Georgetown University Medical Center,Washington,DC 20007,USA)"and unified all names according to"official gene symbol".Then we conducted the bioinformatics analysis of Gene Ontology and KEGG pathway by DAVID Bioinformatics Resources 6.7(National Institute of Allergy and Infectious Diseases,NIH,USA)online tool.Results A total of35 articleswere included.Throughmining,we obtained 764 differentially expressed genes totally,ofwhich 162 were in Group A,423 in Group B and 209 in Group C.In all the 3 groups,there were 21 common reported genes.DESwas reported themost in Group A,which appeared 6 times.ACPPwas reported themost in Group B,which appeared 4 times.ACTN1,HSPB1 and LMNA were reported themost in Group C,which all appeared 3 times.All these genes played important roles in biological processes of regulation of cell death,regulation of cell proliferation,response to wounding,protein transport and homeostatic process(genes count≥72,percentage≥9.4%),as well as in molecular function of nucleotide binding,calcium ion binding,identical protein binding and enzyme binding(genes count≥60,percentage≥7.9%).Their cellular componentsweremainly in extracellular region,membrane-enclosed lumen,cytoskeleton,vesicle and mitochondrion(genes count≥85,percentage≥11.1%).They weremainly involved in the biological pathways like pathways in cancer,focal adhesion,regulation of actin cytoskeleton and MAPK signaling(genes count≥29,percentage≥3.8%).The co-occurrence KEGG pathways from different groups were focal adhesion,complement and coagulation cascade and ECM-receptor interactions.Conclusion We found out there is strong association between DES,ACTN1,ATP5B,TLN1,COL6A2,MYH9,OGN,PGAM1 and PCa butwithout specific relevant reports,whichmeansmore experimental researches are needed to prove that.What'smore,focal adhesion,regulation of actin cytoskeleton and MAPK signaling pathwaymay play important roles in the development of PCa.Further analysiswill provide new targets for clinical prevention and treatment of PCa.
Prostatic neoplasms;Genomics;Proteomics;Bioinformatics
R 737.25 R 394
A
10.3969/j.issn.1007-9572.2015.32.028
063000河北省唐山市,华北理工大学研究生院(陈晨,韩会);天津市滨海新区汉沽第一中学(曹笑歌);华北理工大学附属医院泌尿外科(张立国,么安亮,刘健,康绍叁,高伟兴,曹凤宏);华北理工大学医学实验研究中心,老年医学国际科技合作基地(李治国)
曹凤宏,063000河北省唐山市,华北理工大学附属医院泌尿外科;E-mail:caofenghong@163.com。李治国,063000河北省唐山市,华北理工大学医学实验研究中心,老年医学国际科技合作基地;E-mail:lzg1017@163.com
2015-05-18;
2015-08-28)
