Cloning and characterization of gene-resistant analogs(RGAs) involved in rust(Puccinia psidii)resistance in Eucalyptus grandis
2015-06-05MarceloLuizLaiaAcelinoCoutoAlfenasSergioHermnioBrommonschenkelShinitiroOdaEduardoJosedeMeloInaeMariedeAraujoSilvaJananaFernandesGoncalvesAriadneMarques
Marcelo Luiz Laia•Acelino Couto Alfenas•Sergio Hermı´nio Brommonschenkel•Shinitiro Oda•Eduardo Jose´de Melo•InaeˆMarieˆde Arau´jo Silva•Janaı´na Fernandes Gonc¸alves•Ariadne Marques
Cloning and characterization of gene-resistant analogs(RGAs) involved in rust(Puccinia psidii)resistance in Eucalyptus grandis
Marcelo Luiz Laia1•Acelino Couto Alfenas2•Sergio Hermı´nio Brommonschenkel2•Shinitiro Oda3•Eduardo Jose´de Melo3•InaeˆMarieˆde Arau´jo Silva1•Janaı´na Fernandes Gonc¸alves1•Ariadne Marques1
Disease-resistant genes play an important role in defending againsta variety of pathogens and insectpests in plants.Most of the disease-resistant genes encode proteins with conserved leucine rich repeat and nucleotide binding site domains.In this study,we cloned and characterized gene-resistant analogs(RGAs)from Eucalyptus grandis using degenerate PCR,with primers specifically targeting these two domains.The amplified fragments were cloned into the pGEM-T vector and transformed into Escherichia coli.Among the 90 clones obtained,13 were sequenced and compared with each other and with previously identified gene-resistantdiseases.A BLASTX search in GenBank revealed high similarities among the conserved domains of these cloned genes with RGA genes. Some clones,however,showed no significant similarity with DNA sequences in GenBank.Southern blotting analysis identified several polymorphic RFLP loci between distinct genotypes.However,none of them co-segregated with the Puccinia psidii Winter resistance gene 1(Ppr1)in a population study.
Cloning·Conserved domains·RGAs·Rust· Eucalyptus
Introduction
Plants have evolved a complex defense system to protect themselves from viral,bacterial,and fungalpathogens.The products of disease-resistant genes play a crucial role in pathogen defense by recognizing specific pathogen effectors(McDowell and Woffenden 2003).Most disease-resistantgenes code proteins thathave been classified in the nucleotide binding site–leucine rich repeat(NBS-LRR) family(Ellis and Jones 1998).These proteins have atleast three conserved domains:an NBS region atthe N terminal and an LRR region.In the N terminal,in addition to the NBS region,there may be a second variable region called coiled coil(CC)(Pan et al.2000)or a domain that is homologous to the domain of Drosophila Toll and the mammalian IL-1R,called TIR(Whitham et al.1996; Hammond-Kosack and Jones 1997).
The CC domain is involved in protein–protein interaction and is found in proteins encoded by genes present in both mono and dicots,such as RPS2,Cre3,RPM1,I2C, Prf,XA1,RPP8,RPS5,Mi and RGC2 thatconfer resistance to various pathogens.The TIR domain of proteins found only in monocots seems to be involved in a signal-transduction pathway.The genes L6,N,Rpp5,M and Rpp1 are included in this subclass(Pan et al.2000).
Disease-resistant genes can be found in distantly related plant families.Plant breeding programs involving disease resistant genes have been widely used to control plant diseases since the turn of the century(McDowell and Woffenden 2003).The presence of conserved NBS–LRR domains in products of different resistance genes enablethe use of PCR to identify and clone the gene-resistant analogs(RGAs)by using degenerate primers targeting these conserved regions.Degenerate primer-based strategies have been successfully used to clone NBS–LRR containing RGAs in various crops,including potatoes, soybeans,wheat,banana,pear,and millet(Leister et al. 1996;Kanazin etal.1996;Feng etal.2010;Lu etal.2011; Zhang et al.2011;Ramachandra et al.2011).
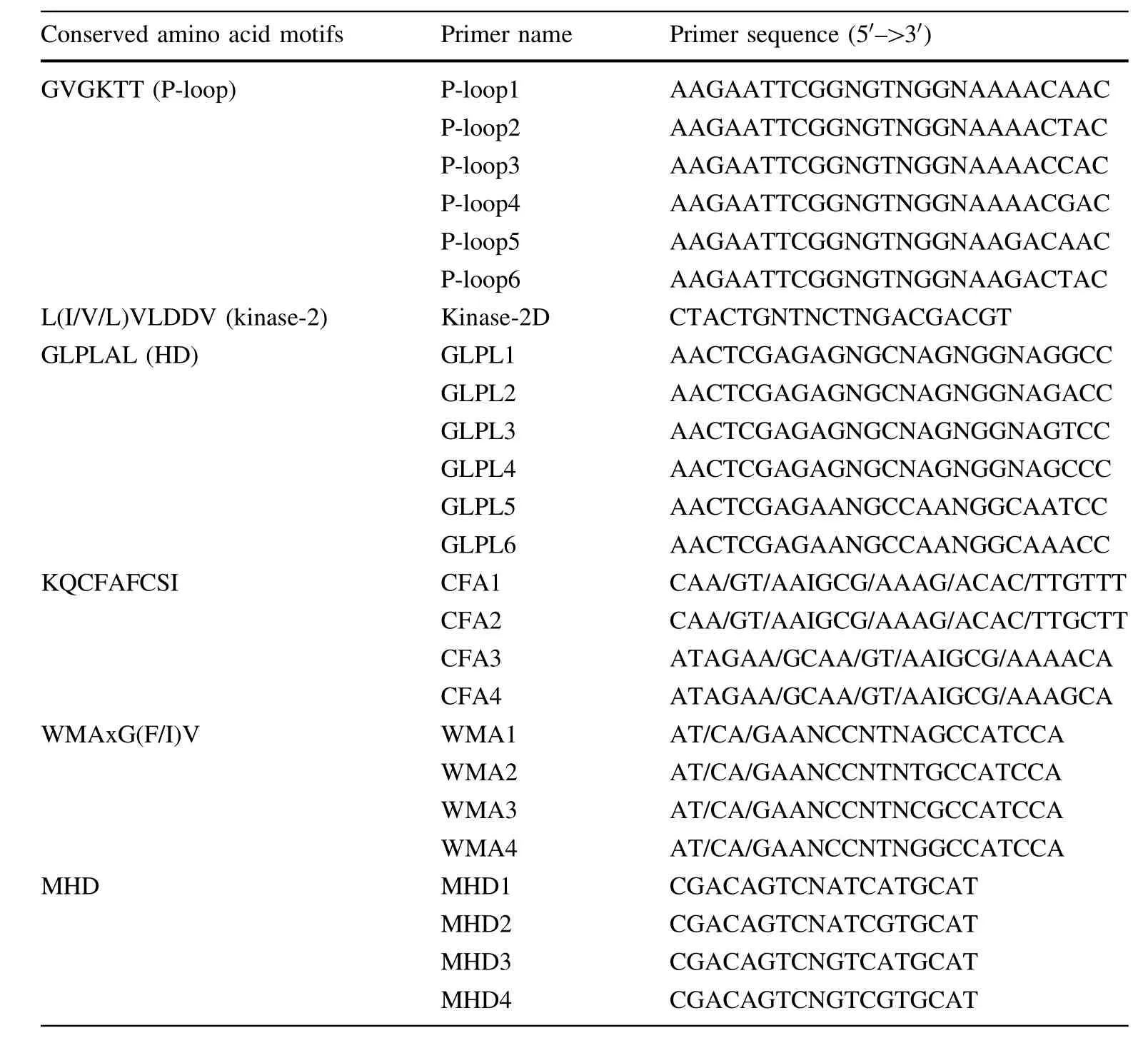
Table 1 Primers used to amplify and tested RGAs via PCR in this study(Collins etal. 1998)
Some of these cloned RGAs were mapped to loci containing disease-resistantgenes in plants,demonstrating the potential application of PCR-based strategies for cloning resistantgenes of NBS–LRR domains.Nevertheless,there are few studies thatdemonstrate the existence of RGAs in tree species and mostof these are limited to poplar(Zhang etal.2006,2008,2010)and pine(Liu and Ekramoddoullah 2004,2007;Jermstad et al.2006).
In Eucalyptus grandis,resistance to the rust,Puccinia psidii Winter,is controlled by a locus,named Ppr1(P. psidii resistance gene 1)(Junghans et al.2003).The cosegregation analysis between the rust-resistance phenotype and RAPD markers allowed us to identify a marker,AT9/ 917,thathas shown to be strongly linked to the Ppr1 locus in a totalof 994 plants,from a cross between a susceptible and a resistant E.grandis.The approximate distance between RAPD marker AT9/917 and the Ppr1 locus is 258,384 bp.Thus,the use of the AT9/917 marker for selection of disease-resistant genotypes may have led to selection of false positives.Therefore,identification of other markers tightly linked to disease-resistant genes can facilitate both positional cloning of the disease-resistant genes and marker-assisted selection of resistance phenotypes. In addition,cloning of the Ppr1 gene can be applied to other genotypes ofcommercialinterestthatare susceptible to this disease.
To further improve genetic breeding of eucalyptus via molecular techniques,this study had two objectives:(1)to amplify,clone,and characterize RGAs in E.grandis;and (2)to determine whether any RGAs and their counterparts co-segregate with Ppr1.
Materials and methods
Plant materials and isolation of genomic DNA
Both a rust resistant(G21)and a susceptible(G38)E. grandis and 41 F1 progeny from the cross between these two parentaltrees were included in this study.The parentaland progeny E.grandis were previously used to study the inheritance and genetic mapping of Ppr1 by Junghans etal. (2003).This previous study showed that recombination events between RAPD markers AC8/1180 and AV10/765 occurred in these F1 progeny(Junghans et al.2003).Genomic DNA was isolated from fresh,young expanding leaves orfrom young leaves stored at-80°C according to the protocoldescribed by Ferreira and Grattapaglia(1995).
Amplification of resistance gene analogs(RGAs)
To potentially amplify disease-resistant genes from E. grandis,degenerate primers were picked up from conserved domains ofotherplantdisease-resistantgenes.First, the selected degenerate primers(Table 1)were used for PCR amplification from E.grandis G21(rust resistant). PCR amplification was then conducted according to the protocoldescribed by Collins etal.(1998).
The expected amplicon size was approximately 600 bp, based on the known DNA sequences of disease-resistant genes in otherplantspecies.To selectappropriate amplicons for cloning and DNA sequencing,the PCR products were subjected to electrophoresis on 1.4%agarose gelin a TBE buffer,containing 0.2 M of ethidium bromide(Sambrook etal.1989).DNAladdersofknown sizewereused to identify thefragmentofinterest.Afterelectrophoresis,thegelimages were photo documented and stored electronically.
Cloning of RGAs
The amplified DNA fragments with sizes close to 600 bp were purified from the agarose gelusing the ConcertRapid Gel Extraction System kit(Life Technologies)and cloned into the pGEM-T Easy vector(Promega),according to the manufacturers’recommendations.The cloned fragments were transformed into competent cells of E.coli DH5-alpha,using the heat-shock method(Sambrook etal.1989). Transformed cells were plated on LB medium containing ampicillin(0.1 mg/ml),IPTG(200 mg/ml),and X-GAL (20 mg/ml)and incubated at 37°C for 12 h.Colonies containing recombinant plasmids were identified by their white color,transferred to tubes containing 3 ml of LB medium with ampicillin(0.1 mg/ml),and incubated at37° C for 12 h,under constantagitation(250 rpm).
Plasmid DNA was isolated by the alkaline lysis method (Sambrook et al.1989)and quantified.The plasmid DNA was then amplified with PCR,using the universal primers, M13F and M13R(GIBCO),or cleaved with enzyme EcoR I to confirm the presence of the fragment of interest.We sought to identify clones with insert size close to 600 bp, since thatis the average size of the fragmentamplified by these primers(Aarts etal.1998;Collins et al.1998).
DNA sequencing and analysis of recombinant plasmids
The nucleotide sequences ofthe inserts were determined in a Perkin-Elmerautomated sequencerABImodel310,using the kit Thermo Sequenase Dye Terminator Cycle Sequencing(Amersham),according to the manufacturer’s instructions.The universalprimers,M13F and M13R,were used for DNA sequencing of inserted fragments,because they are complementary to the plasmid sites flanking the inserted fragments.The obtained nucleotide sequences and the deduced amino acid sequences were aligned using the Clustal W program and subjected to phylogenetic analysis using MEGA,version 2.1(Kumar etal.2001).Finally,the obtained sequences were used to search homologues in the GenBank nr database using the BLASTX algorithm(Altschulet al.1997).
RFLP analysis
Probe preparation
The selected recombinant plasmids described above were subjected to PCR,using the primers,M13F and M13R, under the following reaction conditions:2.5μl of 10×PCR buffer(100 mM Tris–HClpH 8.3,50 mMKCl, 25 mM MgCl2),3.0μlof deoxynucleotidyl(dGTP,dCTP, dATP,dTTP)of 2.5 mM,0.6μl primer M13F of 10 mM, 0.6μl primer M13R of 10 mM,and 0.5μl of Taq DNA polymerase(1 unit),as wellas 30–50 ng of DNA.Doubledistilled sterile water was added to the PCR reaction to reach a volume of 25μl.The PCR amplification program started at 94°C for 1 min,was followed by 30 cycles consisting of30 s each at94°C,1 min at55°C and 1 min at 72°C.At the end of the cycles,a period of 7 min at 72°C was extended.The completed PCR was keptat4°C until removal from the thermal cycler.
The amplified PCR products were separated on 1.4% agarose gelin a TBE buffer solution,containing 0.2 mMof ethidium bromide.DNA ladders of known size were used to identify the fragment of interest.After 90 min of electrophoresis under 90 V,the gelimages were photographed and stored electronically.Then the fragmentofinterestwas purified from the agarose gel,using the Concert Rapid Gel Extraction System kit(Life Technologies),according to manufacturer’s instructions,and quantified.Approximately 25–50 ng DNA was labeled with P32,using the Random Primers DNA Labeling Kit System(Life Technologies) and hybridized with the membranes obtained in the next step,according to the procedures suggested by the manufacturer(Life Technologies and Amersham Pharmacia).
Southern blotting
Genomic DNA of parental and recombinant F1 progeny was digested with the restriction enzymes EcoR I,Dra I, Hind III,Hae III,and EcoR V,according to the procedure recommended by the manufacturer(GIBCO).The digested DNA fragments were separated on 1%agarose gelin TBE buffer at35 V for 12 h.The gelimage was photographed. The gelwas then subjected to depurination in 0.25 M HCl under mild agitation for 10 min.The DNA on the gel was transferred to a hybridization membrane according to the manufacturer’s instructions(Amersham Pharmacia).
The hybridized membranes were placed in contactwith X-ray film in suitable holders and stored at 80°C for 3–10 days,depending on the intensity of the radioactive signal.Afterexposure,the film was developed using the kit GBX(Kodak)and examined visually under a white light transilluminator.
Results and discussion
Due to the highly conserved NBS domain in disease-resistance genes across plantspecies,PCR(using degenerate primers specific for these conserved regions)made it possible to amplify and characterize RGAs in E.grandis (G21).The tested primers(Table 1)generated different amplification patterns.Some primer pairs did not give rise to any amplification from genomic DNA of Eucalyptus.
After testing all possible combinations of primers—P-loop1×GLPL1,P-loop2×GLPL4,P-loop3×GLPL3, P-loop4×GLPL4,P-loop5×GLPL5,P-loop5×GLPL1 and P-loop6×GLPL4—were finally used for PCR amplification of potential RGAs from parental E.grandis (R21)thatwere resistantto rustand generated amplicons of expected size(~600 bp).
Among the 90 recombinant clones obtained,38 were used for extraction of plasmid DNA.Of these,13 were picked randomly(ML 5,ML 21,ML 23,ML 25,ML 27, ML 28,ML 29,ML 31,ML 32,ML 33,ML 34,ML 35, and ML 38)for DNA sequencing.The recombinant plasmids were digested with the restriction enzyme EcoR Iand the size ofthe shorterfragmentwas determined on argarose gelto be about600 bp,suggesting the fragments amplified using degenerate primers may be potential RGAs.
The nucleotide sequences of the 13 selected clones exhibited 32–96%identity when compared with each other. The dendrogram obtained,on the basis of nucleotide sequence,classified these clones into four groups(Fig.1). The clone ML23 was the most diverse in the group,and clones ML31 and ML35 were the closest to each other (Fig.1).The dendrogram obtained on the basis of deduced amino sequence identified five groups(Fig.2).Clones ML27 and ML33 were grouped togetherwith clones ML35 and ML31,unlike the organization found on the basis of the nucleotide sequences.Clones ML 21 and ML23 stood outas the mostdistinctgroup(Fig.2).Itis noteworthy that clones ML35,ML31,ML27,and ML33 are similar to resistance genes N,L6,and M.These genes have a TIR domain at the N-terminal,following the NBS domain.
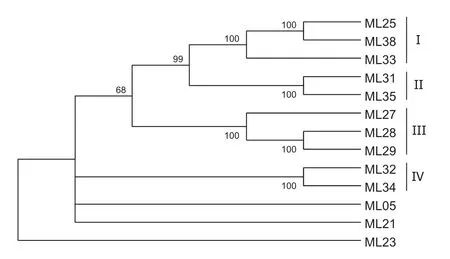
Fig.1 Dendrogram of RGA clones based on DNA sequences.The values at the nodes are bootstrap values supporting the phylogenetic organization.Group I=1,Group II=2,Group III=3,Group IV=4
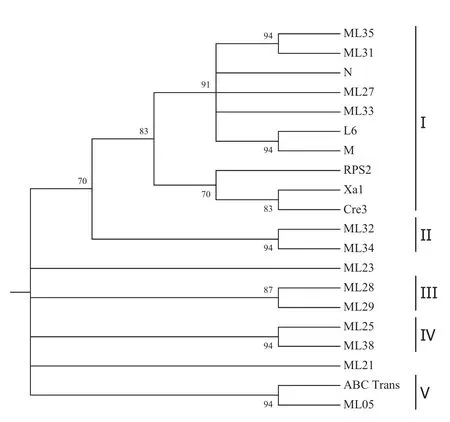
Fig.2 Relationship at the level of amino acid sequences among the recombinant clones from rust resistant E.grandis.The values at the nodes are bootstrap values.C gene conferring resistance to TMV in tobacco;L6 gene conferring resistance to Melampsora lini in linen, M gene conferring resistance to Melampsora lini in linen,RPS2 gene conferring resistance to Pseudomonas syringae pv.tomato in Arabidopsis,XA1 gene conferring resistance to Xanthomonas oryzae pv.oryzae in rice,Cre3 gene conferring resistance to Heterodera avenae in wheat,ABC=ABC transporter of Arabidopsis thaliana. I=Group 1,II=Group 2,III=Group 3,IV=Group 4, V=Group 5
Alignment of amino-acid sequences between cloned RGAs and previously identified and characterized disease-resistant genes showed similarities of conserved domains, demonstrating that the degenerate primers can be used to identify a large number of RGAs,even though they were designed on sequences of other plant species.

Table 2 Genes and/or amino acid sequences conferring to disease resistance found in GenBank thathave significanthomology to recombinant clones
When searched with the BLASTX algorithm(Table 2), some homologs of the Eucalyptus RGAs were found in GenBank.The protein sequence of RGA ML05 shared high similarity with the Arabidopsis ABC transporter.ML25 is a likely homolog of OB8 and LM13,disease-resistant genes found in beans and soybeans,respectively.The BLASTX search also revealed that clones ML27 and ML34 are similar to gene N in Nicotiana glutinosa and gene RPS5 in Arabidopsis thaliana,respectively.In addition,the clone ML27 was similar to the gene N homolog,presentin Pinus radiata.
The difference between the sequences in separate dendrogram groups was confirmed by different hybridization patterns,revealed by Southern blotting,using the clones ML27,ML33,ML34,and ML35,representative of each group as probes(Fig.3).These results also suggestthatthe identified RGAs are likely members of multigene families, as observed in otherstudies(Kanazin etal.1996;Speulman et al.1998;Hulbertetal.2001).
Since RGAs are usually members of multigene families, it is essential to establish the relationship between RGAs and the locus containing the gene of interest.It is then possible to infer the relationship of RGAs with the gene of interestor by using RGAs in marker-assisted selection.
Since Southern blotting has revealed genetic polymorphisms between the parental E.grandis(R21 and R38)that have different phenotypes(resistant and susceptible),this technique was further used to investigate whether any RGA clone was linked with the disease resistance gene Ppr1.For this purpose,four RGA/enzyme combinations(ML27/ Dra I,ML33/Dra I,ML34/EcoR V and ML35/Hae III)were selected for Southern blot analysis in the 41 F1 progeny with recombination events between markers AC8/1180 and AV10/765 to determine if any of the RGAs were linked to the Ppr1 locus.
In Eucalyptus spp.,although several RFLP polymorphism were identified in rust-resistant against rust susceptible trees,no evidence of co-segregation of these RGAs with Ppr1 locus was confirmed(Fig.4),indicating that none of these RGAs is linked to the genomic region containing Ppr1.
Similar results were found in the study conducted by Spielmeyer et al.(2000),using the same strategy with wheat.The authors failed to identify any RGAs linked to the disease-resistance genes and speculated thatthis failure was due to the low number of plants used and the smallnumber of resistance genes cloned.In Eucalyptus spp.,itis possible that the low number of RGAs used may have contributed to the failure of the present study.

Fig.3 RFLP profile of the parental E.grandis(G21 is resistant and G38 is susceptible),by hybridization with probe RGAs:A—ML27 and B—ML34.Genomic DNA was cleaved with restriction enzymes EcoR I,Dra I,Eco RV,Hind III and Hae III.λlambda phage DNA cleaved with the restriction enzyme Hind III,bp base pairs

Fig.4 Segregation analysis of RFLPs detected with the probe from RGA ML34 clone. Genomic DNA was digested with the restriction enzyme EcoR I.G21 and G38,parental E.grandis resistant and susceptible to eucalyptus rust, respectively.λlambda phage DNA digested with the restriction enzyme Hind III,bp base pairs,Underlined resistant to eucalyptus rust,not underlined susceptible to eucalyptus rust
In contrast,other researchers have successfully mapped disease-resistantgenes in plants by using degenerate primers. Feuilletetal.(1997)cloned a resistantgene(Lr10)using the RGAanalysistechniquein Wheatand Collinsetal.(1998)and found two RGAs linked to the disease resistance genes Rp1 and Rp3 in corn.Furthermore,these authorscloned and linked one RGAto the gene Hm1 and 20 quantitative traitloci(QTL), conferring resistance to diseases and pests.
In A.thaliana,Aarts et al.(1998)mapped eight RGA loci,six of which were closely linked to genes RPS5,Rpp4 and Rpp5.Unlike the pathosystem of E.grandis and P. psidii,the high number of resistant genes in maize and Arabidopsis may make iteasier to identify large amountof RGAs.The effectiveness of this technique for mapping resistance genes were developed in severalcrops,including melons,chickpeas,soybeans,and oats have been well documented(Garcia-Mas et al.2001;Huettel et al.2002; Wang et al.2004;Satheeskumar et al.2011;Sanz et al. 2013).
Studies involving a largernumberof RGAs can increase the chances of identifying loci linked to the Ppr1 gene,as well as allow for inferences about the presence or absence of an NBS domain in this gene.However,as NBS domains are found in many protein families,including atpases, elongation factors,and G proteins(Saraste et al.1990),it may be difficult to identify RGAs contributing to disease resistance as mentioned above.It is possible to use a greater number of RGAs as probes to find RGAs that cosegregate with the gene Ppr1.Even if obtaining a link between an RGA and Ppr1 proves futile,construction of a genetic map from NBS probes can be useful,as itcan chart and characterize additional genes of interest in E.grandis genotypes.
However,the identification of candidate genes that confer disease resistance(through RFLP analysis of RGA sequences)may have limitations,because polymorphisms between differentgenotypes can be caused by mutations in pseudogenes.In this case,the sequences are not actually associated with resistant genes,but can be useful to saturate a region near the gene of interest in subsequent positional cloning of a disease-resistance gene.
AcknowledgmentsWe thank the Suzano Celulose S/A for logistical support and the‘‘Conselho Nacional de Desenvolvimento Cientı´fico e Tecnolo´gico’’(CNPq),the‘‘Coordenac¸a˜o de Aperfeic¸oamento de Pessoal de Nı´vel Superior’’(CAPES)and the‘‘Fundac¸a˜o de Amparo a`Pesquisa do Estado de Minas Gerais’’(FAPEMIG)for financial support to this study.
Aarts MG,te Lintel Hekkert B,Holub EB,Beynon JL,Stiekema WJ, Pereira A(1998)Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana.Mol Plant Microbe Interact 11:251–258
Altschul SF,Madden TL,Scha¨ffer AA,Zhang J,Zhang Z,Miller W, Lipman DJ(1997)Gapped BLAST and PSI-BLAST:a new generation of protein database search programs.Nucleic Acids Res 25:3389–3402
Collins NC,Webb CA,Seah S,Ellis JG,Hulbert SH,Pryor A(1998) The isolation and mapping of disease resistance gene analogs in maize.Mol Plant Microbe Interact 11:968–978
Ellis J,Jones D(1998)Structure and function of proteins controlling strain-specific pathogen resistance in plants.Curr Opin Plant Biol 1:288–293
Feng DS,Ma X,Lin AL,Wang HG,Tian JC(2010)Isolation of resistance gene analogues to powdery mildew resistance sequences in hexaploid wheat.Biol Plant54:551–555
Ferreira ME,Grattapaglia D(1995)Introduc¸a˜o ao uso de marcadores RAPD e RFLP em ana´lise gene´tica.Embrapa-Cenargen, Brası´lia,p 220
Feuillet C,Schachermayr G,Keller B(1997)Molecular cloning of a new receptor-like kinase gene encode at the Lr10 disease resistance locus of wheat.Plant J 11:45–52
Garcia-Mas J,Van Leeuwen H,Monfort A,de Vicente MC, Puigdomenech P(2001)Cloning and mapping of resistance gene homologues in melon.Plant Sci 161:165–172
Hammond-Kosack KE,Jones JDG(1997)Plant disease resistance genes.Annu Rev Plant Biol 48:575–607
Huettel B,Santra D,Muehlbauer FJ,Kahl G(2002)Resistance gene analogues of chickpea(Cicer arietinum L.):isolation,genetic mapping and association with a Fusarium resistance gene cluster.Theor Appl Genet 105:479–490
Hulbert SH,Webb CA,Smith SM,Sun Q(2001)Resistance gene complexes:evolution and utilization.Annu Rev Phytopathol 39:285–312
Jermstad KD,Sheppard LA,Kinloch BB,Delfino-Mix A,Ersoz ES, Krutovsky KV,Neale DB(2006)Isolation of a full-length CCNBS-LRR resistance gene analog candidate from sugar pine showing low nucleotide diversity.Tree Genet Genomes 2:76–85
Junghans DT,Alfenas AC,Brommonschenkel SH,Oda S,Mello EJ, Grattapaglia D(2003)Resistance to rust(Puccinia psidii Winter) in eucalyptus:mode of inheritance and mapping of major gene with RAPD markers.Theor Appl Genet 108:175–180
Kanazin V,Marek LF,Shoemaker RC(1996)Resistance gene analogs are conserved and clustered in soybean.Proc Natl Acad Sci USA 93:11746–11750
Kumar S,Tamura K,Jakobsen IB,Nei M(2001)MEGA2:molecular evolutionary genetics analysis software.Bioinformatics 17:1244–1245
Leister D,Ballvora A,Salamini F,Gebhardt C(1996)A PCR based approach for isolating pathogen resistance genes from potato with potential for wide applications in plants.Nat Genet 14:421–429
Liu JJ,Ekramoddoullah AKM(2004)Isolation,genetic variation and expression of TIR-NBS-LRR resistance gene analogs from western white pine(Pinus monticola Dougl.ex.D.Don.).Mol Genet Genomics 270:432–441
Liu JJ,Ekramoddoullah AKM(2007)The CC-NBS-LRR subfamily in Pinus monticol a:targeted identification,gene expression,and genetic linkage with resistance to Cronartium ribicola.Phytopathology 97:728–736
Lu Y,Xu WH,Xie YX,Zhang X,Pu JJ,Qi YX,Li HP(2011) Isolation and characterization of nucleotide-binding site and C-terminal leucine-rich repeat-resistance gene candidates in bananas.Genet Mol Res 10:3098–3108
McDowell JM,Woffenden BJ(2003)Plant disease resistance genes: recent insights and potential applications.Trends Biotechnol 21:178–183
Pan Q,Liu YS,Budai-Hadrian O,Sela M,Carmel-Goren L,Zamir D, Fluhr R(2000)Comparative genetics of nucleotide binding siteleucine rich repeat resistance gene homologues in the genomes of two dicotyledons:tomato and arabidopsis.Genetics 155: 309–322
Ramachandra SB,Sathyanarayana NR,Subramonium S,Shetty SH (2011)Isolation,cloning and characterization of resistance gene analogues in pearlmilletbased on conserved nucleotide-binding sites.J Phytopathol 159:382–389
Sambrook L,Fritsch EF,Maniatis T(1989)Molecular cloning:a laboratory manual.Cold Spring Harbor Laboratory Press,New York,p 956
Sanz MJ,LoarceY Fominaya A,Vossen JH,Ferrer E(2013) Identification of RFLP and NBS/PK profiling markers for disease resistance loci in genetic maps of oats.Theor Appl Genet 126:203–218
Saraste M,Sibbald PR,Wittinghofer A(1990)The P-loop:a commom motif in ATP-and GTP-binding proteins.Trends Biochem Sci 15:430–434
Satheeskumar S,Sharp PJ,Lagudah ES,McIntosh RA,Molnar SJ (2011)Genetic association of crown rust resistance gene Pc68, storage protein lociand resistance gene analogs in oats.Genome 54:484–497
Speulman E,Bouchez D,Holub EB,Beynon JL(1998)Disease resistance gene homologs correlate with disease resistance loci of Arabidopsis thaliana.Plant J 14:467–474
Spielmeyer W,Huang L,Bariana H,Laroche A,Gill BS,Lagudah ES (2000)NBS-LRR sequence family is associated with leaf and stripe rust resistance on the end of homoeologous chromosome group 1S of wheat.Theor Appl Genet 101:1139–1144
Wang B,Wang Y,Wang Q,Luo G,Zhang Z,He C,He SJ,Zhang J, Gai J,Chen S(2004)Characterization of an NBS-LRR resistance gene homologue from soybean.J Plant Physiol 161:815–822
Whitham S,Mccormick S,Baker B(1996)The N gene of tobacco cofers resistance to tobacco mosaic virus in transgenic tomato. Proc Natl Acad Sci USA 93:8776–8781
Zhang Y,Zhang SG,Qi LW,Liu B,Xiong BQ,Gao JM,Chen XQ, Chen CB,Li XL,Song WQ(2006)Cloning and characterization ofdisease resistance gene analogs from poplar(Populus tremula) chromosome 1.Int J Plant Sci 167:403–412
Zhang Q,Zhang ZY,Lin SZ,Zheng HQ,Lin YZ,An XM,Li Y,Li HX(2008)Characterization of resistance gene analogs with a nucleotide binding site isolated from a triploid white poplar. Plant Biol 10:310–322
Zhang Y,Zhang SG,Qi LW,Zhang T,Wang CG,Chen CB,Song WQ(2010)Isolation,characterization and phylogenetic analysis of nucleotide binding site-encoding disease-resistance gene analogues from European Aspen(Populus tremula).Silvae Genet 59:68–77
Zhang H,Wang Y,Zhang C,Wang X,Li H,Xu W(2011)Isolation, characterization and expression analysis of resistance gene candidates in pear(Pyrus spp.).Sci Hortic 127:282–289
11 June 2013/Accepted:11 November 2013/Published online:5 May 2015
©Northeast Forestry University and Springer-Verlag Berlin Heidelberg 2015
The online version is available at http://www.springerlink.com
Corresponding editor:Chai Ruihai
✉Acelino Couto Alfenas aalfenas@ufv.br
1Departamento de Engenharia Florestal,Universidade Federal dos Vales do Jequitinhonha e Mucuri,Diamantina, Minas Gerais 39100-000,Brazil
2Departamento de Fitopatologia,Universidade Federal de Vic¸osa,Vic¸osa,Minas Gerais 36570-900,Brazil
3Suzano Bahia Sul Celulose S/A,Suzano,Sa˜o Paulo,Brazil
杂志排行
Journal of Forestry Research的其它文章
- Management of pests and diseases of tropical sericultural plants by using plant-derived products:a review
- Gamma generalized linear model to investigate the effects of climate variables on the area burned by forest fire in northeast China
- Diversity,abundance,and structure of tree communities in the Uluguru forests in the Morogoro region,Tanzania
- Brazilian savanna re-establishment in a monoculture forest: diversity and environmental relations of native regenerating understory in Pinus caribaea Morelet.stands
- Carbon storage and sequestration rate assessment and allometric model development in young teak plantations of tropical moist deciduous forest,India
- Use of infrared thermal imaging to diagnose health of Ammopiptanthus mongolicus in northwestern China
