Deficiency of rpoS is the major factor leading to attenuation of Salmonellaenterica serovar Choleraesuis vaccine strain C500
2015-03-17XULijuanLIQiuchunLIUJieHUYachenTAOMingxinXIEXiaoleiGENGShizhongJIAOXinan
XU Li-juan,LI Qiu-chun,LIU Jie,HU Ya-chen,TAO Ming-xin,XIE Xiao-lei,GENG Shi-zhong,JIAO Xin-an
Deficiency ofrpoSis the major factor leading to attenuation ofSalmonellaentericaserovar Choleraesuis vaccine strain C500
XU Li-juan,LI Qiu-chun,LIU Jie,HU Ya-chen,TAO Ming-xin,XIE Xiao-lei,GENG Shi-zhong,JIAO Xin-an
Salmonellaentericaserovar Choleraesuis strain C500 is a live attenuated vaccine that has been widely used in China for over 50 years to prevent piglet paratyphoid. However, as C500 is obtained by chemical methods, the genetic background of this strain remained unclear. In this study, we compared the genomic differences between the virulent reference strain C78-2 and C500 by suppression subtractive hybridization combined with the mirror orientation selection method (MOS-SSH). Six genes (asr,ydgF,ydgD,ydgE,rpoS, andptsG) were lost in C500 strain. Using real-time PCR analysis, we demonstrated that the genes regulated byrpoS, a vital transcriptional regulator playing an important role inSalmonellainfection, were downregulated in C500. Additionally, the virulence of therpoSmutant strain C78-2ΔrpoSwas 100 000 times lower than the parental strain in BALB/c mice. So loss ofrpoSgene is the major factor leading to the attenuation of C500 strain.
SalmonellaCholeraesuis; vaccine; attenuation; Mirror Orientation Selection (MOS); Suppression Subtractive Hybridization (SSH)
Salmonellaentericaserovar Choleraesuis (SalmonellaCholeraesuis) is a host-adapted pathogen that causes swine paratyphoid[1]. Because it can also cause septicaemic disease in humans, the reservoir ofSalmonellaCholeraesuis in swine is a concern for the government[2-3]. Vaccination has been proven to be a feasible approach to controlSalmonellain swine, thus lessening the likelihood ofSalmonellatransmission through the food chain to humans. Different vaccines againstSalmonellaCholeraesuis have been developed and utilized in China, the United States, and Europe[1]. Some vaccines were derived by mutation of the genes involved in bacterial survival or virulence, while others were obtained by chemical mutagenesis[4-8]. The basis of attenuation for the Arco vaccine is not understood as it, along with the C500 vaccine, was derived by chemical mutagenesis.
The C500 strain has not only been used as vaccine against swine paratyphoid, but also attracted a great deal of interest as potential live oral vaccine vectors for the delivery of DNA vaccines adapted to swine[9]. Although the vaccine strain has been used in China for over 50 years, the genetic background of the vaccine remained unclear. To develop the C500 strain as a safe and convenient vaccine vector, it is essential to elucidate the attenuation mechanism of the C500 strain at the molecular level. In this study, suppression subtractive hybridization combined with the mirror orientation selection method (MOS-SSH) was applied to detect the genomic changes in C500 with comparison to the virulent strain C78-2. The relationships between genes deleted during chemical mutagenesis and the attenuation of virulence were further identified and evaluated.
Materials and methods
Suppression Subtractive Hybridization (SSH)
SalmonellaCholeraesuis strains C78-2 and C500 were provided by the China Institute of Veterinary Drug Control. SSH was performed usingSalmonellaCholeraesuis C78-2 genomic DNA as the tester and C500 genomic DNA as the driver. The genomic subtraction between C78-2 and C500 was performed using a Clontech PCR-SelectTMBacterial Genome Subtraction Kit (Clontech, Takara, Japan). After two rounds of hybridization, the primary PCR products were collected and saved to eliminate false positive clones by mirror orientation selection.
Mirror orientation selection (MOS) to eliminate false positive clones
The mirror orientation selection (MOS) method was used to eliminate background clones representing non-differentially (redundant) DNA fragments in the subtracted library[10]. Briefly, 10 independent tubes of primary PCR products from SSH were combined and diluted 1 000 fold. Using the same primer and conditions for primary PCR, the secondary PCR products were obtained and adjusted to the concentration of 20-30 ng/μL. The NP1 adapters were then removed from both termini byXmaI, after which 1 μL ofXmaI-digested DNA (5-7 ng) was mixed with 1 μL of 4 × hybridization buffer and 2 μL of driver C500RsaI-digested DNA (300 ng/μL). After incubation in a PCR system at 98 ℃ for 1.5 min and then at 68 ℃ for 3 h, the sample was mixed with 200 μL of dilution buffer, heated at 63 ℃ for 7 min and then 1 μL of diluted DNA was used for subsequent PCR with the adapter-specific primer NP2Rs (5′-GGTCGCGGCC GAGGT-3′). The purified PCR products were cloned into a pMD18®-T cloning vector (Takara, Japan) and transformed intoE.coliDH5α competent cells. The plasmids harbouring inserted DNA fragments were identified by PCR analysis.
Dot-Blot assay, DNA sequencing and Identification of positive clones
A Dot-blot assay was used to screen the specific fragments of C78-2 in the subtracted library according to the protocol of the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche, Germany). Sequencing of the plasmid templates was performed using the Applied Biosystems 3730 DNA sequencer at the Nanjing Genscript Company (http://www. genscript.com.cn/index.html). The DNA sequences were compared with the GenBank databases using BLASTN. According to the sequence of the fragments, primer pairs were synthesized for their identification in strain C78-2 and C500.
Quantitative Real Time PCR analysis
Quantitative real-time PCR (qRT-PCR) was used to compare gene expression between C78-2 and C500. The total RNA of C78-2 and C500 cells cultured in liquid LB for 16 h was extracted using a Bacterial total RNA Extraction Kit (Tiangen, China) following the manufacturer’s instruction. Contaminating genomic DNA was removed from the RNA sample by RNase-Free DNase I (Takara, Japan), and then reverse transcribed into cDNA by a PrimeScriptTMRT reagent kit (Takara, Japan) for qRT-PCR analysis. The qRT-PCR was performed as a relative quantification run using a 7500 system (Applied Biosystems, USA) and a SYBR Premix Ex TaqTMII kit (Takara, Japan). The qRT-PCR reactions were performed in a final volume of 20 μL containing 12 μL Premix Ex Taq II, 0.8 μL of each primer, 0.4 μL ROX Reference Dye II, 2 μL cDNA template, and 6 μL double-distilled water (ddH2O). The protocol for qRT-PCR was as follows:hot start at 95 ℃ for 30 s, followed by 45 cycles at 95 ℃ for 5 s and 60 ℃ for 34 s. The reproducibility of the qRT-PCR reaction was confirmed by running independent reactions from independently grown cultures. All runs were performed three times, and the results for each gene were averaged.
Construction of the C78-2ΔrpoSmutant and complementary strain
To identify the relationship between the deletion ofrpoSand the attenuation of C500, disruption ofrpoSgene was performed in C78-2 strain to give rise to C78-2ΔrpoSmutant. C78-2ΔrpoSwas prepared by homologous recombination using a suicide vector pGMB151 to delete therpoSgene. The 712 bp upstream fragment and 859 bp downstream fragment ofrpoSgene was amplified by PCR and ligated together, and then inserted into the suicide vector pGMB151. The recombinant plasmid was transformed into C78-2 to produce the mutant strain C78-2ΔrpoS.
TherpoSgene from C78-2 was then cloned into the downstream of thelacpromoter in the pBR322 vector (Takara, Japan) to give rise to complementary plasmid prpoS. After identification of the nucleotide sequences in the recombinant plasmids by sequencing analysis using an ABI PRISM 3100-Avant Genetic Analyzer, the recombinant plasmid was then transformed into C78-2ΔrpoSto produce C78-2ΔrpoS/propS.
Animal test
Eight week-old mice were purchased from Experimental Animal Centre of Yangzhou University. The mice were treated according to protocols approved by Yangzhou University’s Institutional Animal Care and Use Committee. The LD50was determined for the three strains (C78-2, C500, C78-2ΔrpoS, C78-2ΔrpoS/propS). Five groups of mice for each strain were challenged intraperitoneal (i.p.) with different amounts of bacteria. The mortality was observed for 30 days, and the LD50was calculated according to a method described by Reed and Muench[11].
Results
MOS-SSH identification of deleted sequences in the vaccine strain C500 compared to C78-2
Both the tester and the driver DNAs were digested byRsaI and used in SSH and MOS procedures. As shown in Figure 1a, the electrophoretic gel of the amplified products showed fragments ranging from 200 to 2 000 bp. The SSH product was an even smear with almost unapparent bands (Figure 1a, lane 1). However, after MOS, the sample contained bright bands with the background level reduced, as shown by the decrease in the smear in lane 2 of Figure 1a.
Seven clones were screened out from the subtracted library containing 240 clones by Dot-blot analysis with the two genomic DNA probes. Sequencing and homology analysis showed that these clones represented five DNA fragments includingasr,ydgF,rpoS,ptsGgenes and CRISPR sequences, as shown in Table 1. As shown in Figure 1b, PCR analysis with specific primers to these DNA fragments confirmed that these genes were lost in C500. Interestingly, theasrgene was close toydgF, so a pair of primers was designed to detect the length of the deleted sequences. Compared to C78-2, sequencing analysis showed there was a 3290 bp fragment deficient in the C500 genome, including theasr,ydgD,ydgEandydgFgenes (Figure 1c).

(a). Isolation ofSalmonellaCholereasuis C78-2 DNA fragments not present in C500 using SSH and MOS-SSH.
Lane 1:Final PCR product of unsubtracted C78-2 DNA fragments. Lane 2:Final PCR product of specific strain C78-2 DNA sequences using SSH. Lane3:Final PCR product of specific strain C78-2 DNA sequences using MOS-SSH. Lane M:DL 2000 marker (Takara, Japan).
(b). Identification of positive clones using PCR analysis. The PCR-amplified fragments were C7003 ( Lanes 1 & 2 ), C7006 ( Lanes 3 & 4 ), C7026 ( Lane 5 & 6 ), C7154 ( Lanes 7 & 8 ), C7625 ( Lanes 9 & 10 ).
Lanes 1, 3, 5, 7, and 9 show the PCR-amplified fragments when using the genomic DNA of strain C78-2 as the template. Lanes 2, 4, 6, and 8 show the PCR-amplified fragments when using the genomic DNA of C500 strain as the template.
(c). Identification of fragments betweenasrandydgFgenes deficient in C500 strain.
Lane 1:The PCR products amplified from C500 genomic DNA; Lane 2:The PCR products amplified from C78-2 genome. Lane M:λ-EcoT14 marker.
Fig.1 Screening and identification of DNA fragments deficient in C500 by SSH and MOS
Differences in gene expression between C500 and C78-2 as determined by quantitative real-time PCR
Based on comparison between the twoSalmonellaCholeraesuis strains, one regulator gene (rpoS) has been confirmed to be an important reg-ulator involved inSalmonellainfection. Seven genes (spvA,spvB,spvC,osmY,glnP,osmEandotsB) regulated byrpoSwere selected for further analysis to monitor changes at the mRNA level using qRT-PCR. The gene forSalmonellaCholeraesuis guanylate kinase (gmk) was used as a reference. The results showed that the expression levels ofrpoS-regulated genesotsB,spvA,spvB,spvC, andosmYin C78-2 were significantly higher than in C500 (Figure 2).

Tab.1 Results of homology search with subtracted DNA fragments.
Note:a:Homology listed is based on nucleotide sequences (BLASTN).
b:As the homology of the sequences increases, the e-value approaches 0.
Role ofrpoSin virulence ofSalmonellaCholeraesuis
For further identification of the role ofrpoSdeficiency in attenuation of C500, we successfully constructed therpoSgene deletion mutant C78-2ΔrpoSfrom C78-2. No obvious difference was observed in theinvitrogrowth curves of the wild-type strain and the mutant strain, but the i.p. LD50of C78-2ΔrpoSto BALB/c mice 4.8 × 107colony forming units (CFU), which represented > 4-log increase in the i.p. LD50of 6.4 × 102CFU that was observed for the wild-type parent C78-2. And overexpression ofrpoSin C78-2ΔrpoS/propScaused the LD50 increased to 6.4 × 101CFU. This result is consistent with the report that mutation ofrpoScaused the dramatic attenuation ofSalmonellaCholeraesuis[12].
To validate the effect ofrpoS-deficient in C500, seven genes (spvA,spvB,spvC,osmY,glnP,osmEandotsB) regulated byrpoSwere selected to monitor their expression difference with C78-2. Total bacterial RNA was extracted from C78-2 or C500 grown in LB broth media was used as the template to assay the expression level of these genes by real-time PCR. The results shown are the mean of three independent experiments (error bars, SD). The data were compared by Student’st-test (*P<0.05, **P<0.01, ***P<0.001).
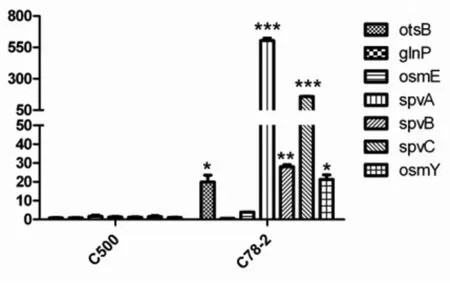
Fig.2 Identification of screened genes andrpoS-regulated genes by Real-time PCR analysis
Discussion
Compared to SSH without MOS, the sample containing bright bands with the background level were highly reduced after MOS (Figure 1a, lane 3). The background level of the PCR products using MOS-SSH was obviously reduced compared to SSH, implying that MOS decreased the portion of false positive clones as has been described previously[10].
Among the screened seven fragments including six genes, the sigma factor RpoS has been identified as an important regulator involved in positively regulating more than 50 genes inE.coli[13]. The RpoS mediates the stationary-phase expression of a wide variety of genes in response to environmental stresses, including nutrient deprivation, heat, oxidative and osmotic shocks[14]. It is also reported that RpoS regulates the expression of theSalmonellavirulence plasmidspvgenes, which control the growth rate ofSalmonellain deep organs and are required for systemic infection[15]. In this study, the expression level of threespvgenes were expressed highly in C78-2 with comparison to C500. Besides, otherrpos-regulated genes includingosmYandostBwere also downregulated in C500. Given the diversity and importance ofrpoS-regulated genes, it is not surprising thatrpoSmutant demonstrate increased susceptibility to the environmental stresses and reduced virulence to host[15-17]. ArpoSmutant ofSalmonellaTyphimurium 14028s has a 1 000-fold increase in the LD50[16], while mutation ofrpoSin C78-2 caused a 10 000-fold decrease in virulence, demonstrating the role ofrpoSin the disease-causing potential ofSalmonella.
In order to develop new carrier vectors highly adapted to the pig,SalmonellaCholeraesuisphoPandrpoSmutant was constructed and evaluated for their attenuation in mice and pigs[18]. Both mutants were extremely attenuated in comparison to wild-type strain and a commercial live vaccine strain SC-54. Further analysis showed thatSalmonellaCholeraesuisrpoSmutant and other attenuated strains could be used as a bactofection vector to deliver foreign genes to the intestinal immune system[9,12]. As therpoShas been confirmed as a very important regulator playing critical roles inSalmonellainfection and pathogenesis, we speculated that the deletion ofrpoSin C500 strain may be the major reason causing the virulence downregulated.
[1]Chiu CH, Su LH, Chu C.Salmonellaentericaserotype Choleraesuis:epidemiology, pathogenesis, clinical disease, and treatment [J]. Clin Microbiol Rev, 2004, 17:311-322.
[2]Blaser MJ, Feldman RA.Salmonellabacteremia:reports to the Centers for Disease Control, 1968-1979[J]. J Infect Dis, 1981, 143:743-746.
[3]Chiu CH, Wu TL, Su LH, et al. The emergence in Taiwan of fluoroquinolone resistance inSalmonellaentericaserotype Choleraesuis[J]. N Engl J Med, 2002, 346:413-419.
[4]Kelly SM, Bosecker BA, Curtiss III R. Characterization and protective properties of attenuated mutants ofSalmonellacholeraesuis[J]. Infect Immun, 1992, 60:4881-4890.
[5]Kennedy MJ, Yancey RJ Jr, Sanchez MS, et al. Attenuation and immunogenicity ofΔcrp-cdtderivatives ofSalmonellacholeraesuisin pigs[J]. Infect Immun, 1999, 67:4628-4636.
[6]Roof MB. Doitchinoff DD. Safety, efficacy, and duration of immunity induced in swine by use of an avirulent liveSalmonellacholeraesuiscontaining vaccine[J]. Am J Vet Res, 1995, 56:39-44.
[7]Fang X, Li Y, Huang C, et al. Live vaccine against swine paratyphoid from the attenuated smooth strain C500 ofSalmone-llacholeraesuis[J]. Chin J Anim Vet Sci, 1981, 12:99-106.
[8]Huang C, Feng W, Xue M, et al. Oral administration of the live vaccine against swine paratyphoid[J]. Sci Agric Sin, 1981, 6:89-94.
[9]Zhao Z, Xue Y, Wu B, et al. Subcutaneous vaccination with attenuatedSalmonellaentericaserovar Choleraesuis C500 expressing recombinant filamentous hemagglutinin and pertactin antigens protects mice against fatal infections with bothS.entericaserovar Choleraesuis andBordetellabronchiseptica[J]. Infect Immun, 2008, 76:2157-2163.
[10]Rebrikov DV, Britanova OV, Gurskaya NG, et al. Mirror orientation selection (MOS):a method for eliminating false positive clones from libraries generated by suppression subtractive hybridization[J]. Nucleic Acids Res, 2000, 28:e90.
[11]Reed LJ, Muench H. A simple method of estimating fifty percent endpoints[J]. Am J Hyg, 1938, 27:493-497.
[12]Bartolome A, Herrero-Gil A, Horcajo P, et al.Salmonellaentericaserovar Choleraesuis derivatives harbouring deletions inrpoSandphoPregulatory genes as vehicles for DNA vaccines[J]. Vet Microbiol, 2010, 141:81-88.
[13]Loewen PC, Hu B, Strutinsky J, et al. Regulation in therpoSregulon ofEscherichiacoli[J]. Can J Microbiol, 1998, 44:707-717.
[14]Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes inEscherichiacoli[J]. J Bacteriol, 2001, 183:5803 5812.
[15]Kowarz L, Coynault C, Robbe-Saule V, et al. TheSalmonellatyphimuriumkatF(rpoS) gene:cloning, nucleotide sequence, and regulation ofspvRandspvABCDvirulence plasmid genes[J]. J Bacteriol, 1994, 176:6852 6860.
[16]Nickerson CA, Curtiss III R. Role of sigma factor RpoS in initial stages ofSalmonellatyphimuriuminfection[J]. Infect Immun, 1997, 65:1814-1823.
[17]Fang FC, Libby SJ, Buchmeier NA, et al. The alternative sigma factor KatF ( RpoS ) regulatesSalmonellavirulence[J]. Proc Natl Acad Sci U S A, 1992, 89:11978-11982.
[18]Dominguez-Bernal G, Tierrez A, Bartolome A, et al.Salmonellaentericaserovar Choleraesuis derivatives harbouring deletions inrpoSandphoPregulatory genes are attenuated in pigs, and survive and multiply in porcine intestinal macrophages and fibroblasts, respectively[J]. Vet Microbiol, 2008, 130:298-311.
Received:2015-05-25;Revision accepted:2015-08-11
(JiangsuKeyLaboratoryofZoonosis/JiangsuCo-InnovationCenterforPreventionandControlofImportantAnimalInfectiousDiseasesandZoonoses,YangzhouUniversity,Yangzhou225009,China)
10.3969/j.issn.1002-2694.2015.10.003
Jiao Xin-an, Email:jiao@yzu.edu.cn

Supported by the National Natural Science Foundation of China (Nos. 31320103907, 31230070, 31201905), the Special Fund of Agro-scientific Research in the Public Interest (No. 201403054), the National High Technology Research and Development Program of China (No. 2011AA10A212), the National Science & Technology Pillar Program (No. 2014BAD13B02), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD)
Xu Li-juan, Li Qiu-chun and Liu Jie contributed equally to this article.
