Biodegradation of microcystin-RR and-LR by an indigenous bacterial strain MC-LTH11 isolated from Lake Taihu
2014-09-17ZhouYuanlongYangFeiLiangGeyuYinLihongPuYuepu
Zhou Yuanlong Yang Fei Liang Geyu Yin Lihong Pu Yuepu
(Key Laboratory of Environmental Medicine Engineering of Ministry of Education, Southeast University, Nanjing 210009, China)
L ake Taihu is situated on the ancient Yangtze River delta with a catchment of 36 500 km2,and surrounded by most heavily industrialized cities with almost the highest population density in China[1].Due to the massive discharge of industrial wastewater into Lake Taihu in recent years,harmful cyanobacterial blooms(HCBs)occurred frequently in Lake Taihu,which gave a threat to residents'lives and health[2].In 2007, the HCBs spread out over 40% of the total area of Lake Taihu,which caused sudden water pollution accidents in Wuxi City[3-4].HCBs also produce many kinds of microcystins(MCs),which can cause illness and death in humans[5].A guideline value of 1.0 μg/L microcystin-LR(MC-LR)in drinking water was set by the World Health Organization[6].
Microcystin-RR(MC-RR)and MC-LR have the same cyclo-(D-Ala-X-D-MeAsp-Z-Adda-DGlu-Mdha-)structure, where X and Z represent variable L-amino acids[7].They are stable and resistant to physicochemical and biological factors such as temperature, pH values, sunlight,and enzymes[8]. Although MCs can be destructed by some conventional and advanced oxidation processes,the biodegradation of MCs by indigenous bacteria can be one of the safest and mildest treatments for removing cyanobacterial toxins from fresh water[8-9].To date, only a few bacteria with the ability to degrade MCs have been reported,and most strains appear to be limited to the familySphingomonas[9].
Bourne et al.[10]first identified a gene cluster,mlrA,mlrB,mlrCandmlrDin strain ACM 3962,which was responsible for the degradation of MC-LR.The most important enzymeMlrAcoding by themlrAgene breaks off the cyclic peptide of MC-LR to linear MC-LR.Moreover, the toxicity of the degradation products of MC-LR,including the linearized MC-LR and the tetrapeptide,was less than that of MC-LR[11].
The objectives of this study are to isolate and identify the indigenous MC-degrading bacteria from Lake Taihu,then explore the mechanism of MCs degradation by the bacteria and investigate the capacity of degrading MC-LR and MC-RR by the indigenous bacteria under different conditions.
1 Materials and Methods
1.1 Preparation and analysis of MCs
Authentic MCs were purchased from the Sigma,and crude MCs were extracted from the lyophilizedM.aeruginosacells.MCs were identified using the Agilent 1100 high performance liquid chromatography(HPLC).The mobile phase was a combination of Milli-Q water(Millipore,Germany)containing TFA and methanol(Dikma Technology Inc., USA)(45∶55, volume ratio), which was set at a flow rate of 1 mL/min.The injection volume was 20 μL at 40 ℃ for analysis and an absorbance at 238 nm was monitored.The MC-RR and MC-LR in the crude extracts were 46.6 mg/L and 23.1 mg/L, respectively.
1.2 Isolation and identification of MC-degrading bacteria
The sediment sample was suspended in sterilized water and shaken at 120 rev/min for 30 min.The supernatant was inoculated into a mineral salt medium(MSM)mixed with crude MC extracts.MSM was composed of 1.0 g/L MgSO4·7H2O,0.5 g/L KH2PO4,4.0 g/L K2HPO4,1.0 g/L NaCl,20 mg/L CaCl2,5 mg/L FeSO4,5 mg/L MnCl2·4H2O,5 mg/L ZnCl2,and 0.5 mg/L CuCl2.Inocula were subcultured for 4 times.Subsequently,the medium diluted gradually was spread onto the MSM agar plates(2%agar),which also contained crude MCs extract. After cultivating, single colonies were transferred to MSM containing crude MC extracts and cultivated.The capability of degrading MCs was identified by monitoring the remaining MCs in the media,and the MC-degrading strain can be screened out.
Bacterial DNA was extracted by using TIANamp Bacteria DNA Kit(TIANGEN Biotech,Beijing,China).The 16S rRNA gene was amplified by polymerase chain reaction(PCR)using forward primer(5’-AGAGTTTGATCMTGGCTCAG-3’)and reverse primer(5’-TACGGYTACCTTGTTACGACTT-3’).The PCR was carried out for 35 cycles of 94℃ for 1 min,55℃ for 1 min and then 72℃ for 2 min.The 16S rRNA gene PCR product was sequenced by the BGI Company,China.The homology of the 16S rRNA gene sequence of the isolates,with reference 16S rRNA gene sequences,was analyzed using the BLAST algorithm in GenBank available in the National Centre for Biotechnology Information(NCBI).
1.3 PCR for mlrA
ThemlrAgene was amplified by PCR using specific forwardprimer(5’-GACCCGATGTTCAAGATACT-3’)and specific reverse primer(5’-CTCCTCCCACAAATCAGGAC-3’).The PCR was carried out for 40 cycles of 98℃ for 10 min,56℃ for 30 s and then 72℃for 30 min,with an initial denaturation at 95℃ for 5 min and a final elongation at 72℃ for 5 min.
1.4 Batch degradation experiments
The ability of the bacterial strains to degrade MCs isolated from Lake Taihu was examined by inoculating them into the different media:MSM with the crude extract,MSM with the authentic MC-LR,and water body samples collected from Lake Taihu.Before inoculating at a constant condition,the strains were cultured for 48 h with shaking in MSM containing MCs,harvested by centrifugation,and washed twice with 20 mmol/L sodium phosphate buffer(pH=7.2).The MSM containing MC extracts was employed for evaluating the effect of temperature,pH value and MCs concentrations on the degradation of MCs.All experiments were carried out in duplicate,and bacterialfree medium was employed as a control.
2 Results and Discussion
2.1 Isolation and identification of MC-degrading bacteria
A sediment sample from cyanobacteria-salvage yards at Fudu Port in Lake Taihu was collected and screened.The bacterial strain MC-LTH11 showed a great ability to degrade MC-RR and MC-LR.This strain was aerobic,gram-negative,rod-shaped and produced white-colored colonies on agar medium.
For bacterial identification,the 16S rDNA gene for the isolate MC-LTH11 was amplified.The obtained DNA nucleotide was sequenced by the BGI Company,China and then analyzed using DNA Blast search(NCBI).The analysis of the 16S rDNA sequence reveals that the bacterium belongs to the genusStenotrophomonasspecies(see Fig.1),and deposited in the Genbank with an accession number of KC734883.Recent investigations have revealed the isolation of some different MC-degrading bacteria isolated from differentChina water bodies[10].Among these bacteria,the strain EMS isolated from Lake Taihu also belongs to theStenotrophomonassp.,which indicates thatStenotrophomonassp.may be a common MC-degrading bacterial strain and play an important role in natural MCs degradation,in Lake Taihu.

Fig.1 Phylogenetic tree of Stenotrophomonas sp.MC-LTH11
2.2 Detection of mlrA gene from MC-degrading bacterium MC-LTH11
DNA extracted from MC-LTH11 and specific oligonucleotide primers were used for PCR targeting themlrAgene with conditions as described before.The amplification product ofmlrA(approximately 850 bp)was obtained(see Fig.2).This result confirms thatmlrAhomologues exist in this bacterial strain.
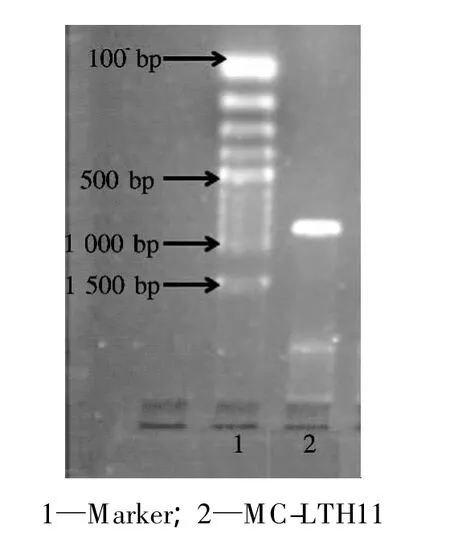
Fig.2 Detection of mlrA gene fragment by PCR in MC-degrading strain MC-LTH11
2.3 MCs degradation in batch experiments
The degradation rates of MCs were significantly affected by temperature.At 37℃,the MC-RR and MC-LR were completely removed at the highest degradation rates of 6.18 mg/(L·d)and 3.08 mg/(L·d),respectively.Meanwhile,the analogues were degraded at 30 ℃and 20℃ in lower rates.The degradation rates of MCs were also affected by pH values.The optimum pH values for degradation were neutral or weak acidic conditions.These results are different from strain EMS,which prefers to degrade MC-RR and MC-LR in alkaline rather than acidic environments[12].

Fig.3 Effect of MCs concentrations on degradation of MCs.(a)Degradation for MC-RR;(b)Degradation for MC-LR
Fig.3 shows the highest initial MCs(37.13 mg/L MCRR,18.49 mg/L MC-LR)were degraded at the highest rates of 6.18 mg/(L·d)and 3.08 mg/(L·d),respectively.When the initial MCs decreased to 18.56 mg/L MC-RR and 9.56 mg/L MC-LR,the degradation rates were 6.19 mg/(L·d)and 3.19 mg/(L·d),respectively.At the lowest initial MCs(9.08 mg/L MC-RR,4.82 mg/L MC-LR),the rates reduced to 4.54 mg/(L·d)and 2.41 mg/(L·d),respectively.Lag periods were evident prior to MC-LTH11 initiating the degradation of both analogues,and gradually shortened with the decreased analogues concentration.
Many studies reported that a lag phase exists before the initiation of degradation,but it is not clear why this lag phase exists.A possible explanation for the lag phase is that the synthesis of MCs degradative enzymes(microcysrinase)is repressed by some catabolite repressor substrates present in theM.aeruginosaextracts[13].In this study,both heavy metals and catabolite repressor substrates may exist in MSM with the crude extract,which can lead to the emergence of the demurrage.Nevertheless,once degradation commences after the lag period,both MCs can complete removal quickly.
Fig.4 shows that authentic MC-LR at 14.41 mg/L was removed directly at a rate of 7.21 mg/(L·d).Compared with the experiment in MSM with the crude extract,the degradation rate increased,and the lag phase disappeared,although the initial concentration of MC-LR was at a higher level.

Fig.4 Degradation of MC-LR in the MSM with the crude extract and authentic MC-LR
MC-LTH11 also performed a good ability of degradation for MCs in the water body collected from Lake Taihu.MC-RR at 3.56 mg/L and MC-LR at 2.04 mg/L were completely removed in 24 h while no losses of MCRR and MC-LR were evident in control(see Fig.5).These results confirm that the MC-LTH11 strain may be actively involved in the degradation of MCs during the disappearance of the HCBs in Lake Taihu.Since the application of the biological sand filter embedded with MC-degrading bacteria MJ-PV successfully removed MC-LR and MC-LA from source water[14],and a bioreactor using the immobilized B-9 completely degraded MCRR after 24 h,an effective biofilter with immobilized MC-LTH11 to degrade MC-LR and MC-RR may be our future research.

Fig.5 Degradation of MC-RR and MC-LR in water body collected from Lake Taihu
3 Conclusion
An indigenous MC-degrading bacterium MC-LTH11 isolated from Lake Taihu is identified asStenotrophomonassp.based on 16S rDNA sequence analysis.AmlrAgene involved in MCs biodegradation is detected in MC-LTH11.MC-LTH11 has a capability of MC-degrading in MSM with MCs and body water of Lake Taihu,and the degradation rates are dependent on temperature,pH,initial MCs concentration and media.Therefore,theStenotrophomonassp.MC-LTH11 has the capacity to bioremediate water bodies contaminated by MCs and may contribute to the degradation of MCs after the outbreak of harmful cyanobacterial blooms in Lake Taihu.
[1]Liu Y,Xie P,Zhang D,et al.Seasonal dynamics of mi
crocystins with associated biotic and abiotic parameters in two bays of Lake Taihu,the third largest freshwater lake in China[J].Bulletin of Environmental Contaminationand Toxicology,2008,80(1):24-29.
[2]Mao X W,Xu F,Xu B,et al.Changes of water quality and eutrophication in Taihu Lake[J].Water Resources Protection,2009,25(1):48-51.
[3]Duan H T,Ma R H,Xu X F,et al.Two-decade reconstruction of algal blooms in China's Lake Taihu[J].Environmental Science and Technology,2009,43(10):3522-3528.
[4]Stone R.China aims to turn tide against toxic lake pollution[J].Science,2011,333(6047):1210-1211.
[5]Li X Q,Yuan J,Yang F,et al.Environmental abundance and microcystin-LR production ability of toxic microcystis in Nanquan region of Lake Taihu[J].Journal of Southeast University:English Edition,2010,26(1):96-99.
[6]World Health Organization.Guidlines for drinking-water quality[S].2nd ed.Geneva,Switzerland:World Health Organization,1998.
[7]Carmichael W W.Cyanobacteria secondary metabolites—the cyanotoxins[J].Journal of Applied Bacteriology,1992,72(6):445-459.
[8]Gagala I,Mankiewicz-Boczek J.The natural degradation of microcystins(cyanobacterial hepatotoxins)in fresh water—the future of modern treatment systems and water quality improvement[J].Polish Journal of Environmental Studies,2012,21(5):1125-1139.
[9]Dziga D,Wasylewski M,Wladyka B,et al.Microbial degradation of microcystins[J].Chemical Research in Toxicology,2013,26(6):841-852.
[10]Bourne D G,Riddles P,Jones G J,et al.Characterisation of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin LR[J].Environmental Toxicology,2001,16(6):523-534.
[11]Tsuji K,Asakawa M,Anzai Y,et al.Degradation of microcystins using immobilized microorganism isolated in an eutrophic lake[J].Chemosphere,2006,65(1):117-124.
[12]Chen J,Hu L B,Zhou W,et al.Degradation of microcystin-LR and RR by astenotrophomonassp.strain EMS isolated from Lake Taihu,China[J].International Journal of Molecular Sciences,2010,11(3):896-911.
[13]Jones G J,Bourne D G,Blakeley R L,et al.Degradation of the cyanobacterial hepatotoxin microcystin by aquatic bacteria[J].Natural Toxins,1994,2(4):228-235.
[14]Bourne D G,Blakeley R L,Riddles P,et al.Biodegradation of the cyanobacterial toxin microcystin LR in natural water and biologically active slow sand filters[J].Water Research,2006,40(6):1294-1302.
杂志排行
Journal of Southeast University(English Edition)的其它文章
- Wavelet transform and gradient direction based feature extraction method for off-line handwritten Tibetan letter recognition
- Analyses of unified congestion measures for interrupted traffic flow on urban roads
- Conditional autoregressive negative binomial model for analysis of crash count using Bayesian methods
- Design and analysis of traffic incident detection based on random forest
- Inverse kinematic deriving and actuator control of Delta robot using symbolic computation technology
- Operation optimization modefor nozzle governing steam turbine unit
