Isolation and Characterization of a Fucoidan-Degrading Bacterium from Laminaria japonica
2014-05-02WANGYingLIBafangZHAOXueandPIAOMeizi
WANG Ying, LI Bafang, ZHAO Xue and PIAO Meizi
1) College of Food Science and Engineering, Ocean University of China, Qingdao 266003, P. R. China
2) College of Food Science and Engineering, Qingdao Agricultural University, Qingdao 266109, P. R. China
Isolation and Characterization of a Fucoidan-Degrading Bacterium from Laminaria japonica
WANG Ying1),2), LI Bafang1),*, ZHAO Xue1), and PIAO Meizi2)
1) College of Food Science and Engineering, Ocean University of China, Qingdao 266003, P. R. China
2) College of Food Science and Engineering, Qingdao Agricultural University, Qingdao 266109, P. R. China
Fucoidan, a polysaccharide containing abundant fucose and sulfate ester group, was prepared fromLaminaria japonica. In order to obtain fucoidan-degrading enzyme, bacteria capable of degrading fucoidan were screened from kelp. A bacterial strain named RC2-3 was obtained, which degraded fucoidan by the maximum extent of 54% ± 1.3%, the highest among all bacterial isolates. High-performance size exclusion chromatography (HPSEC) showed that the molecular weight of fucoidan was gradually reduced by RC2-3 with culturing time, suggesting the production of fucoidan-degrading enzyme by RC2-3. Phylogenetic analysis of partial 16S ribosomal RNA gene (16S rDNA) sequence showed that RC2-3 belonged to the familyFlavobacteriaceae. However, it showed different physiological and biochemical characteristics from the knownFlavobacteriaceaemembers producing fucoidan-degrading enzyme, thus RC2-3 was proposed to be a new member of this family.
fucoidan degradation;Flavobacteriaceae; 16S rDNA; HPSEC
1 Introduction
Fucoidans are a group of marine sulfated polysaccharides in the cell-wall matrix of brown algae, which contain large proportions of L-fucose and sulfate, along with minor amounts of other sugars such as xylose, galactose, glucose, mannose, uronic acids and rhamnose (Berteau and Mulloy, 2003; Mccandless and Craigie, 1979). Previous research demonstrated substantial bioactivities of fucoidan such as gastric ulcer-protective activity (Hwanget al., 2008), antithrombotic activity (Boisson-Vidalet al., 2000), antitumor activity (Maruyamaet al., 2003), and inhibitory activity on viral and bacterial infection (Shibataet al., 2003). Thus, the production and application of fucoidans as therapeutic agents have become hot topics of intensive researchers (Berteau and Mulloy, 2003). However, the application of fucoidan, especially as a therapeutic agent, has been limited due to the large molecular mass and high viscosity (Kimet al., 2008). This problem can be solved with the help of fucoidan-degrading enzymes which can digest fucoidans into oligosaccharides. Such enzymes can also be used to analyze the structures of fucoidan and the mechanisms of its bioactivities (Sakaiet al., 2003a). To prepare the fucoidan-degrading enzymes, fucoidan-degrading bacteria are appropriate resources (Furukawaet al., 1992; Bakuninaet al., 2000; Sakaiet al., 2003b; Descampset al., 2006; Changet al., 2010).
In this study, a bacterium capable of producing fucoidan-degrading enzyme(s) was isolated from kelp. The bacterial isolate was characterized according to its physiological and biochemical properties. The 16S rDNA-based phylogenetic analysis showed that this isolate is a new member of the fucoidanase-producing familyFlavobacteriaceae.
2 Materials and Methods
2.1 Preparation and Chemical Analysis of Fucoidans
Fucoidan extracted fromLaminaria japonicawas purchased from Rizhao Jiejing Ocean Biotechnology Development Co. Ltd., Shandong, China. The fucoidan was preliminarily classified by a Q-sepharose Fast Flow (GE Healthcare, USA) column, and then eluted with 1, 1.5 and 2 mol L−1NaCl solutions separately. The fucoidan content was determined with methylene blue method (Soedjak, 1994) with some modifications. In brief, 9 mL of distilled water was added to 20 μL of the sample, followed by 1 mL of 0.41 mmol L−1methylene blue solution. The absorbance of the reaction system was measured at 559 nm, and the fucoidan content was calculated according to the standard curve plotted with fucoidan (Sigma, USA) as the standard. Total sugar content was quantified with the phenol-H2SO4method (Duboiset al., 1956). The sulfate content was de-termined with BaCl2-gelatin method (Silvestriet al., 1982).
2.2 Isolation of Fucoidan-Degrading Bacteria
In April 2011, kelp samples were collected in the sea area of Rongcheng, Shandong, China. Ten grams of kelp sample was pestled in 10 mL of sterilized 0.9% NaCl and 1 mL of the mixture was pipetted into 15 mL of broth medium (2 g L−1fucoidan, 2 g L−1NH4NO3, pH 7.0). The inoculated medium was incubated at 25℃ on a shaker (180 r min−1) for 5 d, and the culture was transferred into fresh medium (5%) for another 5 d twice. Then, 0.5 mL of the culture was spread onto agar plates (2 g L−1fucoidan, 5 g L−1peptone, 20 g L−1agar) to obtain single colonies. After 3 days of cultivation, individual colonies with different phenotypes were selected and transferred to agar slants (2 g L−1fucoidan, 5 g L−1peptone, 20 g L−1agar) for 3-day incubation.
To test their fucoidan-degrading ability, the bacterial isolates were aerobically cultivated in a liquid medium (2 g L−1fucoidan, 5 g L−1peptone, pH 7.0) on a 180-rmin-1rotary shaker for 72 h. The bacterial isolates that degraded fucoidan to the maximum extent were selected as a microbial source of fucoidan-degrading enzyme(s).
2.3 Confirmation of Fucoidan-Degrading Ability
The selected bacterial isolate was grown in a fucoidancontaining medium (2 g L−1fucoidan, 2 g L−1NH4NO3) for 72 h. Samples aseptically taken at each 24 h were heated at 100℃ for 10 min to inactivate the fucoidan-degrading enzyme. The activity of fucoidan- degrading enzyme was confirmed by the gradually decrease in the molecular weight of fucoidan along with the culturing time. The molecular weight was monitored with the high-performance size exclusion chromatography (HPSEC) method as previously described by Changet al.(2010) with the help of an Aglient 1100 HPLC (Aglient Technologies, USA) equipped with a gel filtration column (STK-gel 3000 PWxl, Tosoh Co., Japan) and a refractive index detector (Aglient Technologies, USA ).
2.4 Gene Sequencing and Phylogenetic Analysis of the Fucoidan-Degrading Bacterium
The 16S rDNA sequence of the selected bacterium was amplified by polymerase chain reaction (PCR) using universal bacterial primers: 27f (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492r (5’-GGCTACCTTGTTACGACTT-3’) (Sariset al., 1990). The PCR products were cloned into pUCm-T vectors (Invitrogen, USA) and sequenced with an ABI 3730 sequencer (Applied Biosystems Inc., USA). The 1512-bp 16S rDNA sequence was compared with those available in the National Center for Biotechnology Information (NCBI) database. Phylogenetic trees were constructed with neighbour-jointing method (MEGA 5.0, Kumaret al., 1994). The morphology of the selected bacterium was observed by microscopy (Olympus CX-41; Olympus Co., Japan). Other physiological characteristics were analyzed as previously described (Kimet al., 2008; Descampset al., 2006).
3 Results
3.1 Preparation and Chemical Properties of Fucoidans
The fucoidan was fractionated by a Q-sepharose Fast Flow column (Fig.1) with fractions I, II and III collected, dialysed (5 kDa cut-off) and lyophilized. The yield and chemical composition of fractions I, II and III are shown in Table 1. Fraction III was further used for the isolations of fucoidan-degrading bacteria and confirmation of the bacterial fucoidan-degrading ability.
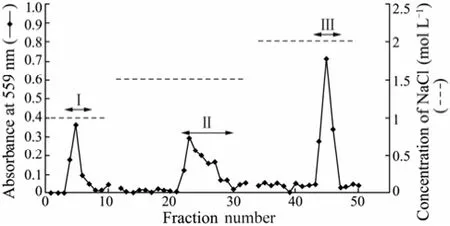
Fig.1 Chromatography of fucoidan on a Q-sepharose Fast Flow column. A Q-sepharose Fast Flow column (2.4 × 30 cm) was equilibrated with distilled water. The volume of each fraction was 5 mL. Fraction numbers 4–7 (Fraction I), Fraction numbers 22–29 (Fraction II) and Fraction numbers 43–46 (Fraction III) were pooled.
3.2 Isolation of Fucoidan-Degrading Bacteria
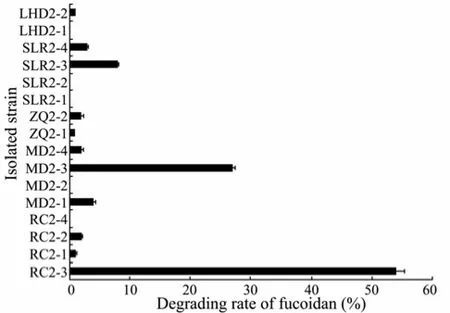
Fig.2 Degrading rate of fucoidan by different bacterial isolates. The assays were performed three times and the bars indicated the standard deviation of the measurements.
A total of 16 single colonies were transferred to agar slants and the fucoidan-degrading rates of bacterial iso-lates are shown in Fig.2. The isolate RC2-3 showed the highest fucoidan-degrading ability (up to 54% ± 1.3%). Hence, RC2-3 was selected for further physiological, biochemical and phylogenetic analysis.
3.3 Confirmation of Bacterial Fucoidan-Degrading Ability
According to the standard curve of molecular mass (data not shown), the peak at 13.64 min of HPSEC data was attributed to the fucoidan (fraction III) which was the only large-molecule component in the culture medium. This peak area gradually decreased with the culturing time (Fig.3) as a result of the fucoidan degradation. These indicated that the isolate RC2-3 produced fucoidan-degrading enzyme(s) and utilized the macromolecular component of fucoidan.
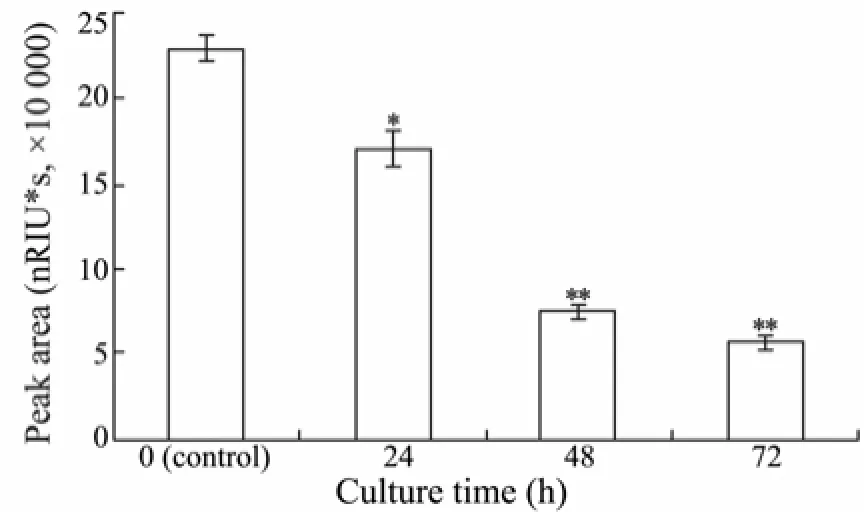
Fig.3 HPSEC results of RC2-3 at different culture time. The mobile phase was used as 0.2 mol L-1NaCl solution with a flow rate of 0.5 mL min-1and the column temperature was at 40℃. The data shown were only the peak area at 13.64 min which was contributed by fraction III. * indicated a significant difference between peak area of different culture time and control. * P < 0.01, ** P < 0.001. Bars indicated standard deviation of three replicates.
3.4 Characterization and Identification of the Fucoidan-Degrading Isolate RC2-3
The fucoidan-degrading isolate RC2-3 was a Gram-negative bacterium, being rod shaped and 1–2 μm in length. It developed yellow colonies on the agar plates and grew on the seawater medium, but not on freshwater medium. The isolate RC2-3 was a facultative anaerobic organism, which did not use oxygen as the hydrogen acceptor in the test of glucose oxidation and fermentation. The bacterium hydrolyzed gelatin but not casein and starch. It did not produce indole. Other physiological and biochemical characteristics of the isolate RC2-3 are listed in Table 2. The partial 16S rDNA sequence of RC2-3 (JQ408440) was closely related to strainFlavobacteriaceaeCZ1127 (FJ526380) andFucobacter marinaSA-0082 (AB057592) (sequence similarity 97%) of familyFlavobacteriaceae. The phenotypic characteristics and 16S rDNA-based phylogenetic analyses suggested that the isolate RC2-3 was a member of the familyFlavobacteriaceae(Fig.4).
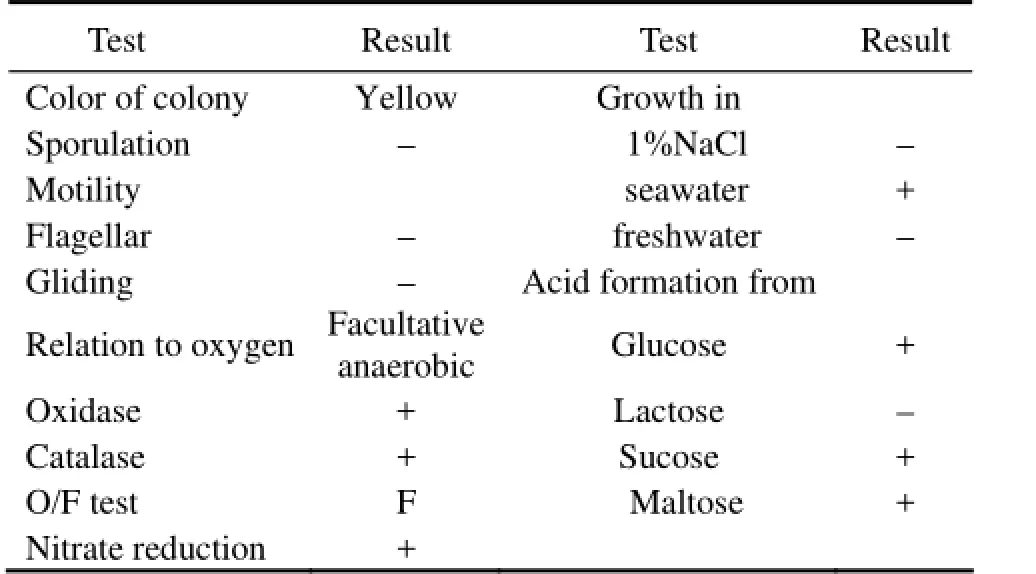
Table 2 Characteristics of the fucoidan-degrading bacterium RC2-3†
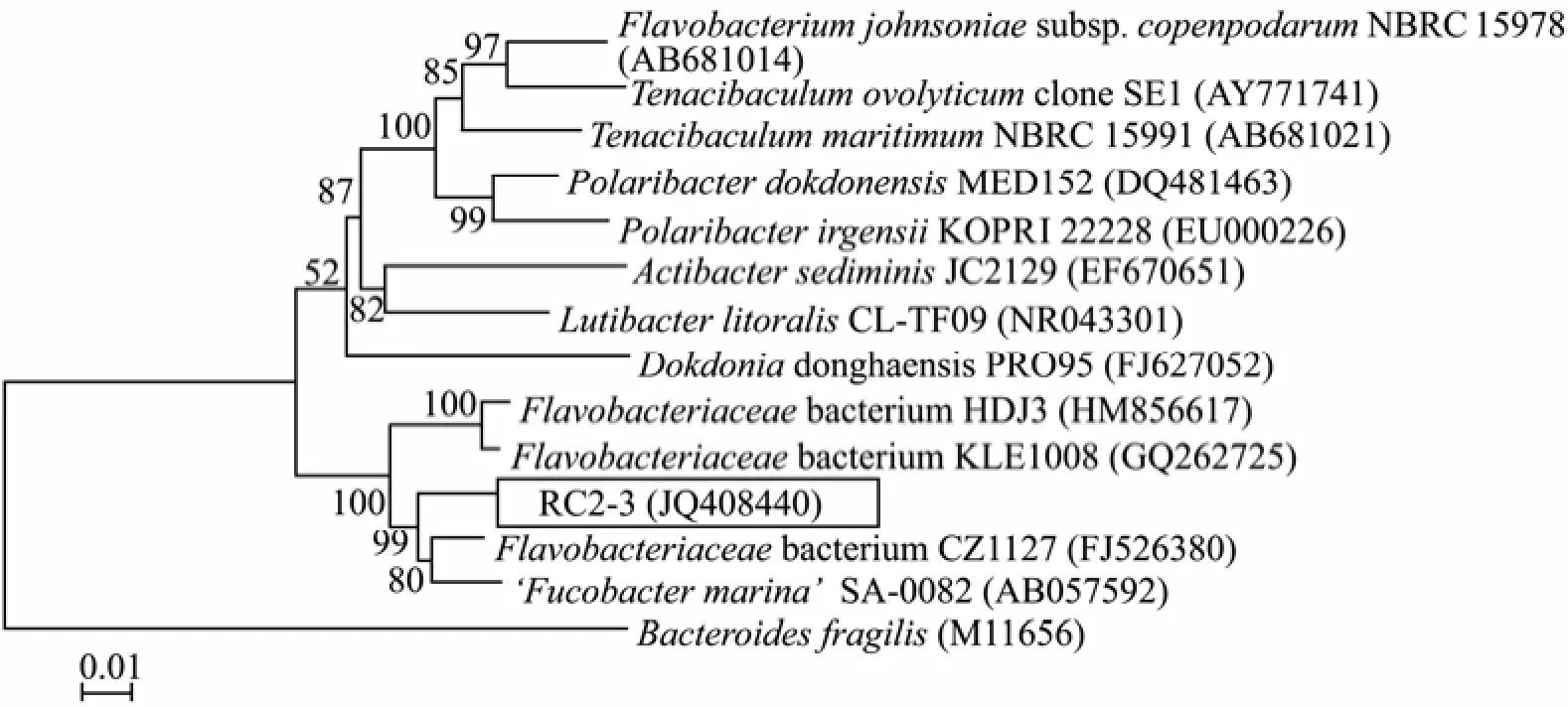
Fig.4 Phylogenetic relationships of the fucoidan-degrading bacterium RC2-3 to other known relatives. The topology shown in the tree was obtained using the neighbour-joining method (Saitou et al., 1987). Numbers at the nodes referred to the bootstrap values (%). The scale bar represented 0.01 substitutions per nucleotide position.
4 Discussion
Descampset al. (2006) found that theMariniflexile fucanivoransstrain SW5 degraded brown alga (Pelvetia canaliculata) fucoidan. In the present study, we found the 16S rDNA sequence of RC2-3 displayed only 89% similarity with that ofMariniflexile fucanivoransSW5, indicating that the isolate RC2-3 andMariniflexile fucanivoransSW5 were different.
A number of bacterial strains of familyFlavobacteriaceaeare able to digest fucoidan and produce fucoidan-degrading enzyme(s), such asFucobacter marinaSA-0082 (Sakaiet al., 2002),FlavobacteriaceaeCZ1127 (Changet al., 2010), andMesonia algaeKMM3909 (Urvantsevaet al., 2006). Compared with these known bacteria, our fucoidan-degrading isolate RC2-3 showed differences in physiological and biochemical characteristics to some extent. The isolate RC2-3 was a facultative anaerobic organism, which did not use oxygen as the hydrogen acceptor in the test of glucose oxidation and fermentation. We considered that the isolate RC2-3 was different from known members of familyFlavobacteriaceae. Further studies are required to clearly identify the isolate RC2-3.
After 72 h, the isolate RC2-3 digested 54% ± 1.3% of fraction III, with residual fucoidans barely declined later. This indicated that the bacterium produced enzyme(s) to degrade fraction III specifically by recognizing its original structure. The isolate RC2-3 also digested the other two components purified from fucoidan,i.e., fractions I and II by 46% and 38%, respectively. It was likely that the enzyme produced by RC2-3 recognized the structural differences among the three fractions. Further studies are needed to confirm this hypothesis.
In order to assess the application potential of the isolate RC2-3, its fucoidan-degrading ability was verified by HPSEC. The peak area showed gradual decrease at 13.64 min along with the culturing time, suggesting the bacterial fucoidan-degrading activities in the culture medium.
In conclusion, a novel fucoidan-degrading bacterium of familyFlavobacteriaceae(designated RC2-3) was isolated in this study, which promises to be applied in fucoidan degradation.
Acknowledgements
We acknowledged the financial support from the National Natural Science Foundation of China (No. 30800858).
Bakunina, I. Y., Shevchenko, L. S., Nedashkovskaya, O. I., Shevchenko, N. M., Alekseeva, S. A., Mikhailov, V. V., and Zvyagintseva, T. N., 2000. Screening of marine bacteria for fucoidanases. Microbiology, 69: 370-376.
Berteau, O., and Mulloy, B., 2003. Sulfated fucans, fresh perspectives: structures, functions, and biological properties. Glycobiology, 13: 29R-40R.
Boisson-Vidal, C., Frederic, C., Lionel C., Corinne, S., Jocelyne, T., Jean, M., Claude, S., Barbara, M., and Anne-Marie, F., 2000. Relationship between antithrombotic activities of fucans and their structure. Drug Development Research, 51: 216-224.
Chang, Y., Xue, C., Tang, Q., Li, D., Wu, X., and Wang, J., 2010. Isolation and characterization of a sea cucumber fucoidan utilizing marine bacterium. Letters in Applied Microbiology, 50: 301-307.
Descamps, V., Colin, S., Lahaye, M., Jam, M., Richard, C., Potin, P., Barbeyron, T., Yvin, J. C., and Kloareg, B., 2006. Isolation and culture of a marine bacterium degrading the sulfated fucans from marine brown algae. Marine Biotechnology, 58: 27-39.
Dubois, M., Gilles, K. A., Hamilton. J. K., Rebers, P. A., and Smith, F., 1956. Colorimetric method for determination of sugars and related substances. Analytical Chemistry, 28: 350-356.
Furukawa, S., Furukawa, T., Koga, D., and Ide, A., 1992. Purification and some properties of exo-type fucoidanases from Vibiro sp. N-5. Bioscience Biotechnology and Biochemistry, 56: 1829-1834.
Hwang, H. J., Kwon, M. J., Kim, I. H., and Nam, T. J., 2008. The effect of polysaccharide extracted from the marine alga Capsosiphon fulvescens on ethanol administration. Food and Chemical Toxicology, 46: 2653-2657.
Kim, W. J., Kim, S. M., Lee, Y. H., Kim, H. G., Kim, H. K., Moon, S. H., Suh, H. H., Jang, K. H., and Park, Y. I., 2008. Isolation and characterization of marine bacterial Strain degrading fucoidan from Korean Undaria pinnatifida Sporophylls. Journal of Microbiology and Biotechnology, 18: 616-623.
Kumar, S., Tamura, K., and Nei, M., 1994. Mega: molecular evolutionary genetics analysis software for microcomputers. Bioinformatics, 10: 189-191.
Maruyama, H., Tamauchi, H., Hashimoto, M., and Nakano, T., 2003. Antitumor activity and immune response of Mekabu fucoidan extracted from sporophyll of Undaria pinnatifida. In Vivo, 17: 245-249.
Mccandless, E. L., and Craigie, J. S., 1979. Sulfated polysaccharides in red and brown algae. Annual Review of Plant Physiology, 30: 41-53.
Saitou, N., and Nei, M., 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4: 406-425.
Sakai, T., Kimura, H., and Kato, I., 2002. A marine strain of a flavobacteriaceae utilize brown seaweeds fucoidan. Marine Biotechnology, 4: 399-405.
Sakai, T., Ishizuka, K., and Kato, I., 2003a. Isolation and characterization of a fucoidan-degrading marine bacterium. Marine Biotechnology, 5: 409-416.
Sakai, T., Kimura, H., and Kato, I., 2003b. Purification of sulfated fucoglucuronomannan lyase from bacterial strain of Fucobacter marina and study of appropriate conditions for its enzyme digestion. Marine Biotechnology, 5: 380-387.
Saris, P. E., Lars, P., and Uhlén, M., 1990. Direct amplication of DNA from colonies of Bacillus subtilis and Escherichia coli by the polymerase chain reaction. Journal of Microbiological Methods, 11: 121-126.
Shibata, H., Iimuro, M., Uchiya, N., Toshihiko Kawamori1, Nagaoka, M., Ueyama, S., Hashimoto, S., Yokokura, T., Sugimura, T., and Wakabayashi, K., 2003. Preventive effects of Cladosiphon fucoidan against Helicobacter pylori infection in mongolian gerbils. Helicobacter, 8: 59-65.
Silvestri, L. J., Hurst, R. E., Simpson, L., and Settine, J. M., 1982. Analysis of sulfate in complex carbohydrates. Analytical Biochemistry, 123: 303-309.
Soedjak, H. S., 1994. Colorimetric determination of carrageenans and other anionic hydrocolloids with methylene blue. Analytical Chemistry, 66: 4514-4518.
Urvantseva, A. M., Bakunina, I. Y., Nedashkovskaya, O. I., Kim, S. B., and Zvyagintseva, T. N., 2006. Distribution of intracellular fucoidan hydrolases among marine bacteria of the Family Flavobacteriaceae. Applied Biochemistry and Microbiology, 42: 484-491.
(Edited by Qiu Yantao)
* Correspondending author. Tel: 0086-532-82031852
E-mail: bfli@ouc.edu.cn
(Received April 17, 2012; revised June 4, 2012; accepted February 21, 2013)
© Ocean University of China, Science Press and Spring-Verlag Berlin Heidelberg 2014
杂志排行
Journal of Ocean University of China的其它文章
- Properties of Klebsiella Phage P13 and Associated Exopolysaccharide Depolymerase
- Comparative Study on the Allergenicity of Different Litopenaeus vannamei Extract Solutions
- Toxicity of Five Phenolic Compounds to Brine Shrimp Artemia sinica (Crustacea: Artemiidae)
- Application of CFD Modeling to Hydrodynamics of CycloBio Fluidized Sand Bed in Recirculating Aquaculture Systems
- DNA Barcoding Assessment of Green Macroalgae in Coastal Zone Around Qingdao, China
- QSAR for Photodegradation Activity of Polycyclic Aromatic Hydrocarbons in Aqueous Systems
