DNA Barcoding Assessment of Green Macroalgae in Coastal Zone Around Qingdao, China
2014-05-02DUGuoyingWUFeifeiMAOYunxiangGUOShenghuaXUEHongfanandBIGuiqi
DU Guoying, WU Feifei, MAO Yunxiang, GUO Shenghua, XUE Hongfan, and BI Guiqi
Bioengineering Department, College of Marine Life Science, Ocean University of China, Qingdao 266003, P. R. China
DNA Barcoding Assessment of Green Macroalgae in Coastal Zone Around Qingdao, China
DU Guoying, WU Feifei, MAO Yunxiang*, GUO Shenghua, XUE Hongfan, and BI Guiqi
Bioengineering Department, College of Marine Life Science, Ocean University of China, Qingdao 266003, P. R. China
An assessment with assistance of DNA barcoding was conducted on green macroalgae in coastal zone around Qingdao, China, during the period of April − December, 2011. Three markers were applied in molecular discrimination, including the plastid elongation factortufA gene, the internal transcribed spacer (ITS) region of the ribosomal cistron and rubisco large subunit gene 3’regions (rbcL-3P). DNA barcoding discriminated 8 species, excluding species of genusCladophoraandBryopsisdue to failures in amplification. We ascertained and corrected 4 species identified by morphological methods for effectively assisting the classification. The genetufA presented more advantages as an appropriate DNA marker with the strongest amplification success rate and species discrimination power than the other two genes. The poorest sequencing success largely handicapped the application of ITS. Samples identified bytufA andrbcL asUlvaflexuosawere clustered into the clade ofU.proliferaby ITS in the neighbor-joining tree. Confusion with discrimination of the complex ofU. linza,U.proceraandU.prolifera(as the LPP complex) still existed for the three DNA markers. Based on our results,rbcL is recommended as a preferred marker for assistingtufA to discriminate green macroalgae. In distinguishing green-tide-formingUlvaspecies, the free-floating sample collected from the green tide in 2011 was proved to be identical withU. proliferain Yellow Sea for ITS andrbcL genes. This study presents a preliminary survey of green macroalgae distributed in the coastal area around Qingdao, and proves that DNA barcoding is a powerful tool for taxonomy of green macroalgae.
green macroalgae; DNA barcoding;tufA; ITS;rbcL
1 Introduction
Like most marine algae, green macroalgae are notoriously difficult to identify with certainty due to their simple morphologies, phenotypic plasticity and convergent evolution. Species identification would, therefore, benefit greatly from the application of molecular tools. DNA barcoding is an excellent powerful method to assist taxonomy of macroalgae (Hebert and Gregory, 2005; Saunders, 2005; Robbaet al., 2006; Medlinet al., 2007; McDevit and Saunders, 2009). Following red and brown macroalgae, marine green macroalgae are increasingly developed DNA barcoding in recent few years (Nakazawaet al., 2004; Loughnaneet al., 2008; O’kellyet al., 2010; Saunders and Kucera, 2010; Buchheimet al., 2011).
Considering the essentials of molecular markers for DNA barcoding, viz. discrimination on species level and universality of PCR primers (Hebertet al., 2003; Meieret al., 2008), several markers have been applied to green macroalgae (Saunders and Kucera, 2010; Mareset al., 2011; Wanget al., 2010a; Liuet al., 2012). These markers include the plastid rubisco large subunit (rbcL), elongation factortufA, universal plastid amplicon (UPA), the nuclear D2/D3 region of the large ribosomal subunit (LSU), the internal transcribed spacer of the ribosomal cistron (ITS), and 5S rDNA spacer region. There has been also an attempt using the secondary structure information of ITS2 to overcome some of the limitations of ITS2 as DNA barcode (Buchheimet al., 2011). Seldom could any markers be successful for all taxonomy of green macroalgae. Each of them has its advantages and disadvantages for barcoding. Some of them, such astufA, show satisfactory amplification and sequencing success, but still fail for the Cladophoraceae of green algae; ITS andrbcL even have high variability of species level resolution, and are not effective for amplification and sequencing due to the presence of introns (Famaet al., 2002; Clarkston and Saunders, 2010; Saunders and Kucera, 2010). Therefore, two or more markers have been commonly used for DNA barcoding rather than relying on a single marker (Hallet al., 2010; Saunders and Kucera, 2010).
Since 2007, especially during the 2008 Summer Olympic Games, large-scale green tides caused by algae bloom constantly occured in Qingdao costal areas, making green algae attacting high society attention (Liuet al., 2009; Huet al., 2010; Zhaoet al., 2012). With the assistance ofDNA barcoding, researchers identified the species causing these green tides as mainlyUlva prolifera, and found their sources off the southern coast of Jiangsu province (Wanget al., 2010a, b; Panget al., 2010; Liuet al., 2012). These researches strongly promoted a molecular system for identifying green algae based on both morphological and molecular assessments. However, there is still a short age of molecular system and a thorough investigation on green macroalgae in coastal areas of China. On the other hand, along with decreasing biodiversity of macroalgae in the coastal area of Qingdao (Liu and Zhang, 1994; Liuet al., 1999; Yanget al., 2009), it is essential to record the present species for long-term monitoring on green macroalgae. Only depending on this database would it be possible to uncover those invasive or cryptic species.
Our study aims to investigate the green macroalgae in coastal zone of Qingdao, identify the distributed species with assistance of DNA barcoding, evaluate the appropriate markers for green macroalgae, and distinguish the localUlvaspecies with the green-tide-formingUlvaspecies.
2 Materials and Methods
2.1 Sample Collections and Identification
Forty-four individuals were collected from April, 2011 to December, 2011 in the intertidal zone along the coast of Northwest Yellow Sea around Qingdao, China (Three of them were collected as references in Rizhao, south of Qingdao (Fig.1)). After cleaning, each sample was morphologically identified (according to Tseng, 1983 and Tsenget al., 2009). Microscopic morphology of the fresh fronds, including the color, texture and branching of the thalli, as well as the cell arrangements and shapes were observed under an Olympus CX31microscope (Olympus Co., Japan). Subsamples were stored in liquid N2(− 80℃) for DNA extraction.

Fig.1 Map of sampling sites. a) western coast of Yellow Sea around Qingdao; b) detailed sites around Qingdao.
2.2 DNA Extraction and Sequencing
DNA was extracted from about 100 mg subsample using DNeasy Plant Kit (TIANGEN Biotech, Beijing, China) following the manufacturer’s specifications. The plastid elongation factortufA gene, nuclear internal transcribed spacer (ITS) region of the ribosomal cistron, and plastid Rubisco large subunit gene 3’ regions (rbcL-3P) were amplified using the published primers,i.e.: the forward primer oftufA(tufGF4) 5’ GGNGCNGCNCAAATGGA YGG 3’) from Saunders and Kucera (2010), the reverse primer oftufA(tufA R) 5’ CCTTCNCGAATMGCRAAW CGC 3’ from Famaet al. (2002); for therbcL-3P, the forward primer GrbcLFi (5’ TCTCARCCWTTYATG CGTTGG 3’) from Saunders and Kucera (2010); the reverse primer is 1385R (5’ AATTCAAATTTAATTTCTT TCC 3’) as published by Manhart (1994); the ITS primers were designed by Haydenet al.(2003) (18S1505, F: 5’TCTTTGAAACCGTATCGTGA 3’; ENT26S, R: 5’GCT TATTGATATGCTTAAGTTCAGCGGGT 3’).
PCR amplification was carried out using the PCR amplification reactor (Mycylcer thermal cycler, BIO-RAD, US) with the PCR Master Mix (TIANGEN Biotech, Beijing, China) to a final volume of 20 μL per reaction according to the manufacturer’s recommendations. PCR profiles referred to Saunders and Kucera (2010) were: tufA− an initial denaturation cycle at 94℃ for 4 min, 38 cycles at 94℃ for 1 min, 45 ℃ annealing for 30 s, 72℃extension for 1 min, followed by 72℃ final extension step performed for 7 min; ITS − an initial 3 min denaturation at 94℃, 38 cycles of 94℃ for 30 s, 54℃ annealing for 40 s, 72℃ extension for 1.5 min, followed by 72℃ final extension for 7 min. rbcL-3P − an initial 2 min denaturation at 95 ℃, 35 cycles of 93℃ for 1 min, 50℃ annealing for 45 s, 72℃ extension for 2 min, followed by 72 ℃ final extension for 7 min. All PCR products were held at 4℃ following amplification till the samples were processed.
Amplification products were checked by 1.0% agarose gel electrophoresis. Fragments oftufA, ITS region andrbcLwere cut from the gel and purified using a TIANgel midi DNA purification Kit (TIANGEN, Beijing, China).
The amplified DNA samples were sequenced by procedures specified by BGI Biotechnology Co. LTD (Shenzhen, China).
2.3 Data Analyses
Multiple sequence alignment was conducted using the CLustal X 1.83 (UCD, Bublin, Ireland). The evolutionary distances were calculated with the neighbor-joining (NJ) method using Mega 5.0 (Tamuraet al., 2007). Additional sequences oftufA, ITS andrbcLgene of green macroalgae were downloaded from GenBank for phylogenetic analysis. The evolutionary divergences of NJ tress were computed using Kimura 2-parameter method (Tamaruet al., 2004). Robustness of the NJ trees was tested with 1000 replicates of the data by bootstrapping.
3 Results
3.1 Morphological Identification
By traditional morphological methods, totally 44 collected individuals were identified as 12 species,i.e.,Ulva pertusa,U. linza,U. intestinalis,U.lactuca,U.compre-ssa,U. prolifera,Monostromanitidum,Codiumfragile,Cladophorafascicularis,C.utriculosa,Bryopsispennate, andB.corticulans.
3.2 DNA Barcoding Identification
There were 8 collected samples that failed to be amplified in molecular identification, including 4 ofCladophorafascicularis, one ofC.utriculosa, one ofBryopsis pennate, and two ofB.corticulans. A total of 9 species were identified by the three gene barcodes (Table 1).
Thirty-sixtufA sample sequences were obtained, and were sorted into 8 distinct groups in the NJ tree, which was composed of 13 groups with 27 additional sequences downloaded from the GenBank (Fig.2) Based on the sequence matching, these specimens were assigned to 8 species,i.e.,Ulva flexuosa,U.californica,U.procera/linza,U.laetevirens,U.compressa,U.australis,Monostromasp.,Codiumfragile; there was an unkown species of Ulvales (close toBligingiamarginata). ForUlva, intraspecific divergences were 0−0.6%, while interspecificdivergences were 1.0%−10.3%. The barcoding gap betweenU.linza/proceraandU.proliferawas 2.4% fortufA. ForMonostromaspp., the evolutionary distance was 2.3%. The intraspecific variation ofCodium fragilewas 1.8%. Among all tested genera (Ulva,Monostroma,BlidingiaandCodium), the evolutionary divergences ranged from 22.4% to 34.2%.
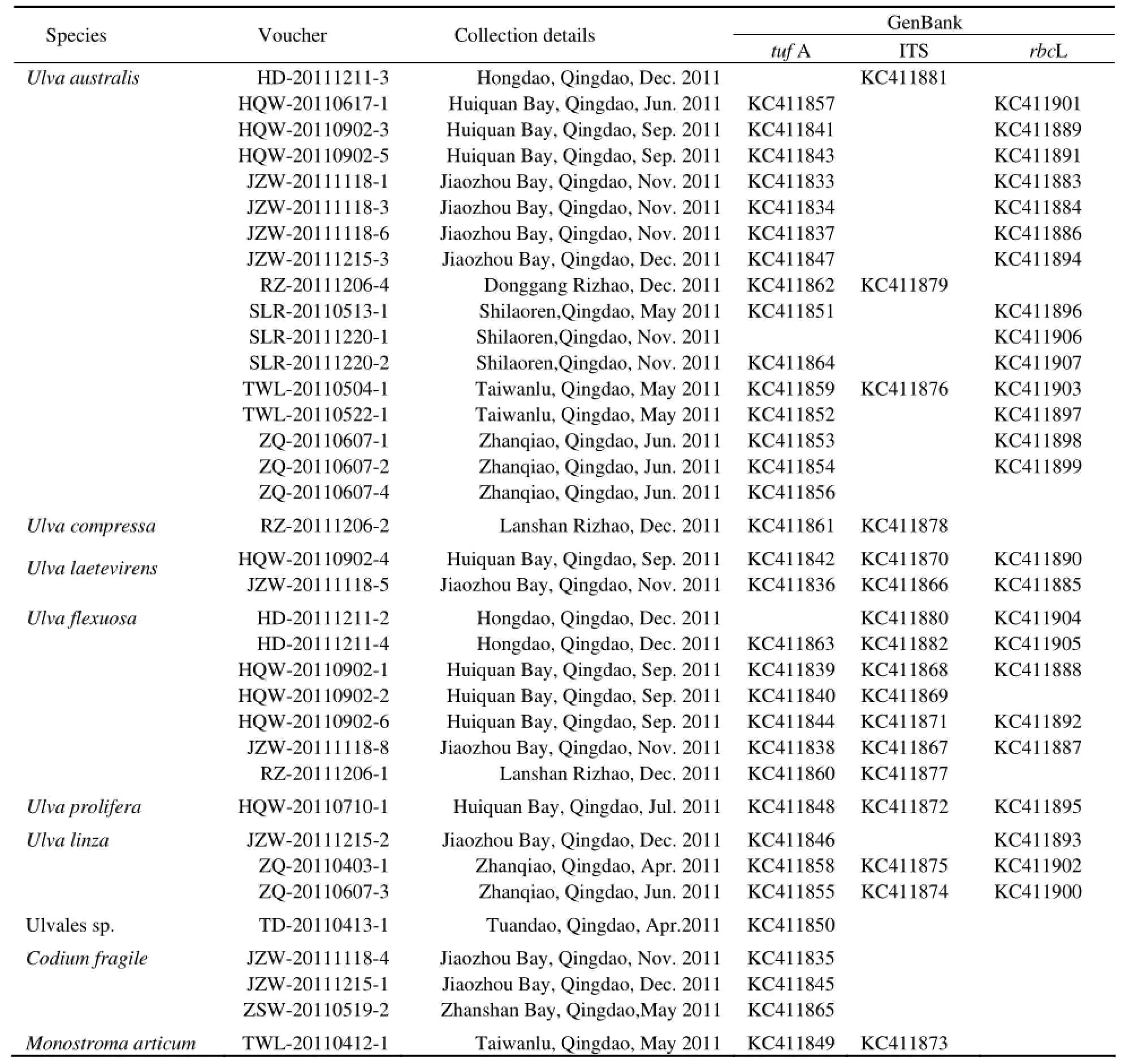
Table 1 List of samples identified by tufA, ITS and rbcL sequences
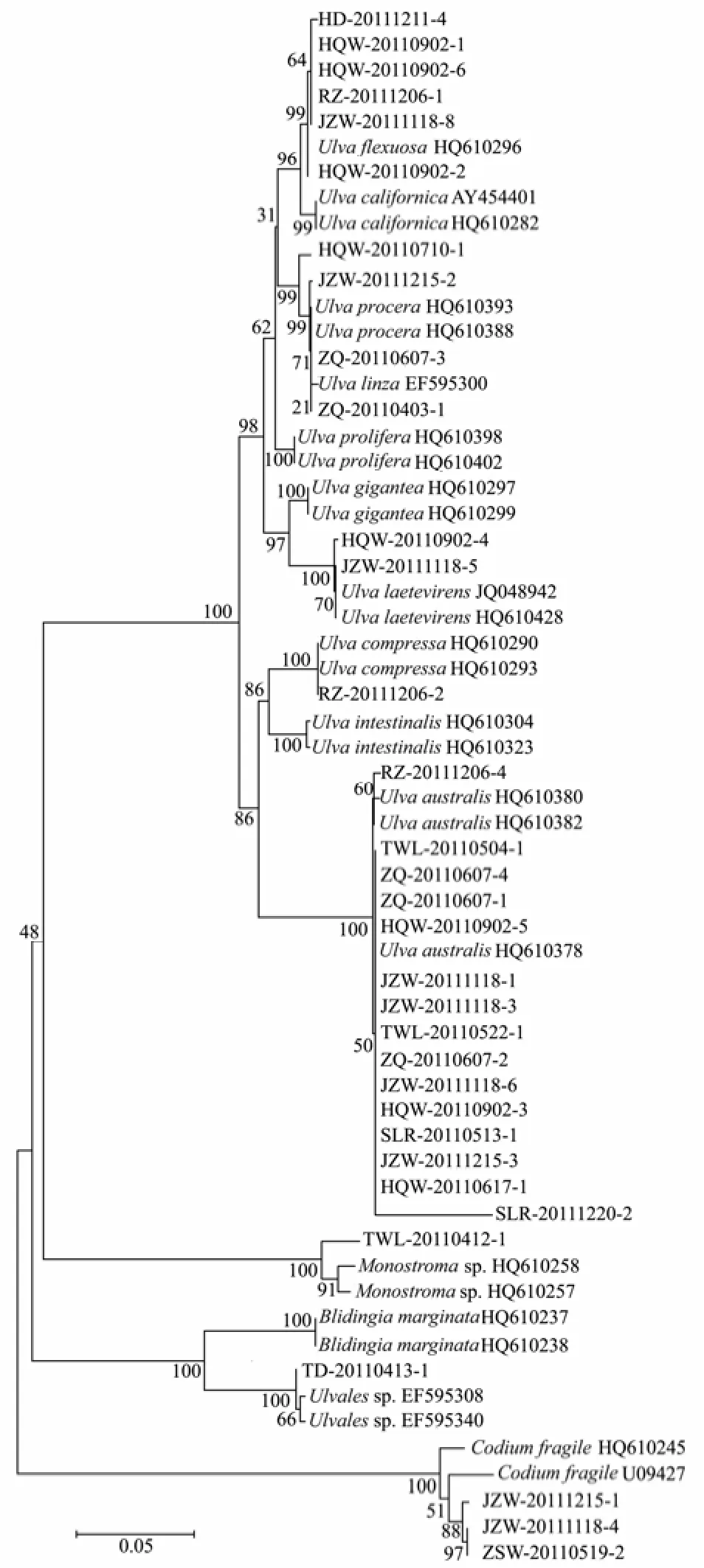
Fig.2 Neighbor-joining tree using tufA sequences of 36 collected samples and 27 additional sequences.
Sequencing success rate was the lowest for the ITS with only 21 sequences obtained. With 27 additional downloaded sequences, totally 48 ITS sequences were grouped into 12 distinguishable clades in the NJ tree. Those 21 sample sequences were resolved into 6 clades, belonging to 6 species,i.e.,U.prolifera,U.linza/prolifera,U.laetevirens,U.compressa,U.australis/pertusa,Monostromaarcticum(Fig.3). ForUlva, the barcoding gaps of intraspecies ranged from 0 to 0.7%, and those of interspeciese were 1.2%−14.5%. ForMonostroma, the intraspecific variations were 0 and 3.0% forM.arcticumandM.grevillei, respectively. The interspecific variation forMonostromawas 3.4%. The evolutionary divergences between species of genusUlvaandMonostromawere larger than 34.6%.
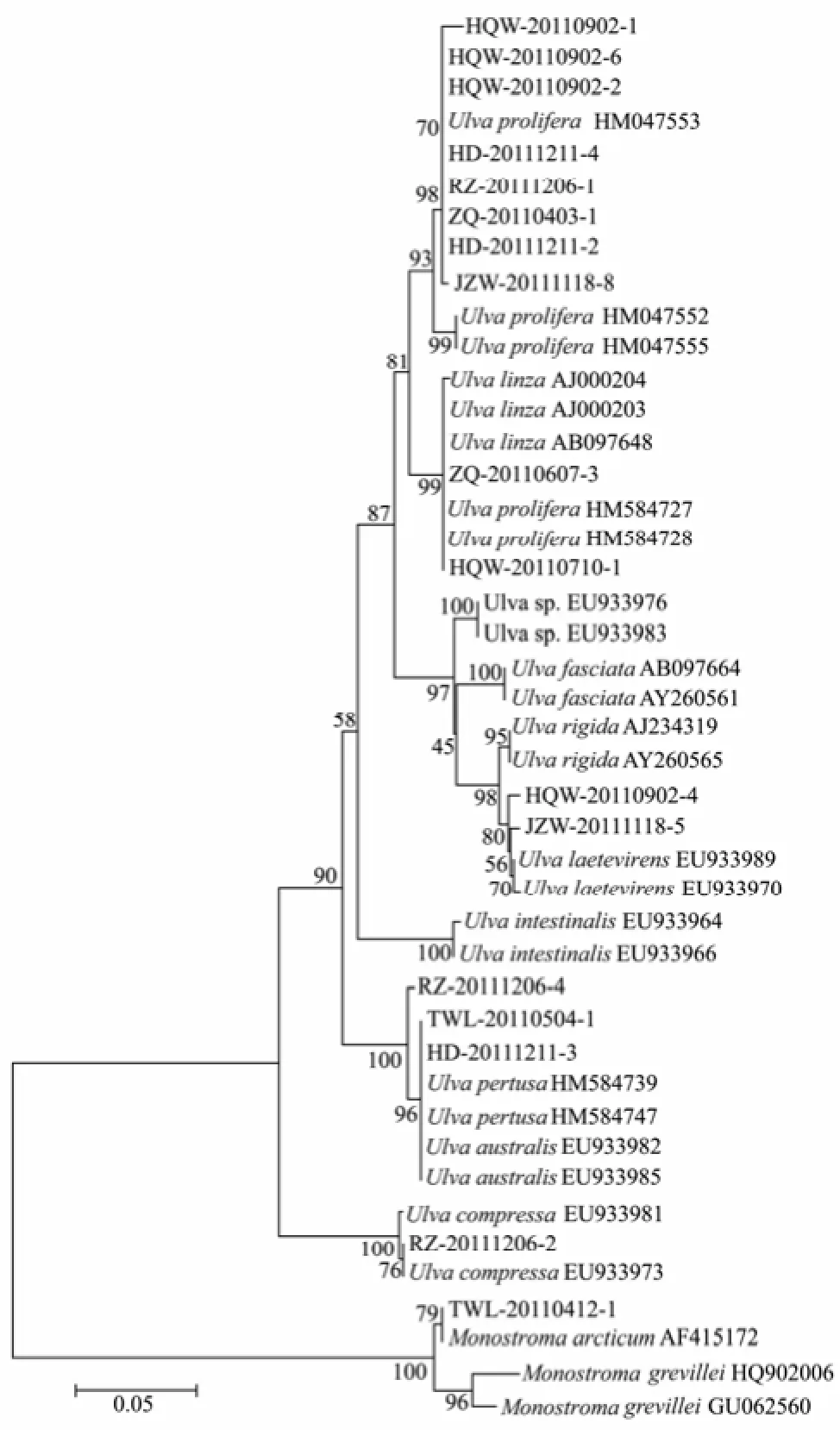
Fig.3 Neighbor-joining tree using ITS sequences of 21 collected samples and 27 additional sequences.
TherbcLsequences of 28 samples were successfully amplified and sequenced, but were only identified to 4−5 species. They were assigned to species ofU.procera/prolifera,U.procera,U.flexuosa,U.laetevirensand U.australis/pertusa(Fig.4). The intraspecific divergences ofUlvawere less than 0.18%. ForMonostroma grevillei, its intraspecific divergences were 0−0.98%. The interspecific divergences ofUlvavaried from 0.98% to 4.4%. The evolutionary divergences between species of genusUlvaandMonostromawere 17.3%−18.8%.
When excluding genusCladophoraandBryopsis, the rates of successful amplification of the three markers were 91.7%, 77.8% and 66.7% fortufA, ITS andrbcL, respectively. If including these two genera, they would decrease to 75%, 63.3% and 54.5%, respectively. Therates of successful sequencing of the three markers were 100%, 57.1% and 100%, respectively.
Based on the data for genusUlvawhich showed the largest most species diversity in this study, the barcode gaps (difference between maximal intra- and minimal inter-specific divergence) were 1.69%, 0.49% and 0.31% fortufA, ITS andrbcL, respectively.

Fig.4 Neighbor-joining tree using rbcL sequences of 28 collected samples and 26 additional sequences.
4 Discussions
4.1 Applicability of the Markers
All three markers failed to amplify the 5 samples fromCladophoraand the 3 samples fromBryopsis. When excluding the two genera,tufA showed the highest amplification success rate of 91.7%, sequencing success rate of 100%, and the largest barcode gap of 1.69%. ThetufA showed the strongest species-discrimination power by sorting out 8 species of the remaining 36 samples, includingCodium fragileandMonostromasp. Till to date,tufA has been known to lack introns for the variety of green algal taxa (De Clercket al., 2008; Verbruggenet al., 2009; Zuccarelloet al., 2009; Händerleret al., 2010). Generally, thetufA has the best combination of universality, sequence success and discriminatory power. Therefore,tufA is the most appropriate marker among all the candidates for DNA barcoding of green macroalgae, and has been strongly recommend by Saunders and Kucera (2010) after evaluating five markers of ITS, LSU, rbcL,tufA and UPA for marine green macroalgae.
Although ITS had relatively higher PCR success thanrbcL in this study, its lowest sequencing success rate might be due to the contaminants or multiple PCR products (Saunders and Kucera, 2010). The ITS discriminated 7 species excludingCodium, and had a medium barcode gap (0.49%). However, in the identification ofU. flexuosaandU. linza-procera-prolifera(LPP, Shimadaet al., 2008), the ITS showed large discrepancy with the other two markers, and classified most of those samples toU.proliferaorU.linza/prolifera(Table 1). Considering its handicap to sequencing and high levels of variation that make such above disarray on molecular concepts, ITS is not recommended as a suitable DNA marker for green macroalgae barcoding, even though ITS was used in most of previous studies on green tide species in Yellow Sea (Liuet al., 2009; Huet al., 2010; Zhaoet al., 2012).
Due to its utility of DNA barcoding,rbcL has formed the basis of several taxonomic and phylogenetic studies in maine green macroalgae to resolve taxonomic issues of the genusUlva(Hayden and Waaland, 2002, 2004; Haydenet al., 2003). On the other hand, the presence of introns inrbcL still negatively affects its universality as a barcode (Hanyudaet al., 2000; Saunders and Kucera, 2010). Pursuing a higher barcode gap ofrbcL-3P thanrbcL-5P (refer to Saunders and Kucera, 2010), the authors used 3P region ofrbcL in this study, obtaining 100% sequencing success with shorter sequence fragments. However, the lowest amplification success also exhibited in our study due to the obstacle of introns. On the contrary, it was deduced that there were no introns inrbcL-5P region in the same study of Saunders and Kucera (2010). Given the fact thatrbcL is an important chloroplast gene for all photosynthetic organisms (Hollingsworthet al., 2009), the applicability of this marker might be extended by choosing a more suitable fragment in this gene. On the background of unsettlement of DNA barcodes for macroalgae, we recommend use ofrbcL as an assistant marker coupled withtufA, and endeavor further to make it more universal.
4.2 Species Discrimination
The molecular methods show their advantages on species identification, such as certainty and accuracy (Valentiniet al., 2008; Raduloviciet al., 2010). In this study, DNA barcoding greatly assisted ascertaining and correcting morphological identification. For example, one sample which was hard to be classified morphologically, was sorted into Ulvales bytufA, close toBlidingia marginatain evolutionary distance (0.087); 9 samples identified by morphological methods asU. intestinaliswere confirmed belonging toU. australis/pertusaandU. flexuosa, respectively. Moreover, 2 samples ofU. lactucamorphologi-cally were distinctly ascertained by three markers asU. laetevirens. And one sample ofMonostramamorphologically identified asM. nitidumin fact molecularly matched toM.arcticumby ITS.
In our NJ trees based on ITS and rbcL sequences,Ulva australiswas proved to be identical withU.pertusa. FortufA discrimination, due to the absence ofU. pertusa tufA gene in NCBI Gene Bank (although it was listed in the paper by Saunders and Kucera (2010), it was presented in NCBI Gene Bank asU. australis), 15 amplified samples were directly identified asU. australis. Therefore, totally 17 samples were discriminated as the same species ofU. australisby at least one of the three genes (tufA, ITS orrbcL).
In previous studies, it wasUlva prolifera(Müller) J. Agardh that was identified as the causative species for green tides happening in successive years since 2007 in coastal areas of Yellow Sea (Sunet al., 2008; Leliaertet al., 2009). Because of the lacking ofU. proliferatufA gene in Gene Bank in china, we analyzed the gene of our sample (HQW-20110710-1, which was collected from the green tide in 2011) with those ITS andrbcL genes of green algae from Yellow Sea of China. It indicated that our sample was identical with HM 584727 and HM 584728 for ITS gene, and with HM 584765 and HM 584769 forrbcL gene. However, in the three gene’s NJ trees, this freefloating sample was grouped into different clades asU. procera/linzabytufA,U. Linza/proliferaby ITS, andU. procera/proliferabyrbcL, respectively. This discrepancy is, on the one hand, owing to the uncompleted database of DNA barcode, and, on the other hand, to the complex of so calledUlva linza-procera-proliferacomplex (LPP clade, Shimadaet al., 2008, Leliaertet al., 2009). For the latter case, we suggest thatU. proceraandU. linzabe unified asU. linza, not only because of the fact that theU. procera(K. Ahlner) is currently regarded as a taxonomic synonym ofU. linzaLinnaeus (Haydenet al., 2003) in Algaebase (http://www.algaebase.org/), and both species were clustered together intufA NJ tree in our study, but also the fact that phylogeneticallyU. linzaandU. proceracould be clearly distinguished fromU. proliferaby 5s rDNA spacer (Shimadaet al., 2008, 2010; Liuet al., 2012). In addition, either morphologically or molecularly, further discrimination or rectification needs to be carried on samples from different regions in the world.
The remarkable benefits of DNA barcoding have already been widely accepted; however, for green macroalgae, their convergence of morphological characters is strong but the number of synapomorphic characters is few, and conflicts between traditional phonetic and phylogenentic classifications would inevitably exist for a period (Medlinet al., 2007; Pröschold and Leliaert, 2007; Saunders and Mcdevit, 2012). In our study, the confusion in molecular identification of some samples also implied that further work needs to be done in several ways, such as establishing appropriate DNA markers, completing the gene database on consistency of traditional and molecular concepts, as well as combining molecular and morphological methods together when identifying species with extreme morphological plasticity (Mareset al., 2011).
Acknowledgements
This research was financially supported by the Public Science and Technology Research Funds Projects of Ocean (201105021 and 201305030), National Natural Science Foundation China (41276137), Shandong Provincial Natural Science Foundation, China (ZR2011CM 018), and Qingdao Municipal Science and Technology Program, China (09-2-5-3-hy).
Buchheim, M. A., Keller, A., Koetschan, C., Förster, F., Merget, B., and Wolf, M., 2011. Internal transcribed spacer 2 (nu ITS2 rRNA) sequence-structure phylogenetics: Towards an automated reconstruction of the green algal tree of life. Plos one, 6 (2): 1-10.
Clarkston, B. E., and Saunders, G. W., 2010. A comparison of two DNA barcode markers for species discrimination in the red algal family Kallymeniaceae (Gigartinales) with a description of Euthora timburtonii. Botany, 88: 119-131.
De Clerck, O., Verbruggen, H., Huisman, J. M., Faye, E. J., Leliaert, F., Schils, T., and Coppejans, E., 2008. Systematics and biogeography of the genus Pseudocodium (Bryopsidales, Chlorophyta), including the description of P. natalense sp. nov. from South Africa. Phycologia, 47: 225-235.
Fama, P., Wysor, B., Kooistra, W., and Zuccarello, G. C., 2002. Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene. Journal of Phycology, 38: 1040-1050.
Hall, J. D., Fučíková, K., Lo, C., Lewis, L. A., and Karol, K. G., 2010. An assessment of proposed DNA barcodes in freshwater green algae. Cryptogamie Algologie, 31 (4): 529-555.
Händeler, K., Wägele, H., Wahrmund, U., Rüdinger, M., and Knoop, V., 2010. Slugs’ last meals: Molecular identification of sequestered chloroplasts from different algal origins in Sacoglossa (Opisthobranchia, Gastropoda). Molecular Ecology Resources, 10: 968-978.
Hanyuda, T., Arai, S., and Uedak, K., 2000. Variability in the rbcL introns of Caulerpalean algae (Chlorophyta, Ulvophyceae). Journal of Plant Research, 113: 403-413.
Harper, J. T., and Sauders, G. W., 2001. The application of sequences of the ribosomal cistron to the systematics and classification of the florideophyte red algae (Florideophyceae, Rhodophyta). Les cahiers de Biologie Marine, 42: 25-38.
Hayden, H. S., Blomster, J., Maggs, C. A., Silva, P. C., Stanhope, M. J., and Waaland, J. R., 2003. Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. European Journal of Phycology, 38: 277-294.
Hayden, H. S., and Waaland, J. R., 2002. Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences. Journal of Phycology, 38: 1200-1212.
Hayden, H. S., and Waaland, J. R., 2004. A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific. Phycologia, 43: 364-382.
Hebert, P. D. N., Cywinska, A., Ball, S. L., and Deward, J. R., 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London Biology, 270: 313-321.
Hebert, P. D. N., and Gregory, T. R., 2005. The promise of DNAbarcoding for taxonomy. Systematic Biology, 54: 852-859.
Hu, C., Li, D., Chen, C., Ge, J., Muller-Karger, F. E., Liu, J., Yu, F., and He, M. X., 2010. On the recurrent Ulva prolifera blooms in the Yellow Sea and East. Journal of Geophysical Research, 115, C05017, DOI: 10.1029/2009JC005561.
Leliaert, F., Zhang, X., Ye, N., Malta, E., Engelen, A. H., Mineur, F., Verbruggen, H., and Clerck, O. D., 2009. Identity of the Qingdao algal bloom. Phycological Research, 57: 147-151.
Liu, D. Y., Wang, Z. Y., Sun, J., Huang, Z. Y., and Qian, S. B., 1999. Study of the benthic algae in the littoral of Qingdao coast. Transactions of Oceanology and Limnology, 3: 35-40.
Liu, D. Y., Keesing, J. K., Xing, Q. G., and Shi, P., 2009. World’s largest macroalgal bloom caused by expansion of seaweed aquaculture in China. Marine Pollution Bulletin, 58: 888-895.
Liu, F., Pang, S. J., Zhao, X. B., and Hu, C. M., 2012. Quantitative molecular and growth analyses Ulva propagules in sediment of Jiangsu initially green tides. Marine Environmental Research, 74: 56-63.
Loughnane, C. J., McIvor, L. M., Rindi, F., Stengel, D. B., and Guiry, M. D., 2008. Morphology, rbcL phylogeny and distribution of distromatic Ulva (Ulvophyceae, Chlorophyta) in Ireland and southern Britain. Phycologia, 47: 416-429.
Manhart, J. R., 1994. Phylogenetic analysis of green plant rbcL sequences. Molecular Phylogenetics and Evolution, 3: 114-127.
Mares, J., Leskinen, E., Sitkowska, M., Skácelová, O., and Blomster, J., 2011. True identity of the European freshwater Ulva revealed by molecular and morphological methods. Journal of Phycology, 47: 1177-1192.
Mcdevit, D. C., and Saunders, G. W., 2009. On the utility of DNA barcoding for species differentiation among brown macroalgae (Phaeophyceae) including a novel extraction protocol. Phycological Research, 57: 131-141.
Medlin, L. K., Metifies, K., John, U., and Olsen, J. L., 2007. Algal molecular systematic: A review of the past and prospects for the future. In: Unravelling the Algae-the Past, Present and Future of Algal Systematic. Brodie, J., and Lewis, J., eds., CRC Press, Boca Raton, FL, 341-353.
Meier, R., Zhang, G., and Ali, F., 2008. The use of mean instead of smallest interspecific distances exaggerates the size of the‘Barcoding Gap’ and leads to misidentification. Systematic Biology, 57: 809-813.
Nakazawa, A., Yamada, T., and Nozaki, H., 2004. Taxonomic study of Asterococcus (Chlorophyceae) based on comparative morphology and rbcL gene sequences. Phycologia, 43: 711-721.
O’Kelly, C. J., Kurihara, A., Shipley, T. C., and Sherwood, A. R., 2010. Molecular assessment of Ulva spp. (Ulvophyceae, Chlorophyta) in the Hawaiian Islands. Journal of Phycology, 46: 728-735.
Pang, S. J., Liu, F., Shan, T. F., Xu, N., Zhang, Z. H., Gao, S. Q., Chopin, T., and Sun, S., 2010. Tracking the algal origin of the Ulva bloom in the Yellow Sea by a combination of molecular, morphological and physiological analyses. Marine Environmental Research, 69: 207-215.
Pröschold, T., and Leliaert, F., 2007. Systematics of the green algae: Conflict of classic and modern approaches. In: Unravelling the Algae − the Past, Present and Future of Algal Systematic. Brodie, J., and Lewis, J., eds., CRC Press, Boca Raton, FL, 123-153.
Robba, L., Russell, S., Baker, G., and Brodie, J., 2006. Assessing the use of the mitochondrial COX I marker for use in DNA barcoding of red algae (Rhodophyta). American Journal of Botany, 93 (8): 1101-1108.
Shimada, S., Yokoyama, N., Arai, A., and Hiraoka, M., 2008. Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. Journal of Applied Phycology, 20: 979-989.
Saunders, G. W., 2005. Applying DNA barcoding to red macroalgae a preliminary appraisal holds promise for future applications. Philosophical Transactions of the Royal Society B, 360: 1879-1888.
Saunders, G. W., and Kucera, H., 2010. An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie Algologie, 31 (4): 487-528.
Sun, S., Wang, F., Li, C., Qin, S., Zhou, M., Ding, L., Pang, S., Duan, D., Wang, G., Yin, B., Yu, R., Jiang, P., Liu, Z., Zhang, G., Fei, X., and Zhou, M., 2008. Emerging challenges: Massive green algae blooms in the Yellow Sea. Nature Precedings, hdl: 10101/ npre.2008.2266.1.
Tamura, K., Nei, M., and Kumar, S., 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences of USA, 101: 11030-11035.
Tamura, K., Dudley, J., Nei, M., and Kumar, S., 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software Ver. 4.0. Molecular Biology and Evolution, 24: 1596-1599. Tseng, C. K., 1983. Common Seaweeds of China. Science Press, Beijing, 25-33.
Tseng, C. K., Xia, B. M., and Zhou, X. T., 2009. Seaweeds in Yellow Sea and Bohai Sea of China. Science Press, Beijing, 254pp (in Chinese).
Valentini, A., Pompanon, F., and Taberlet, P., 2008. DNA barcoding for ecologists. Trends in Ecology and Evolution, 24 (2): 110-117.
Verbruggen, H., Tyberghein, L., Pauly, K., Vlaeminck, C., Van Nieuwenhuyze, K., Koositra, W., Leliaert, F., and De Clerck, O., 2009. Macroecology meets macroevolution: Evolutionary niche dynamics in the seaweed Halimeda. Global Ecology and Biogeography, 18: 393-405.
Wang, J. F., Li, N., Jiang, P., Boo, S. M., Lee, W. J., Cui, Y., Lin, H., Zhao, J., Liu, Z., and Qin, S., 2010a. Ulva and Enteromorpha (Ulvaceae, Chlorophyta) from two sides of the Yellow Sea: Analysis of nuclear rDNA ITS and plastid rbcL sequence data. China Journal of Oceanology and Limnology, 28: 763-768.
Wang, J. F., Jiang, P., Cui, Y. L., Li, N., Wang, M. Q., Lin, H. Z., Hee, P., and Qin, S., 2010b. Molecular analysis of green-tideforming macroalgae in Yellow Sea. Aquatic Botany, 93: 25-31.
Yang, Z., Wang Y., Dong, K. S., Tang, X. X., and Zhao, X., 2009. The survey on the community of benthic marine macroalgae. Periodical of Ocean University of China, 39 (4): 647-651.
Zhao, J., Jiang, P., Liu, Z. Y., Wei, W., Lin, H. Z., Li, F. C., Wang, J. F., and Q, S., 2012. The Yellow Sea green tides were dominated by one species, Ulva (Enteromorpha) prolifera, from 2007 to 2011. Chinese Science Bulletin, DOI: 10.1007/ s11434-012-5441-3.
Zuccarello, G., Price, N., Verbruggen, H., and Leliaert, F., 2009. Analysis of a plastid multigene data set and the phylogenetic position of the marine macroalga Caulerpa filiformis (Chlorophyta). Journal of Phycology, 45: 1206-1212.
(Edited by Ji Dechun)
* Corresponding author. Tel: 0086-532-82032789
E-mail: yxmao@ouc.edu.cn
(Received October 30, 2012; revised January 4, 2013; accepted March 20, 2013)
© Ocean University of China, Science Press and Springer-Verlag Berlin Heidelberg 2014
杂志排行
Journal of Ocean University of China的其它文章
- Properties of Klebsiella Phage P13 and Associated Exopolysaccharide Depolymerase
- Comparative Study on the Allergenicity of Different Litopenaeus vannamei Extract Solutions
- Isolation and Characterization of a Fucoidan-Degrading Bacterium from Laminaria japonica
- Toxicity of Five Phenolic Compounds to Brine Shrimp Artemia sinica (Crustacea: Artemiidae)
- Application of CFD Modeling to Hydrodynamics of CycloBio Fluidized Sand Bed in Recirculating Aquaculture Systems
- QSAR for Photodegradation Activity of Polycyclic Aromatic Hydrocarbons in Aqueous Systems
