循环微小RNA与乳腺癌的研究进展
2014-02-08李雅楠邢邯英郝玉宾
李雅楠,邢邯英,郝玉宾
乳腺癌是女性最常见的恶性肿瘤之一。2012年,乳腺癌占美国女性新确诊肿瘤的首位,占女性肿瘤相关病死率的第二位[1]。乳腺癌的发生发展是涉及多因素的复杂过程,2005年,Iorio等[2]首次提出人乳腺癌组织中存在微小RNA(microRNAs,miRNAs)的表达改变。随着对miRNAs研究的深入,人们发现miRNAs广泛而稳定地存在于血清、血浆、组织液及各类体液等细胞外液中,即循环miRNAs[3]。在疾病状态如肿瘤、免疫性疾病、循环系统疾病等情况下,miRNAs的表达谱发生异常改变。现就循环miRNAs与乳腺癌的相关研究进展做一综述。
1 循环miRNAs
1.1 简介 miRNAs是一类广泛存在于动植物体内的长度为18~25个核苷酸的内源性非编码单链微小RNA,具有高度的遗传稳定性。miRNAs主要通过与靶mRNA 3′非翻译区(3′-UTR)特异性结合,引起靶mRNA降解或翻译抑制,发挥对基因表达的转录后调节作用。1993年,Lee等[4]在秀丽隐杆线虫体内发现了miRNAs家族的第一个成员lin-4,其能在翻译水平抑制核蛋白表达,从而调控幼虫的发育进程。随后,科学家们通过随机克隆和高通量测序、微阵列芯片、生物信息学预测等方法在动植物中发现了多种miRNAs。据miRBase数据库(http://www.mirbase.org/)统计,至2012年8月已发现miRNAs 21 264个。据预测,50%以上的mRNA是miRNAs的靶点,并且每个miRNA可调控高达数百个靶基因[5]。2008年,Lawrie等[6]首次报道在B细胞淋巴瘤患者血清中发现miR-155、miR-210及miR-21表达水平升高。之后大量实验证实miRNAs可以广泛而稳定地存在于细胞外液,如血清、血浆、组织液及唾液、鼻腔分泌物、尿液等各种体液[7-9],统称为循环miRNAs。循环miRNAs对核糖核酸酶(RNase)、极端pH、温度不敏感,不受反复冻融、长期保存、酸碱处理等影响[10]。良好的稳定性及肿瘤相关性使得循环miRNAs成为肿瘤等疾病的非创伤性新型生物标志物,是肿瘤诊断与防治的研究热点。
1.2 来源 研究发现,循环miRNAs或存在于外体(exosomes)、外来体样囊泡(exosome-like vesicles)、微粒体(microparticles)和凋亡小体(apoptotic bodies)等微囊结构中[11],或包裹于高密度脂蛋白和RNA结合蛋白(如Ago2、核仁磷酸蛋白1)之中[12-13]。Turchinovich等[14]发现,血浆或细胞培养基中的miRNAs能完全通过0.22 μm的滤器,在110 000×g超速离心后miRNAs仍存在于上清液中,这提示循环miRNAs并非来自于微囊。免疫印迹分析显示,超滤液中的细胞外miRNAs与RNA沉默复合体中的96 kD的Ago2相结合,血浆与培养基中的miRNAs在无清洁剂条件下能与抗Ago2抗体发生免疫共沉淀反应。这首次证明细胞外miRNAs并非来自外体或微泡,而是与Ago2相关。故Turchinovich等推测循环miRNAs大部分来源于死细胞,由于Ago2和Ago2-miRNA复合体的高度稳定性,使得miRNAs能稳定存在于细胞外液,但并不排除有些循环miRNAs与外体相关的可能性。
2 循环miRNA与乳腺癌
目前,人们对循环miRNAs的来源和功能还不十分明确,但大量研究证明在某些病理过程中其表达谱发生改变(见表1)。自2005年Iorio等[2]首次发现乳腺癌组织中存在miRNAs改变,人们发现多种miRNAs以癌基因或抑癌基因的角色参与了乳腺癌的发生发展过程,大量实验证实乳腺癌组织与正常组织相比,miRNAs表达谱不同(见表2)。有人提出是循环miRNAs表达谱而非肿瘤组织中的miRNAs表达谱反映乳腺癌的存在情况[55]。循环miRNAs涉及乳腺癌诊断、病理分级、药物抵抗、转移等多方面(见表3)。
2.1 循环miRNAs与乳腺癌的早期诊断 Cuk等[59]利用TaqMan低密度芯片检测10例早期乳腺癌患者与10例健康人血浆miRNAs表达谱,筛选出7个miRNAs;并利用实时荧光定量聚合酶链式反应(RT-qPCR)在127例乳腺癌患者与80例健康人血浆中进行验证,发现前者miR-148b、miR-409-3p、miR-801表达上调,可能有助于乳腺癌的早期诊断。Schrauder等[62]发现早期乳腺癌患者全血中miR-202表达上调,miR-718表达下调。Si等[63]对比乳腺癌组织、原发性乳腺癌患者血清及健康对照者血清,发现乳腺癌患者血清miR-92a表达下调,且与肿瘤大小、淋巴结转移相关,提示血清miR-92b对乳腺癌具有潜在的预测价值。
有研究发现血浆miR-145和miR-451联合检测乳腺癌的阳性预测值为88%,阴性预测值为92%;在乳腺癌早期血浆中已存在miR-145和miR-451改变,在乳腺导管原位癌中其阳性预测值高达96%[64]。乳腺癌患者血清miR-181a水平下调,受试者工作特征(ROC)曲线分析显示,在早期乳腺癌诊断中,miR-181a的敏感度为55.28%,而糖类抗原15-3(CA15-3)和癌胚抗原(CEA)的敏感度分别为8.13%和7.32%。相比之下,miR-181a用于乳腺癌早期诊断有更高的敏感度[65]。
2.2 循环miRNAs与乳腺癌的组织病理分级 Wang等[66]发现乳腺癌患者癌组织与血清中miR-21、miR-106a、miR-126、miR-155、miR-199a、miR-335的表达变化趋势一致,血清miR-155在良性疾病乳腺组织表达水平升高1.7倍,Ⅱ级乳腺癌组织升高2.7倍,而Ⅲ级乳腺癌组织中升高4.0倍。血清miR-21、miR-106a、miR-155水平随肿瘤恶性程度的增加也相应升高,且在雌激素、孕激素阴性的患者中其表达也增高,证明循环miRNAs与乳腺癌组织病理分级及激素受体表达相关,可能成为乳腺癌诊断、组织病理分级和判断预后的分子标志物[66]。
2.3 循环miRNAs与乳腺癌的药物抵抗 癌症的化疗药物抵抗常导致癌症的复发和转移,因此迫切希望找到能够预测辅助化疗治疗效果的分子标志物来指导临床工作。在浸润性导管癌患者血清中,miR-125b表达水平上调,MCF-7、MDA-MB-231等乳腺癌细胞系中,其过表达能显著抑制5-氟尿嘧啶诱导的细胞毒作用,敲低之后细胞毒作用增强。这提示循环miR-125b与乳腺癌化疗抵抗相关[67]。
Schwarzenbach等[60]发现乳腺癌患者血清miR-214表达上调,术后则显著下降,且与局部淋巴结转移相关。Razis等[68]发现磷酸酶基因(PTEN)的缺失与曲妥珠单抗治疗人表皮生长因子受体2(HER2)阳性乳腺癌患者的低生存率有关,而PTEN是miR-214的重要靶点,是否miR-214的上调导致靶基因PTEN降低或缺失,从而间接导致曲妥珠单抗耐药,尚需实验证明。
2.4 循环miRNAs与乳腺癌转移 Chen等[69]发现淋巴结转移的乳腺癌患者血浆中miR-10b和miR-373水平升高,两者联合检测诊断乳腺癌的敏感度为72.0%,特异度达94.3%,是检测乳腺癌淋巴结转移情况的潜在标志物。Zhao等[70]检测到血清miR-10b过表达与原发乳腺癌的骨转移有关,敏感度为64.8%,特异度为69.5%,且其升高水平与临床阶段的恶性程度相一致,提示其可能为新的治疗靶点。
Sun等[58]发现乳腺癌患者血清miR-155的表达水平是正常人的2.94倍,且与乳腺组织中miR-155的表达变化一致;术后血清miR-155表达水平明显下降,这提示其可能来源于肿瘤组织。有趣的是,化疗之后其水平也降至基础水平。研究还发现乳腺癌患者发生肺转移时,miR-155水平显著升高,当转移灶控制以后,miR-155水平又回到术前水平,这提示miR-155可能是肿瘤细胞的生长刺激因子。
Wu等[71]研究了26例乳腺癌分级在Ⅱ~Ⅲ级的患者血清,其中包括8例2年后出现转移复发的病例,发现miR-122在复发病例中的表达显著升高,其预测乳腺癌转移的敏感度为88%,特异度为78%。
3 循环miRNAs检测
3.1 检测方法 循环miRNAs的检测方法主要有高通量测序法、miRNAs微阵列芯片和RT-qPCR法。高通量测序法能在序列未知情况下研究miRNAs表达谱变化,可发现和鉴定新的miRNAs分子,但检测费用昂贵,不适合临床检测。miRNAs微阵列芯片能进行miRNAs表达谱分析和多种miRNAs同时检测,鉴于miRNAs微阵列芯片价格较贵,且结果不稳定,只能用于miRNAs初筛。RT-qPCR是最常用的检测手段,能对miRNAs定性或定量研究,可作为miRNAs微阵列芯片检测结果的确证实验,有较高的敏感度和特异度,但无法发现新的miRNAs分子,且研究内容不如miRNAs微阵列芯片技术全面[3]。
近几年,很多新型循环miRNAs检测技术出现。Broyles等[72]设计用荧光基团/淬灭基团标记的寡核苷酸探针同时检测血清多种miRNAs。目的基因不存在时,荧光基团与淬灭基团互补结合,导致以荧光共振能量转移为基础的荧光信号的淬灭;当存在目的基因时,其与淬灭探针杂交,随着淬灭探针被目的基因遮蔽超过福斯特半径,荧光强度逐渐增强。此方法的优点在于可直接检测而无需进行血清RNA提取,可用于定量分析,检测限低,能检测液相DNA或RNA靶基因。非标记电化学基因传感器技术可直接检测miRNAs,无需进行聚合酶链式反应(PCR)和标记反应,可用于血清等生物样品miRNAs常规检测[73]。基于α溶血素的纳米孔传感器,利用可编程序的寡核苷酸探针,产生靶点特异性信号,可定量亚皮克水平的肿瘤相关miRNAs,可辨别miRNAs家族成员之间单核苷酸差异,无需标记和扩增[74]。

表1 疾病与循环miRNAs表达谱变化
注:PBMCs=外周血单个核细胞,CA19-9=糖类抗原19-9,ALT=丙氨酸氨基转移酶

表2 乳腺癌组织miRNAs表达谱的改变及作用
注:NRF2=核因子红系2相关因子,FFPE=甲醛固定石蜡包埋,HOXA10=同源盒基因A10,MIM=转移消失蛋白,RhoA=大鼠肉瘤同系物家族成员A,TGF-β=转化生长因子β,VHL=希佩尔林道抑癌基因,KLF4=Kruppel样因子4,PTGS2=前列腺素合酶2,PKCε=蛋白激酶Cε,BCL2=B细胞淋巴瘤/白血病-2,NF-κB=核因子κB,ERK=胞外信号调节激酶,FAK=局部黏着斑激酶
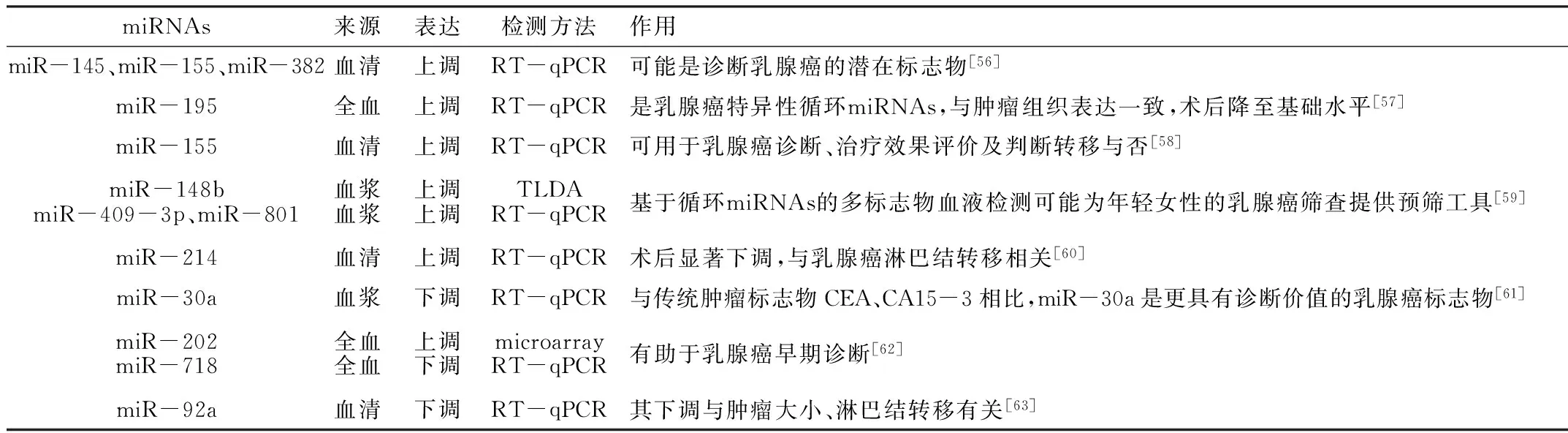
表3 循环miRNAs在乳腺癌诊断、病理分级、药物抵抗、转移等中的作用
注:RT-qPCR=实时荧光定量聚合酶链式反应,TLDA=TaqMan低密度芯片,CEA=癌胚抗原,CA15-3=糖类抗原15-3,microarray=微阵列芯片
3.2 循环miRNAs检测面临的问题 目前,找到像U6一样恒定表达的内源性miRNAs作为内参,仍是临床检测循环miRNAs标志物和实验室之间进行结果对比的主要障碍。Hu等[75]采用Solexa测序和TaqMan低密度芯片两种技术在10例患者血清中系统筛查miRNAs,并用RT-qPCR在50例血清中进行验证,通过两级病例对照分析筛选出2个候选miRNAs,即miR-191和miR-484,敏感度和特异度均为91.9%。还可以通过以下两种方法来减少实验变异:使用相同体积的血清;使用人工合成的非内源性miRNAs,如在提取RNA开始即加入秀丽隐杆线虫的miRNAs。
尽管循环miRNAs能用于预测乳腺癌转移,但其检测并未考虑到以下两点:第一,并不是肿瘤组织中的所有细胞都具有转移能力,具有转移能力的肿瘤细胞在遗传组成上也可能不同;第二,在发生转移时,残留的或复发的肿瘤细胞的遗传特征与原发肿瘤可能不同,因为肿瘤出现转移灶可能发生在很多年后,且已接受过多种抗肿瘤疗法。这些因素都可能导致原发肿瘤与转移灶之间的基因和表观遗传组成的差异[76]。
4 展望与小结
传统的肿瘤标志物,如CEA、CA15-3对乳腺癌的预测有诊断价值,但血液蛋白质组成复杂、水平低、稳定性相对差、敏感度低等缺点,在检测早期癌症方面意义不大[77]。循环miRNAs具有高度保守性、高度稳定性、疾病特异性、无创或微创、检测敏感度高等特点,弥补了蛋白质标志物的不足,有替代传统肿瘤标志物成为新型生物标志物的潜能。目前,循环miRNAs研究结果有一定的局限性:第一,研究样本量小,不能兼顾到所有乳腺癌类型,得不出确切的结果;第二,研究发现有些循环miRNAs在手术前后并未明显变化,可能是由于取样时间与手术的时间间隔较短;第三,大部分研究为横向观察,缺乏对疾病复发、无病生存期等预后效果的长期随访,缺乏对这期间循环miRNAs表达变化的研究。因此,对大规模不同类型的乳腺癌样本和术后不同时间点的样本中循环miRNAs的观测来证实前人得出的结果,是今后研究的重点。
1 Singer A,Tresley J,Velazquez-Vega J,et al.Unusual aggressive breast cancer:metastatic malignant phyllodes tumor[J].J Radiol Case Rep,2013,7(3):24-37.
2 Iorio MV,Ferracin M,Liu CG,et al.MicroRNA gene expression deregulation in human breast cancer[J].Cancer Res,2005,65(16):7065-7070.
3 Reid G,Kirschner MB,van Zandwijk N.Circulating microRNAs:association with disease and potential use as biomarkers[J].Crit Rev Oncol Hematol,2011,80(2):193-208.
4 Lee RC,Feinbaum RL,Ambros V.The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14[J].Cell,1993,75(5):843-854.
5 Gurtan AM,Sharp PA.The role of miRNAs in regulating gene expression networks[J].J Mol Biol,2013,425(19):3582-3600.
6 Lawrie CH,Gal S,Dunlop HM,et al.Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma[J].Br J Haematol,2008,141(5):672-675.
7 Lässer C,O′Neil SE,Ekerljung L,et al.RNA-containing exosomes in human nasal secretions[J].Am J Rhinol Allergy,2011,25(2):89-93.
8 Weber JA,Baxter DH,Zhang S,et al.The microRNA spectrum in 12 body fluids[J].Clin Chem,2010,56(11):1733-1741.
9 Wang G,Kwan BC,Lai FM.et al.Expression of microRNAs in the urinary sediment of patients with IgA nephropathy[J].Dis Markers,2010,28(2):79-86.
10 Mitchell PS,Parkin RK,Kroh EM,et al.Circulating microRNAs as stable blood-based markers for cancer detection[J].Proc Natl Acad Sci U S A,2008,105(30):10513-10518.
11 van der Pol E,Boing AN,Harrison P,et al.Classification,functions,and clinical relevance of extracellular vesicles[J].Pharmacol Rev,2012,64(3):676-705.
12 Arroyo JD,Chevillet JR,Kroh EM.et al.Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma[J].Proc Natl Acad Sci U S A,2011,108(12):5003-5008.
13 Vickers KC,Palmisano BT,Shoucri BM,et al.MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins[J].Nat Cell Biol,2011,13(4):423-433.
14 Turchinovich A,Weiz L,Langheinz A,et al.Characterization of extracellular circulating microRNA[J].Nucleic Acids Res,2011,39(16):7223-7233.
15 Glowacki F,Savary G,Gnemmi V,et al.Increased circulating miR-21 levels are associated with kidney fibrosis[J].PLoS One,2013,8(2):e58014.
16 Zhao A,Li G,Peoc′h M,et al.Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma[J].Exp Mol Pathol,2013,94(1):115-120.
17 Luo Y,Wang C,Chen X,et al.Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome[J].Clin Chem,2013,59(4):658-666.
18 Rotkrua P,Shimada S,Mogushi K,et al.Circulating microRNAs as biomarkers for early detection of diffuse-type gastric cancer using a mouse model[J].Br J Cancer,2013,108(4):932-940.
19 Wu J,Yang T,Li X,et al.Alteration of serum miR-206 and miR-133b is associated with lung carcinogenesis induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone[J].Toxicol Appl Pharmacol,2013,267(3):238-246.
20 Kaduthanam S,Gade S,Meister M,et al.Serum miR-142-3p is associated with early relapse in operable lung adenocarcinoma patients[J].Lung Cancer,2013,80(2):223-227.
21 Fan KL,Zhang HF,Shen J,et al.Circulating microRNAs levels in Chinese heart failure patients caused by dilated cardiomyopathy[J].Indian Heart J,2013,65(1):12-16.
22 Dickinson BA,Semus HM,Montgomery RL,et al.Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure[J].Eur J Heart Fail,2013,15(6):650-659.
23 Gidlöf O,Smith JG,Miyazu K,et al.Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction[J].BMC Cardiovasc Disord,2013,13:12.
24 Li YQ,Zhang MF,Wen HY.et al.Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction[J].Clinics(Sao Paulo),2013,68(1):75-80.
25 Kawaguchi T,Komatsu S,Ichikawa D,et al.Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer[J].Br J Cancer,2013,108(2):361-369.
26 Wang WS,Liu LX,Li GP,et al.Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer[J].Cancer Prev Res(Phila),2013,6(4):331-338.
27 van der Meer AJ,Farid WR,Sonneveld MJ,et al.Sensitive detection of hepatocellular injury in chronic hepatitis C patients with circulating hepatocyte-derived microRNA-122[J].J Viral Hepat,2013,20(3):158-166.
28 Zhang Q,Kandic I,Faughnan ME,et al.Elevated circulating microRNA-210 levels in patients with hereditary hemorrhagic telangiectasia and pulmonary arteriovenous malformations:a potential new biomarker[J].Biomarkers,2013,18(1):23-29.
29 Ragusa M,Caltabiano R,Russo A,et al.MicroRNAs in vitreus humor from patients with ocular diseases[J].Mol Vis,2013,19:430-440.
30 Kan CW,Hahn MA,Gard GB,et al.Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer[J].BMC Cancer,2012,12:627.
31 Carlsen AL,Schetter AJ,Nielsen CT,et al.Circulating microRNA expression profiles associated with systemic lupus erythematosus[J].Arthritis Rheum,2013,65(5):1324-1334.
32 Geekiyanage H,Jicha GA,Nelson PT,et al.lood serum miRNA:non-invasive biomarkers for Alzheimer′s disease[J].Exp Neurol,2012,235(2):491-496.
33 Roderburg C,Luedde M,Vargas Cardenas D,et al.Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis[J].PLoS One,2013,8(1):e54612.
34 Jia SZ,Yang Y,Lang J,et al.Plasma miR-17-5p,miR-20a and miR-22 are down-regulated in women with endometriosis[J].Hum Reprod,2013,28(2):322-330.
35 Zhang WH,Gui JH,Wang CZ,et al.The identification of miR-375 as a potential biomarker in distal gastric adenocarcinoma[J].Oncol Res,2012,20(4):139-147.
36 Rhodes CJ,Wharton J,Boon RA,et al.Reduced microRNA-150 is associated with poor survival in pulmonary arterial hypertension[J].Am J Respir Crit Care Med,2013,187(3):294-302.
37 Han HS,Yun J,Lim SN,et al.Downregulation of cell-free miR-198 as a diagnostic biomarker for lung adenocarcinoma-associated malignant pleural effusion[J].Int J Cancer,2013,133(3):645-652.
38 de Boer HC,van Solingen C,Prins J,et al.Aspirin treatment hampers the use of plasma microRNA-126 as a biomarker for the progression of vascular disease[J].Eur Heart J,2013,[Epub ahead of print].
39 Zhi F,Cao X,Xie X,et al.Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia[J].PLoS One,2013,8(2):e56718.
40 Fayyad-Kazan H,Bitar N,Najar M.et al.Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia[J].J Transl Med,2013,11:31.
41 Nguyen HC,Xie W,Yang M,et al.Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk,localized prostate cancer[J].Prostate,2013,73(4):346-354.
42 Singh B,Ronghe AM,Chatterjee A,et al.MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis[J].Carcinogenesis,2013,34(5):1165-1172.
43 Ozgun A,Karagoz B,Bilgi O,et al.MicroRNA-21 as an indicator of aggressive phenotype in breast cancer[J].Onkologie,2013,36(3):115-118.
44 Goldberger N,Walker RC,Kim CH,et al.Inherited variation in miR-290 expression suppresses breast cancer progression by targeting the metastasis susceptibility gene Arid4b[J].Cancer Res,2013,73(8):2671-2681.
45 Le Quesne J.UTRly malignant:mRNA stability and the invasive phenotype in breast cancer[J].J Pathol,2013,230(2):129-131.
46 Chen Y,Zhang J,Wang H.et al.miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10[J].BMC Cancer,2012,12:111.
47 Lei R,Tang J,Zhuang X,et al.Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis[J].Oncogene,2013,[Epub ahead of print].
48 Plummer PN,Freeman R,Taft RJ,et al.MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells[J].Cancer Res,2013,73(1):341-352.
49 Mulrane L,Madden SF,Brennan DJ,et al.miR-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro[J].Clin Cancer Res,2012,18(24):6702-6713.
50 Kong W,He L,Richards EJ,et al.Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer[J].Oncogene,2013,[Epub ahead of print].
51 Okuda H,Xing F,Pandey PR,et al.miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4[J].Cancer Res,2013,73(4):1434-1444.
52 Li J,Kong X,Zhang J,et al.Correction:miRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer[J].Cancer Cell Int,2013,13(1):7.
53 Korner C,Keklikoglou I,Bender C,et al.MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon)[J].J Biol Chem,2013,288(12):8750-8761.
54 Ryu S,McDonnell K,Choi H,et al.Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration[J].Cancer Cell,2013,23(1):63-76.
55 Cookson VJ,Bentley MA,Hogan BV,et al.Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours[J].Cell Oncol(Dordr),2012,35(4):301-308.
56 Mar-Aguilar F,Mendoza-Ramírez JA,Malagón-Santiago I,et al.Serum circulating microRNA profiling for identification of potential breast cancer biomarkers[J].Dis Markers,2013,34(3):163-169.
57 Heneghan HM,Miller N,Kelly R,et al.Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease[J].Oncologist,2010,15(7):673-682.
58 Sun Y,Wang M,Lin G,et al.Serum microRNA-155 as a potential biomarker to track disease in breast cancer[J].PLoS One,2012,7(10):e47003.
59 Cuk K,Zucknick M,Heil J,et al.Circulating microRNAs in plasma as early detection markers for breast cancer[J].Int J Cancer,2013,132(7):1602-1612.
60 Schwarzenbach H,Milde-Langosch K,Steinbach B,et al.Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients[J].Breast Cancer Res Treat,2012,134(3):933-941.
61 Zeng RC,Zhang W,Yan XQ,et al.Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer[J].Med Oncol,2013,30(1):477.
62 Schrauder MG,Strick R,Schulz-Wendtland R,et al.Circulating micro-RNAs as potential blood-based markers for early stage breast cancer detection[J].PLoS One,2012,7(1):e29770.
63 Si H,Sun X,Chen Y,et al.Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer[J].J Cancer Res Clin Oncol,2013,139(2):223-229.
64 Ng EK,Li R,Shin VY,et al.Circulating microRNAs as specific biomarkers for breast cancer detection[J].PLoS One,2013,8(1):e53141.
65 Guo LJ,Zhang QY.Decreased serum miR-181a is a potential new tool for breast cancer screening[J].Int J Mol Med,2012,30(3):680-686.
66 Wang F,Zheng Z,Guo J,et al.Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor[J].Gynecol Oncol,2010,119(3):586-593.
67 Wang H,Tan G,Dong L,et al.Circulating MiR-125b as a marker predicting chemoresistance in breast cancer[J].PLoS One,2012,7(4):e34210.
68 Razis E,Bobos M,Kotoula V,et al.Evaluation of the association of PIK3CA mutations and PTEN loss with efficacy of trastuzumab therapy in metastatic breast cancer[J].Breast Cancer Res Treat,2011,128(2):447-456.
69 Chen W,Cai F,Zhang B,et al.The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer:potential biomarkers[J].Tumour Biol,2013,34(1):455-462.
70 Zhao FL,Hu GD,Wang XF,et al.Serum overexpression of microRNA-10b in patients with bone metastatic primary breast cancer[J].J Int Med Res,2012,40(3):859-866.
71 Wu X,Somlo G,Yu Y,et al.De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer[J].J Transl Med,2012,10:42.
72 Broyles D,Cissell K,Kumar M,et al.Solution-phase detection of dual microRNA biomarkers in serum[J].Anal Bioanal Chem,2012,402(1):543-550.
73 Lusi EA,Passamano M,Guarascio P,et al.Innovative electrochemical approach for an early detection of microRNAs[J].Anal Chem,2009,81(7):2819-2822.
74 Wang Y,Zheng D,Tan Q,et al.Nanopore-based detection of circulating microRNAs in lung cancer patients[J].Nat Nanotechnol,2011,6(10):668-674.
75 Hu Z,Dong J,Wang LE,et al.Serum microRNA profiling and breast cancer risk:the use of miR-484/191 as endogenous controls[J].Carcinogenesis,2012,33(4):828-834.
76 Mostert B,Sieuwerts AM,Martens JW,et al.Diagnostic applications of cell-free and circulating tumor cell-associated miRNAs in cancer patients[J].Expert Rev Mol Diagn,2011,11(3):259-275.
77 Uehara M,Kinoshita T,Hojo T,et al.Long-term prognostic study of carcinoembryonic antigen(CEA) and carbohydrate antigen 15-3(CA 15-3) in breast cancer[J].Int J Clin Oncol,2008,13(5):447-451.
