Biogeographic and Phylogenetic Relationships of Some Scincella (Squamata: Scincidae) from China and North America Inferred from 12S rRNA Gene Sequences of Mitochondrial DNA
2013-10-28QINGNingLINHungduTONGRanranZHANGXiaoqiLUWenhuaLAZELLJames
QING Ning, LIN Hungdu, TONG Ranran, ZHANG Xiaoqi, LU Wenhua, LAZELL James
(1. Key Laboratory of Ecology and Environment Sciences in Guangdong Higher Education, Guangdong Provincial Key Laboratory for Healthy and Safe Aquaculture, School of Life Science, South China Normal University, Guangzhou 510631, China; 2. Department of Physical Therapy, Shu Zen College of Medicine and Management, Kaohsiung 821, Taiwan; 3. The Conservation Agency, 6 Swinburne Street, Jamestown, RI 02835, USA.)
BiogeographicandPhylogeneticRelationshipsofSomeScincella(Squamata:Scincidae)fromChinaandNorthAmericaInferredfrom12SrRNAGeneSequencesofMitochondrialDNA
QING Ning1*, LIN Hungdu2, TONG Ranran1, ZHANG Xiaoqi1, LU Wenhua3, LAZELL James3
(1. Key Laboratory of Ecology and Environment Sciences in Guangdong Higher Education, Guangdong Provincial Key Laboratory for Healthy and Safe Aquaculture, School of Life Science, South China Normal University, Guangzhou 510631, China; 2. Department of Physical Therapy, Shu Zen College of Medicine and Management, Kaohsiung 821, Taiwan; 3. The Conservation Agency, 6 Swinburne Street, Jamestown, RI 02835, USA.)
Grayian distribution denotes the biological similarities between southeastern North America and East Asia. Using 12S rRNA gene sequences of mitochondrial DNA, we present evidence that the North American ground skinks inScincella(Squamata: Scincidae),Sc.lateralisand relatives, exhibit classic Grayian distribution, emerging genetically from within the Chinese group ofScincellathat includesSc.modestaandSc.tsinlingensis; all derivatives from ChineseSc.reevesii. A superficially similar undescribed species, as yet known only from Dinghushan, Guangdong Province, China, also arises within this group but lacks the lower eyelid “spectacle” usually thought diagnostic of the genusScincella. Our molecular analyses confirm previous work indicating that AsianSphenomorphusis paraphyletic with respect toScincella, confirm that the AmericanScincellainclude “Sphenomorphus”cherriei, necessitating that the spectacle scale has been independently either developed or lost in separate lineages of Scincidae, and provide further evidence for the separation ofKaestleatravancoricafromScincella. The separation time of North AmericanScincellafrom their Chinese congeners dates from the Miocene in Tertiary about 7.3-21.6 million years ago when Beringia was extant and mesic. Our studies contribute further to phylogenetics and biogeography ofScincellafrom North America and China and call for further international collaboration on resolution of taxonomic problems among lygosomine species.
Keywords:Scincella;Sphenomorphus; Grayian distribution; genetic distance; separation time; 12S rRNA gene sequences
Genetic investigation of potentially related plants and animals in southeastern North America and East Asia (mainly China and Japan), including mosses, magnolias, beetles, and alligators, may elucidate the origins and relationships of biodiversity in these two widely disjunct continental regions. This disjunct geographic pattern, often termed “Grayian” distribution, was popularized by botanist Asa Gray in the mid-1800s and has been observed for more than two centuries[1-6,56]. The wealth of examples showing this pattern provides unparalleled opportunities for international collaboration in genetics, molecular biology, paleontology, ecology, and climatology.
A striking example of Grayian distribution is the nominal genusScincella(Squamata: Scincidae) of small, brown, ground skinks.Scincellais largely Asian with >10 species, of which more than half are Chinese[7-10], three Mexican and Central American species[11], and one,Sc.lateralis, found throughout much of the eastern half of the United States down into northern Mexico[12]. Eremchenko and Das[13]separated several Indian species from nominalScincellaas a new genus,Kaestlea, based on the presence of prominent, elongate postoculars (granular or absent inScincellasensostricto). Their work has refined and tightened the generic definition ofScincella.
Boulenger[14]suggested, and later Van Denburgh[15]agreed, that the widespread American speciesSc.lateraliswas conspecific with the Chinese speciesSc.reevesiiandSc.modesta. Schmidt[16]agreed thatSc.lateralisandSc.modestawere conspecific, but retainedSc.reevesiias a valid species. Pope[17]acknowledged close relationships of the Chinese and American species but did not regard them as conspecific. Recently, Honda et al.[18-19]used molecular evidence to confirm the basic Grayian distribution hypothesis forScincellausing largely South Asian species. Using mtDNA, Macey et al.[6]included two Chinese species,Sc.potaniniandSc.tsinlingensis, finding that both were more closely related toSc.lateralisthan to the South AsianSc.rupicolaused by Honda et al.[18-19]. Nevertheless, the hypothesis going back a century-and-a-quarter that AmericanSc.lateralisand ChineseSc.modestaandSc.reevesiiare close relatives has yet to be tested phylogenetically. Furthermore,Sc.modestaandSc.reevesiiare widespread species and their relationships with each other, or others, remain unclear.
While not addressing the relationships ofScincellaandSphenomorphusdirectly, Honda et al.[18-19]found some AmericanSphenomorphuswere nested within AmericanScincella, not related to AsianSphenomorphus, thus renderingSphenomorphuspolyphyletic, and that AsianSphenomorphus, as currently conceived, is paraphyletic. ThusScincellaMittleman (1950) would be a junior synonym ofSphenomorphusFitzinger. However, we follow Nguyen et al.[20]in deferring generic name changes until the entire complex of lygosomine scincid lizards can be reassessed. Most members ofScincellaare semifossorial and have a large transparent scale in the lower eyelid, the “window” or “palpebral disk” of Ouboter[7], or “spectacle,” which allows them to see with their eyes closed[21-22]. Most members ofSphenomorphus, including the Central American “Sp.”cherrieiclosely related toSc.lateralis, lack this scale. Furthermore, a new species (Scincellaindet.) from the Dinghushan Man and Biosphere Reserve in Guangdong Province, South China[23]lacks the spectacle. On the other hand, some AmericanSphenomorphus(assatus,incertus,rarus) have the spectacle scale in the lower eyelid. The Dinghushan “Scincellaindet.” may be related to or a range extension of a newly described species from Northern Vietnam and Hainan Island placed inSphenomorphus[24]based on our morphological comparison.
The use of molecular data to estimate the divergence times of clades has been a topic of intense research interest in recent years. Lazell and Lu[3-4]suggest that many species pairs or sets demonstrating Grayian distribution may have evolved in the Pleistocene, but ancestralSc.lateralismay have dispersed from Asia via the Bering Land Bridge (BLB or Beringia) earlier in the Tertiary[18-19]. In his overview of the family Scincidae, Greer[25]recognized close relationships withinScincellaand suggested the BLB as the dispersal route for the congeners. In his revision of the nominal genusScincella, Ouboter[7]noted this route but also went so far as to say that post-glacial transport by humans was “not improbable.” These differences in hypothesized separation time between Asian and North AmericanScincellarequire further investigation. Our initial hypothesis is that the American representatives of nominalScincellaare derived from the richer Chinese species pool, arriving between the Pleistocene and now.
The specific objectives here were to test whetherSc.lateraliswas derived from Chinese congeners and when that divergence occurred. In addition, we sought to clarify the phylogenetic relationships of the newly discoveredScincellaindet. from Dinghushan. We addressed these three issues using 12S rRNA gene sequences of mitochondrial DNA (mtDNA) fromSc.lateralisfrom Mississippi and Texas,Sc.modesta,Sc.reevesii,Sc.tsinlingensis, andScincellaindet. from South China (Hong Kong’s Lantau Island, Guangdong and Sichuan Provinces). We have limited taxon sampling ofScincellaspecies and a limited amount of data, but provide evidence supporting previous work by Honda et al.[18-19]on theScincellaandSphenomorphusrelationship and by Eremchenko and Das[13]on the distinctiveness of East Indian species in addition to testing our hypotheses. Our results call for more work internationally to gain an unbiased outcome of systematics and evolution of scincid lizards and contribute to the understanding of Grayian distribution.
1 Materials and methods
1.1 Sample collection and DNA extraction
A total of 7 species (29 specimens) were collected from Nan Ao Island, Shantou Prefecture; Dinghushan Man and Biosphere Reserve, Zhaoqing Prefecture; South China Normal University campus, Guangzhou Prefecture (all three are in Guangdong Province); Sichuan University campus, Chengdu Prefecture, Sichuan Province; Lantau Island, Hong Kong Special Administrative Region; Dallas, Dallas County, Texas; and Clinton, Hinds County, Mississippi (Table 1). They were hand-caught or noosed, fixed in 95% ethanol immediately, and replenished with it as necessary. Specimens were deposited at South China Normal University (SCNU), Chengdu Institute of Biology (CIB), and Museum of Comparative Zoology (MCZ), Harvard University. We also incorporated published sequence data of 13 species (13 specimens) from India, Indochina, the Philippines, Borneo, New Guinea, and some Pacific islands available from Genbank (Table 1), for a total of 20 species. A Genbank sample, AY308451, labeled “Scincellareevesii,” clustered withScincellarupicola. We believe it is a misidentification, but we have not examined the specimen and omitted it from our analyses. The mtDNA from liver or muscle tissue was extracted by standard proteinase K digestion followed by phenol/chloroform extraction[26].
Table 1 Localities (for 7 skink species used in DNA sequencing) and geographic areas (for 13 species downloaded from Genbank), with SCNU=South China Normal University campus, and SU=Sichuan University campus
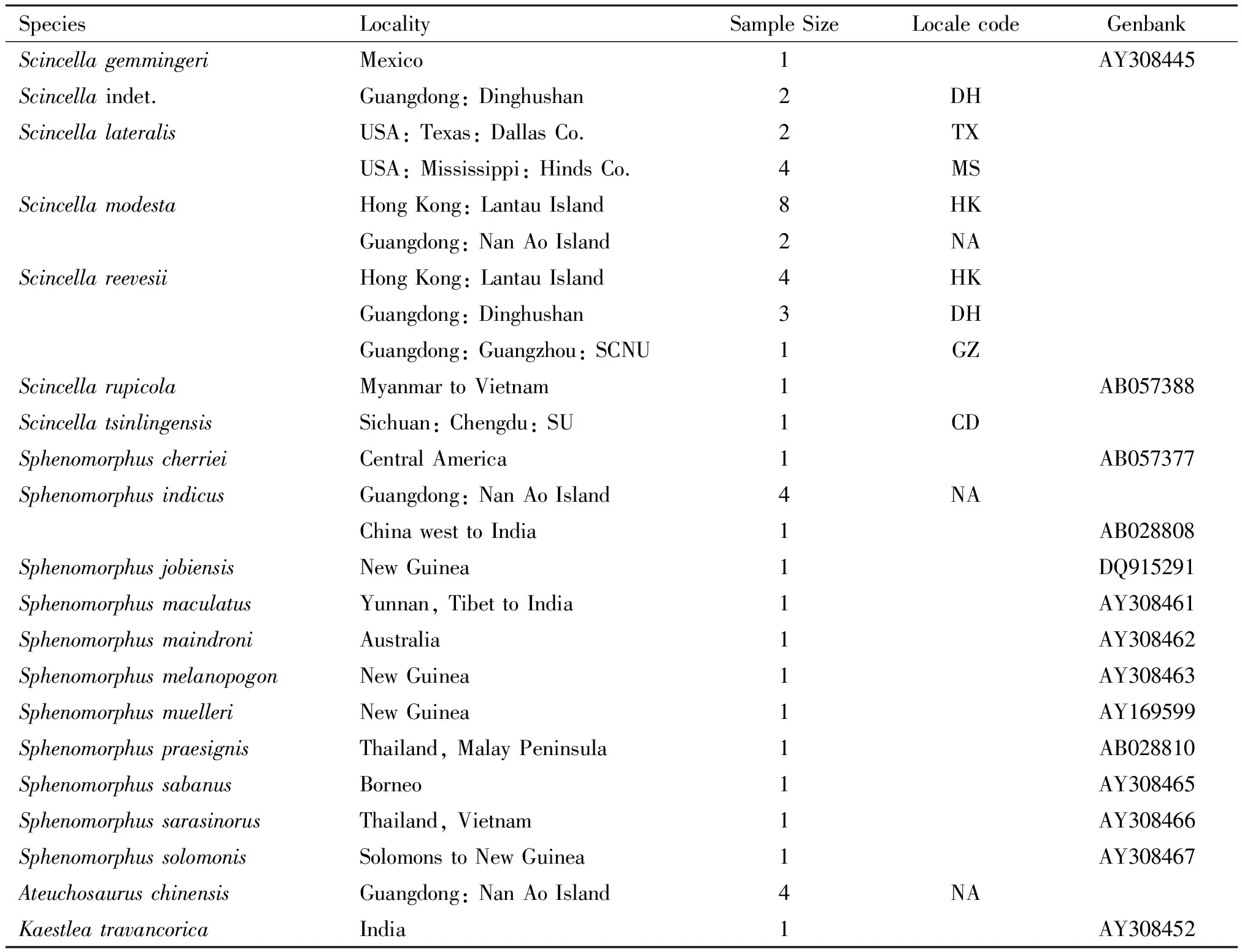
SpeciesLocalitySampleSizeLocalecodeGenbankScincellagemmingeriMexico1AY308445Scincellaindet.Guangdong:Dinghushan2DHScincellalateralisUSA:Texas:DallasCo.2TXUSA:Mississippi:HindsCo.4MSScincellamodestaHongKong:LantauIsland8HKGuangdong:NanAoIsland2NAScincellareevesiiHongKong:LantauIsland4HKGuangdong:Dinghushan3DHGuangdong:Guangzhou:SCNU1GZScincellarupicolaMyanmartoVietnam1AB057388ScincellatsinlingensisSichuan:Chengdu:SU1CDSphenomorphuscherrieiCentralAmerica1AB057377SphenomorphusindicusGuangdong:NanAoIsland4NAChinawesttoIndia1AB028808SphenomorphusjobiensisNewGuinea1DQ915291SphenomorphusmaculatusYunnan,TibettoIndia1AY308461SphenomorphusmaindroniAustralia1AY308462SphenomorphusmelanopogonNewGuinea1AY308463SphenomorphusmuelleriNewGuinea1AY169599SphenomorphuspraesignisThailand,MalayPeninsula1AB028810SphenomorphussabanusBorneo1AY308465SphenomorphussarasinorusThailand,Vietnam1AY308466SphenomorphussolomonisSolomonstoNewGuinea1AY308467AteuchosauruschinensisGuangdong:NanAoIsland4NAKaestleatravancoricaIndia1AY308452
1.2 PCR amplification and sequencing
Total genomic DNA was extracted from muscle tissue with commercial kits in protocols of manufacturers (Sangon Biotech Co., Shanghai). Part of the 12S rRNA gene, approximately 348 base pairs (bp), ranging from 342 to 346 in the seven different species, were amplified by polymerase chain reaction (PCR) with the primers L1091 and H1478[27]. We ran PCR on PTC100 or PTC200 (MJ Research, Waltham, MA, USA) at least once for each specimen in a 50-μL PCR mixture that contained 100 ng template DNA, 5 μL 10×PCR buffer (MgCl220 mmol/L), 5 μL dNTP mix (10 mmol/L), 2 UTaqpolymerase (Promega), and 5 μL of each primer (55 ng/μL), using one cycle of denaturation at 95 ℃ for 4 min, 35 cycles of denaturation at 95 ℃ for 40 sec, annealing at 55 ℃ for 40 sec, and extension at 72 ℃ for 1 min (final extension for 8 min). The PCR products were stored at 4 ℃ for later purification and sequenced by the Yingjun DNA Biotechnologies Company (Guangzhou, Guangdong Province, China) on an automated sequencer (Applied Biosystems ABI 377, Foster City, CA, USA).
1.3 Alignment and phylogenetic analysis
Multiple haplotypic sequence alignments were performed with CLUSTALX 1.81[28]. Ribosomal DNA generally exhibit length variation (gaps in DNA sequences due to insertion or deletion), and therefore pose difficulty for alignment. The problematic sites were compared with closely related species (the same fragments from other scincid species deposited in Genbank) and then corrected manually. The pairwise 2-parameter genetic distance[29]was calculated by software MEGA 4[30]to estimate between-species variation, and the subsequent pairwise percentage of sequence divergence was calculated by hand, based on the pairwise number of mutations/total number of base pairs.
Two different phylogenetic programs were performed to infer relationships: maximum parsimony (MP) and Bayesian inference (BI)[31-33]. The sequence gaps were treated as missing data. For outgroups we choseAteuchosauruschinensis, sympatric with our ChineseScincellaandSphenomorphusand often thought to be in Lygosominae[34]andKaestleatravancorica, formerly placed inScincella. This latter form was recently proven distinct from, but closely related to, and not paraphyletic with, bothScincellaandSphenomorphus[13]; its DNA is available from Genbank. We therefore used bothA.chinensisfor its distant and more primitive relationship with the ingroup andK.travancoricafor its close but non-paraphyletic relationship with the ingroup.
MP analyses were performed with PAUP 4.0b10[35]based on p-distances. We conducted heuristic searches to estimate nodes and branches and assessed statistical support for the robustness of branches of all lineages with nonparametric bootstrap analysis, using tree-bisection-reconnection branch swapping for MP, with 500 random stepwise addition sequence replicates, 10 cladograms in memory in each step. When equally parsimonious trees were found, a strict consensus tree summarized resultant topologies. Tree topologies of phylograms with bootstrap values 70% or greater are regarded as sufficiently resolved[36].
Bayesian analyses were performed with MRBAYES 3.1[37]. The posterior distributions were obtained by the Markov Chain Monte Carlo (MCMC) analyses with one cold chain and three heated chains (4 MCMC chains) from a total of 106MCMC generations; samples of trees and parameters were drawn at every 100 steps of the 106MCMC generations. We selected the HKY85 model in MRBAYES that best fit our data for this analysis. The first 25% of the sampled trees and estimated parameters were discarded as part of the burn-in. From the remaining trees (75%), a majority-rule consensus tree was subsequently produced. Bayesian probabilities based on the posterior distributions by MCMC are considered strongly supportive to split nodes if >0.95.
It was used a Bayesian coalescent analysis in BEAST 1.4.7[38]with the MCMC procedure, to estimate lineage ages of the most recent common ancestor (TMRCA)[39]. Divergence times were calculated at 95% highest posterior density (HPD) intervals on a time-measured phylogeny. The 95% HPD is the shortest interval that contains 95% of all values sampled from the posterior. In all calibrations, it was allowed BEAST to conduct at least two independent MCMC analyses of 10×106to generate the posterior distribution, with the first 106generations discarded as the burn-in, and parameter values sampled every 1,000 generations. All resultant Bayesian parameters can be estimated by TRACER 1.4[40]for convergence to highest values, even with relatively low information content in the sequences and the small age range of the sequences. The posterior estimates of parameters were all distinctly unimodal even with wide 95% HPD; we checked for a highest coalescent value (θ). Because there is no available fossil calibration point for DNA sequences ofScincella, we estimated the divergence time by using a range of neutral mutation rates (μ) in proxy species. Maximum and minimum μ (0.65 and 0.22%, respectively) previously estimated rates were adopted (0.65%: Macey et al., 1998 for lizard; 0.22%: Emerson et al., 2000 for frog)[41-42].
2 Results
2.1 Mitochondrial 12S rRNA variation
Length polymorphism of the 12S rRNA gene sequences occurred among the 7 species, indels ranging from 2 forSc.reevesiito 6 forSc.lateralis. A total of 348 bp of nucleotide sequences were amplified with 146 variable sites (V) in the total dataset (including outgroups), of which 24 were singleton polymorphic sites (S) and 122 were parsim-informpolymorphic sites (Pi). Between-species variation in nucleotide divergence and genetic distance among all eight species ofScincellaincluding “Sphenomorphus”cherrieiwere respectively between 0.071-0.152 and 0.079-0.183 (Table 2); among them, interspecific nucleotide replacements varied from the lowest, 26 bp (0.071:Sc.modestavs.Sc.indet.), to the highest, 45 bp (0.152:Sc.rupicolavs. “Sp.”cherriei). Variations in nucleotide divergence and genetic distance between AmericanSc.lateralisand the five AsianScincellaspanned 0.100-0.123 and 0.113-0.145, respectively; here the divergence ofS.lateralisfromSc.tsinlingensiswas the least and fromSc.reevesiithe greatest. The divergences ofSc.indet. fromSc.modestaorSc.rupicolawere equally low, 0.071-0.080 and 0.079-0.080, respectively.
Consistently, in contrast to between-species variation (Table 2), there was little within-species variation among individuals or populations ofScincella. Within-species variation in genetic distance was all small, 0.001 amongSc.reevesii, 0.002 amongSc.indet., 0.005 amongSc.modesta, and 0.006 amongSc.lateralis. However, within-species variation ofS.modestain nucleotide sequences was relatively high at V sites (1 S and 7 Pi sites) between the Lantau (HK) and Nan Ao (NA) populations, consisting of about 2.2% of the total nucleotides. The greatest within-species variation in nucleotide sequences was inS.lateralis; the two geographically distant populations (Texas and Mississippi) had high V sites (1 S and 7 Pi sites), also consisting of about 2.2% of the total nucleotides. This striking difference has been explained by Jackson and Austin[12]as resulting from rivers as barriers separating glacial maximum refugia in southern North America. The Guangdong and Hong KongSc.reevesiishowed the least within-species variation, with only one V site (1 Pi site) among its three populations.
Table 2 Nucleotide divergence (upper matrix) and genetic distance (lower matrix) of mitochondrial 12s rRNA sequences among North American and AsianScincellaspecies, with Sc.=Scincella, Sp.=Sphenomorphus, andAteuchosaurusandSphenomorphusas Outgroups
Variation betweenScincellaand the two sympatric nominal genera was strikingly different (Table 2). Nucleotide divergence and genetic distance betweenAteuchosauruschinensisandSphenomorphusexcluding “Sp.”cherrieiwere great, 0.155-0.169 and 0.197-0.215, respectively; those betweenAteuchosauruschinensisandScincellawere also great, 0.158-0.195 and 0.197-0.257, respectively. However, nucleotide divergence and genetic distance betweenSphenomorphusindicusandScincella(including “Sp.”cherriei) were much smaller, only between 0.100-0.149 and 0.114-0.179, respectively; these were well within the variation amongScincellaspecies. The smallest variation betweenSp.indicusandScincellawas remarkably between it andSc.indet.; the nucleotide divergence of 0.100 and genetic distance of 0.114 were comparable to the variation amongScincellaspecies, not as great as expected for different genera.
Interestingly, variation betweenScincellaand the allopatricKaestleawas large (Table 2);Scincellawas much more distant genetically from the newly separatedK.travancorica, in the same subfamily, than fromAteuchosauruschinensis, whose subfamilial status remains unclear.
2.2 Phylogenetic analyses
Both the MP and BI analyses presented an identical topology of phylograms (Figure 1, showing only BI values), resulting in a well-supportedScincellagroup (node a) including “Sp.”cherrieiand all otherScincellaspecies. This node emerges fromSphenomorphusand is closest toSp.indicusandSp.maculatus. It divides into two lineages; one contains clade C with onlySc.reevesii. The other lineage contains two clades: A and B. Clade A contains four species,Sc.modesta,Sc.tsinlingensis,Sc.indet., andSc.rupicola; clade B includes all the New World species available to us:Sc.lateralis, “Sp.”cherriei, and Sc.gemmingeri. Monophyly of each of theScincellaspecies for which we have a series of specimens was supported with bootstrap values and Bayesian posterior probabilities of 100%, exceptSc.modesta(89%). Although theScincellataxa fell into clades corresponding to their geographical distributions, the North American clade B separates notably the East Asian clades A and C.
Clade A (4 species) branches into two subclades,S.modestaandS.tsinlingensisin one andSc.indet. andS.rupicolain the other; notably, populations ofSc.modestashared no haplotypes between Lantau (HK) and Nan Ao (NA) Islands (Figure 1). Clade B (3 species) notably contains “Sp.”cherriei, which is taxonomically classified in the genusSphenomorphus, as in Honda et al.[18-19]; however, phylogenetically it clusters with the AmericanScincellaspecies (Figure 1). Clade C has onlySc.reevesii; its populations shared one haplotype among Lantau Island (HK), Dinghushan (DH), and Guangzhou (GZ); another haplotype occurs only on Lantau (Figure 1).
2.3 Estimates of divergence times
We estimated divergence times of the 12S rRNA gene among species at two distinct nodes; the highest value ofθ=45.32 for node a andθ=33.07 for node b; these BI analyses under different evolutionary models estimate consistent mean rates of evolution (μ) for the 12S rRNA gene, given the range from 2.2×10-9[42]to 6.5×10-9[41]substitutions /site/year. Node a represents the origin ofScincellawith a mean age from 10.0 Mya (95% HPD=34.77: 8.8-11.2 Mya) to 29.6 Mya (95% HPD=49.16: 26.2-33.1 Mya), meaning the age of TMRCA of the genusScincellawas in the lower Miocene of Tertiary. Node b represents the separation of the East Asian clade A from the North American clade B at a mean age from 7.3 Mya (95% HPD=25.20: 6.3-8.2 Mya) to 21.6 Mya (95% HPD=36.51: 18.8-24.2 Mya), meaning the time of isolation of the East Asian species from the North American species includingSp.cherrieioccurred only slightly later in the Miocene.
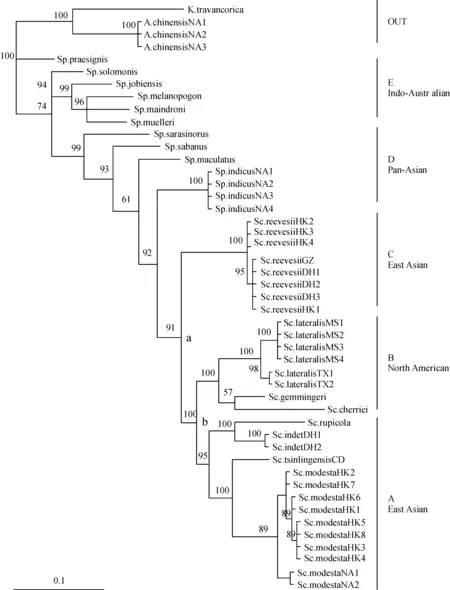
Figure 1 Bayesian phylograms of 20 skink species collected from America and South China (7 species with locale codes and individual identification number) and downloaded from Genbank (13 species without locale codes), based on mitochondrial 12S rRNA sequences, with Bayesian posterior probabilities shown above internode branches, Sc.=Scincella, Sp.=Sphenomorphus, A.=Ateuchosaurus, K.=Kaestlea, and locale codes CD=Chengdu, DH=Dinghushan, GZ=Guangzhou, HK=Hong Kong, MS=Mississippi, NA=Nan Ao, TX=Texas
3 Discussion
3.1SeparationofSc.lateralisanditsAsianrelatives
Our principal hypotheses, the Chinese origin and Grayian distribution of AmericanScincella, are strongly supported. The phylograms not only show monophyly for theScincellaspecies tested, but also support an American lineage and two Asian lineages, withSc.reevesiiancestral to both. The North American species ofScincellaare a monophyletic lineage nested within Asian taxa, indicating a single dispersal event.
The Beringian land bridge (BLB) and the putative North Atlantic Land Bridge (NALB) have been postulated as routes of floristic interchange between Eurasia and North America in the Tertiary that have contributed to modern global floral and faunal disjunctions[43]. The BLB was a determining factor in the structure and biogeography of terrestrial faunas across the Nearctic and Neotropical regions during the Pliocene and Quaternary[44]. Because there are no relevant species in Europe, we reject the NALB and postulate the BLB route for ancestralSc.lateralis. Several North American lineages may have entered this region from Asia during the Miocene; particularly important was a mid-Miocene connection of continuous temperate deciduous forest providing habitats for amphibians and reptiles otherwise excluded from high latitudes[6]. Our molecular clock estimate, 7.3-21.6 Mya, agrees with this mid-Miocene timing. The BLB existed through most of the Miocene, being first interrupted around 4.8-7.4 Mya but still existing intermittently until 11 000 years ago[45-46]. Other disjunct taxa should be tested genetically and compared to climatic and sea level data to verify separation times.
3.2 Systematic position of Scincella indet
Recognition ofScincellarenders AsianSphenomorphusparaphyletic. Although Macey et al.[6]considered only one species ofSphenomorphus,Sp.indicus, their phylogram suggests the generic paraphyly as well. Our results show that the morphological basis for generic separation ofScincellaandSphenomorphusis compromised by not only the ChineseSc.indet., but also the Central American “Sp.”cherriei, both of which lack the spectacle scale. The presence of a spectacle scale has previously resulted in the placement of the Indian speciestravancoricainScincella. It is now placed in the genusKaestleaby Eremchenko and Das[13]based on the prominent elongate postoculars that are either granular or absent inScincellaandSphenomorphus. Our only Genbank sample ofKaestleatravancoricaemerged nearAteuchosaurus. Since the spectacle scale is presumably a derived character, the obvious implication is that this spectacle has been independently either developed or lost in separate lineages of Scincidae. Close relationship ofSc.indet. withSc.rupicolaconfirmed by our mtDNA assessment adds to the difficulties of generic definition ofScincellaandSphenomorphusbut reinforces our view that this character cannot be definitive for the genusScincella. Ouboter[7]notes a specimen ofSc.reevesiiwith a spectacle on one side but a pair of scales on the other. The problems involved in framing definitions in this complex of small brown skinks are exemplified in Shea and Greer[47], where species were reshuffled among different genera. Addressing the taxonomy requires consideration of many characters and lies far beyond our essentially biogeographical purview.
Scincellaindet. andSc.rupicolaof Thailand are so similar that we would have suspected them conspecific.Sc.indet. is known from at least five specimens collected from 1984 to 2006[23,48-51]. Except for missing a spectacle scale, it appears to be aScincella. Allen Greer, then of Australian Museum at Sydney, prophetically opined that it was similar toSc.rupicolain most respects, despite the generic character discrepancy (pers. comm. to JL, 1998). The history of its recognition and other details are provided by Li et al.[23]. AlthoughSc.indet. andSc.rupicolaare closely related, as revealed by our molecular analysis, the two species do not share haplotypes of the 12S rRNA gene (Figure 1), and the variation between them (genetic distance=0.089) is greater than the expected variation among populations within a species (largest=0.006 withinSc.lateralis). All this makes it doubtful that Sc.indet. is just a new distribution record or range extension ofSc.rupicola. However, its relationship with the new species of Nguyen et al. (2011)[24]from Vietnam and Hainan Island needs evaluation.
The relationship ofSc.indet. withSc.modestais interesting.Sc.indet. is the closest relative ofSc.modestaon mainland Guangdong (Figure 1).Sc.modestaranges from east of the Guangdong border into Fujian Province on the mainland[8], but the Guangdong provincial records are only for islands such as Nan Ao and Hong Kong’s Lantau; these two populations do not share haplotypes of the 12S rRNA gene (Figure 1), suggesting relict populations and possible replacement on most of the Guangdong mainland by the abundantSc.reevesi. Comparisons between the island and mainland populations ofSc.modestamay shed light on their biogeography.Sc.modestaandSc.tsinlingensisare the most recent species, deriving from the same common ancestor. Based on our data,Sc.modestadivides into two populations: Nan Ao and Lantau, but our sample size for Nan Ao is very small. We did, in addition, note a striking color difference in the field: Nan AoSc.modestahad brilliant red undersides of their tails, as opposed to the dull rosaceous tints of Lantau individuals.
3.3RelationshipsofAteuchosauruswithSphenomorphusandScincella
Ateuchosaurusis a genus of skinks containing only two Asian species:A.chinensisin North Vietnam (most recently rediscovered by Truong et al., 2008)[52]and South China, andA.pellopleurusin the Ryukyus. Karyotypic study of both species[53]suggests that the genus is closer to theEugongylusgroup of Greer[54]than to theSphenomorphusgroup. Austin and Arnold[55]moved it from Lygosominae to Acontinae based on molecular evidence. Our evidence, surprisingly, indicates thatAteuchosaurusis closer toSphenomorphusandScincellathan is the recently described genusKaestlea, whose species were formerly placed inScincellaof Lygosominae[13]. The subfamilial status of the two genera,AteuchosaurusandKaestlea, needs further examination.
4 Conclusions
North AmericanSc.lateralisand its relatives are close to theSc.modestagroup and derived from ancestral ChineseSc.reevesiiin a single trans-Beringian dispersal event, a typical demonstration of Grayian distribution. The timing of separation ofSc.lateralisfrom its Chinese congeners is calculated as about 7.3 to 21.6 Mya in the Miocene of Tertiary when the Beringian land bridge existed. Our DNA data and absence of the spectacle scale in both ChineseScincellaindet. and Central American “Sphenomorphus”cherrieiprove that this character has arisen or been lost independently and cannot be used for generic definition ofScincella.Sc.indet. is both morphologically and molecularly a new species different fromSc.rupicola, but phylogenetically closer toSc.rupicolathan to any other Indochinese species tested.
AcknowledgmentsWe are indebted to Enge K, Hou M, Huang S H, Kolby J, Krysko K, Mann T, Moore J, Su Z P, Watkins-Colwell G, Willard T, and Xiong J L for specimens. Brandley M provided critical insight.
[1] BOUFFORD D E, SPONGBERG S A. Eastern Asian-Eastern North American phytogeographical relationships-A history from the time of Linnaeus to the twentieth century[J]. Ann Missouri Bot Gard, 1983, 70: 423-439.
[2] QIAN H, RIKLEFFS R E. Large-scale processes and the Asian bias in species diversity of temperate plants[J]. Nature, 2000, 407: 180-182.
[3] LAZELL J, LU W H. Grayian distribution and the herpetofaunas of East Asia and eastern North America[C]∥Fourth Asian Herpetological Conference. Chengdu, Sichuan, China, 2000: 104.
[4] LAZELL J, LU W H. Grayian distributions: The China-America biogeographic connection[C]∥ PANG X F. Proceedings of the Biodiversity of Guangdong Nanling National Nature Reserve. Guangzhou: Guangdong Science and Technology Press, 2003: 65-88.
[5] LAZELL J, LU W H. Grayian trans-Beringian distributions: new twists to the old tale[C]∥Proceedings of the XIX International Congress of Zoology. Beijing, China, 2004: 16-18.
[6] MACEY J R, SCHULTE J A, STRASBURG J L,et al. Assembly of the eastern North American herpetofauna: New evidence from lizards and frogs[J]. Biol Letters, 2006, 2(3): 388-392.
[7] OUBOTER P E. A revision of the genus Scincella (Reptilia: Sauria: Scincidae) of Asia, with some notes on its evolution[J]. Zool Verh, 1986, 229: 1-66.
[8] ZHAO E M, ADLER K. Herpetology of China[M]. Oxford, OH, USA: Society for the study of Amphibians and Reptiles, 1993.
[9] ZHAO E M, ZHAO K T, ZHOU K Y. Fauna Sinica, Reptilia 2, Squamata, Lacertilia[M]. Beijing: Science Press, 1999.
[10] EREMCHENKO V K. Generic and specific redefinition and redescription of the North-Vietnam skink Scincella melanosticta (Boulenger, 1887)[J]. Izvestiya Vuzov, Bishkek, 2003: 1-2, 20-28.
[11] GARCIA-VAZQUEZ U O, CANSECO-MARQUEZ L, NIETO-MONTES DE OCA A. A new species of Scincella (Squamata: Scincidae) from the Cuatro Cienegas Basin, Coahuila, Mexico[J]. Copeia, 2010, 3: 373-381.
[12] JACKSON N D, AUSTIN C C. The combined effects of rivers and refugia generate extreme cryptic fragmentation within the common ground skink (Scincella lateralis)[J]. Evolution, 2010, 64: 409-428.
[13] EREMCHENKO V K, DAS I. Kaestlea: A new genus of scincid lizards (Scincidae: Lygosominae) from the Western Ghats, south-western India[J]. Hamadryad, 2004, 28: 43-50.
[14] BOULENGER A. Lacertidae, Gerrhosauridae, Scincidae, Anelytropsidae, Dibamidae, Chamaeleontidae[J]. Catalogue of the Lizards in the British Museum (Nat. Hist.),1887, 3: 1-575.
[15] VAN DENBURGH J. Concerning certain species of reptiles and amphibians from China, Japan, the Loo Choo Islands, and Formosa[J]. Proceedings of the California Academy of Sciences, 1912, 4: 187-258.
[16] SCHMIDT K P. The reptiles of Hainan[J]. Bulletin of the American Museum of Natural History, 1927,54: 467-551.
[17] POPE C H. The Reptiles of China[M]. American Museum of Natural History, New York, NY, USA. 1935.
[18] HONDA M, OTA H, KOHLER G, et al.Phylogeny of the lizard subfamily Lygosominae (Reptilia: Scincidae), with special reference to the origin of the New World taxa[J]. Genes and Genetic Systems, 2003, 78(1): 71-80.
[19] HONDA M, OTA H, MURPHY R W, et al. Phylogeny and biogeography of water skinks of the genus Tropidophorus (Reptilia: Scincidae): A molecular approach[J]. Zoologica Scripta, 2006, 35(1): 85-95.
[20] NGUYEN Q T, NGUYEN V S, BOHME W, et al. A new species of Scincella (Squamata: Scincidae) from Vietnam[J]. Folia Zool, 2010, 59(2): 115-121.
[21] PALMER W M, BRASWELL A L, KUHLER R. Reptiles of North Carolina[M]. Chapel Hill, NC, USA: University of North Carolina Press, 1995.
[22] BEANE J. Love Skinks[J]. Wildlife in North Carolina, 2006, 70: 14-19.
[23] LI Z C, XIAO Z, QING N, et al. Amphibians and reptiles of Dinghushan in Guangdong Province, China’s oldest nature reserve[J]. IRCF Reptiles & Amphibia, 2009, 16: 130-151.
[24] NGUYEN T Q, SCHMITZ A, NGUYEN T T, et al. Review of the genus Sphenomorphus Fitzinger, 1843 (Squamata: Sauria: Scincidae) in Vietnam, with description of a new species from northern Vietnam and southern China and the first record of Sphenomorphus mimicus Taylor, 1962 from Vietnam[J]. J Herpetol, 2011, 45(2): 145-154.
[25] GREER A E. The generic relationships of the scincid lizard genus Leiolopisma and its relatives[J]. Australian Journal of Zoology Supplemental Series, 1974, 22(31): 1-67.
[26] SAMBROOK J, FRITSCH E F, MANIATIS T. Molecular Cloning[M]. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press, 1989.
[27] KOCHER T D, THOMAS W K, MEYER A, et al. Dynamics of mitochondrial DNA evolution in mammals: Amplification and sequencing with conserved primers[J]. PNAS, 1989, 86(16): 6169-6200.
[28] THOMPSON J D, GIBSON T J, PLEWNIAKl F, et al. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucleic Acid Research, 1997,25(24): 4876-4882.
[29] KIMURA M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences[J]. Journal of Molecular Evolution, 1980, 16(2): 111-120.
[30] TAMURA K, DUDLEY J, NEI M, et al. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0[J]. Molecular Biology and Evolution, 2007, 24(8): 1596-1599.
[31] MAU B. Bayesian phylogenetic inference via Markov chain Monte Carlo methods[D]. USA: University of Wisconsin, Madison, 1996.
[32] RANNALA B, YANG Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference[J]. Journal of Molecular Evolution, 1996, 43(3): 304-311.
[33] LARGET B, SIMON D. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees[J]. Molecular Biology and Evolution, 1999, 16: 750-759.
[34] GREER A E. A subfamilial classification of scincid lizards[J]. Bulletin of the Museum of Comparative Zoology, 1970, 139: 151-183.
[35] SWOFFORD D L. PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods)[EB/OL]. Sinauer Associates, Sunderland, MA, USA.(2002)[2013-8-31]. http:∥paup.csit.fsu.edu.
[36] HILLIS D M, BULL J J. An empirical test of bootstrapping as a method assessing confidence in phylogenetic analysis[J]. Systematic Biology, 1993, 42(2): 182-192.
[37] RONQUIST F, HUELSENBECK J P. MRBAYES 3: Bayesian phylogenetic inference under mixed models[J]. Bioinformatics, 2003, 19(12): 1572-1574.
[38] DRUMMOND A J, RAMBAUT A. BEAST: Bayesian evolutionary analysis by sampling trees[J]. BMC Evolutionary Biology, 2007, 7: 214.
[39] RUTCHMAN F. Molecular dating of phylogenetic trees: a brief review of current methods that estimate divergence times[J]. Diversity and Distributions, 2006, 12(1): 35-48.
[40] RAMBAUT A, DRUMMOND A J. TRACER 1.4.[CP/OL].(2007)[2013-8-31]. http:∥beast.bio.ed.ac.uk/tracer
[41] MACEY J R, SCHULTE II J A, ANANJEVA N B, et al. Phylogenetic relationships among agamid lizards of the Laudakia caucasia species group: Testing hypotheses of biogeographic fragmentation and an area cladogram for the Iranian Plateau[J]. Molecular Phylogenetics and Evolution, 1998, 10(1): 118-131.
[42] EMERSON S B, INGER R F, ISKANDAR D T. Molecular systematics and biogeography of the fanged frogs of Southeast Asia[J]. Molecular Phylogenetics and Evolution, 2000, 16(1): 131-142.
[43] EUGENIA Y Y L, STEFANOVIC S, CHRISTENSEN K, et al. Evidence for genetic association between East Asian and western North American Crataegus L. (Rosaceae) and rapid divergence of the eastern North American lineages based on multiple DNA sequences[J]. Molecular Phylogenetics and Evolution, 2009, 51(2): 157-168.
[44] COOK J A, HOBERG E P, KOEHLER A, et al. Beringia: Intercontinental exchange and diversification of high latitude mammals and their parasites during the Pliocene and Quaternary[J]. Mammal Science, 2005, 30(sp1): S33-S44.
[45] MARINCOVICH L J, GLADENKOV A Y. Evidence for an early opening of the Bering strait[J]. Nature, 1999, 397: 149-151.
[46] SHER A. Traffic lights at the Beringian crossroads[J]. Nature, 1999, 397: 103-104.
[47] SHEA G M, GREER A E. From Sphenomorphus to Lipinia: generic reassignment of two poorly known New Guinea skinks[J]. Journal of Herpetology, 2002, 36(2): 148-156.
[48] LAZELL J, LIAO W P. Contribution to the herpetofauna of Dinghushan, Guangdong[J]. Acta Herpetologica Sinica, 1986, 5: 70-71.
[49] LAZALL J. Herpetology in South China[J]. Herpetological Review, 1988, 19: 49-51.
[50] LAU M. Amphibians and reptiles, Dinghushan 3[J]. Porcupine(University of Hong Kong), 1996, 14: 19.
[51] LAU M. Cebaling herpetofauna[J]. Porcupine(University of Hong Kong), 1996, 15: 28.
[52] TRUONG N Q, TUNG T T, NGOC H V, et al. Rediscovery and redescription of Ateuchosaurus chinensis Gray, 1845 (Squamata: Sauria: Scincidae) from northeastern Vietnam[J]. Herpetology Notes, 2008, 1: 17-21.
[53] OTA H, LINi J T, BOGADEK A, et al. Karyotype of the Lygosomine genus Ateuchosaurus from East Asia[J]. Journal of Herpetology, 1997, 31(4): 604-607.
[54] GREER A E. A phylogenetic subdivision of Australian skinks[J]. Records of the Australian Museum, 1979, 32: 339-371.
[55] AUSTIN J J, AMOLD E N. Using ancient and recent DNA to explore relationships of extinct and endangered Leiolopisma skinks (Reptilia: Scincidae) in the Mascarene Islands[J]. Molecular Phylogenetics and Evolution, 2006, 39(2): 503-511.
[56] NALEPA C A, LI L, LU W H, et al. Rediscovery of the wood-eating cockroach Cryptocercus primarius (Dictyoptera: Cryptocercidae) in China, with notes on ecology and distribution[J]. Acta Zootaxonomica Sinica, 2001, 26: 184-190.
2013-06-30
广东省自然科学基金项目(06025054);美国Falconwood Foundation资助项目
1000-5463(2013)06-0129-11
Q349
A
10.6054/j.jscnun.2013.09.017
基于线粒体DNA的12S rRNA基因序列推断中国及北美部分滑蜥属物种的生物地理和亲缘关系
庆 宁1*, 林弘都2, 仝冉冉1, 张小奇1, 卢文华3, LAZELL James3
(1.华南师范大学生命科学学院,广东省水产健康安全养殖重点实验室,广东省高等学校生态与环境科学重点实验室,广东广州 510631; 2. 树人医护管理专科学校, 台湾高雄821; 3.美国罗德岛州生物保护所,美国詹姆斯镇)
许多东亚和北美的生物类群之间呈现较近的亲缘关系,格雷分布即指这种生物洲际间断分布的现象.本文通过线粒体DNA (mtDNA) 12S rRNA基因序列分析,证明北美的滑蜥属Scincella(Squamata: Scincidae)物种Sc.lateralis与中国的滑蜥属南滑蜥种组呈现典型的格雷分布. 南滑蜥种组包括宁波滑蜥Sc.modesta,秦岭滑蜥Sc.tsinlingensis,南滑蜥Sc.reevesii,和一个采自广东鼎湖山自然保护区的未命名种Scincellaindet..这个鼎湖山未命名种并不具备滑蜥属所特有的鉴别性特征“睑窗”,但显示它与Sc.modesta,Sc.tsinlingensis都是由Sc.reevesii演化而来的近缘种.证实了之前认为亚洲的蜓蜥属Sphenomorphus与滑蜥属Scincella是复系的观点,目前美洲物种Sp.cherriei应隶属于滑蜥属Scincella,具睑窗或不具睑窗的物种分别出现在系统树的不同分支中.研究结果支持前人将Kaestleatravancorica从滑蜥属中划分出去的观点.根据分子钟估算,北美的滑蜥从它们的中国祖先分化出来的时间可追溯到第三纪中新世,约7.3-21.6百万年前,当时白令陆桥还露出海平面而且气候湿润. 该研究为洲际分布的物种的系统发育和生物地理研究提供了理论依据.
Scincella;Sphenomorphus; 格雷分布; 遗传距离; 分化时间; 12S rRNA基因序列
*通讯作者:庆宁,教授,Email:qingn@scnu.edu.cn.
【中文责编:成文 英文责编:李海航】
