Association of RELN promoter SNPs with schizophrenia in the Chinese population
2011-12-25CHANGHuaLIMingLUOXiongJianLIUXingYanYINLiDeYANGShunYingDIAOHongBoSUBingPUXingFu
CHANG Lü-Hua, LI Ming, LUO Xiong-Jian, LIU Xing-Yan, YIN Li-De, YANG Shun-Ying, DIAO Hong-Bo, SU Bing, PU Xing-Fu,*
(1. The First Affiliated Hospital of Kunming Medical College, Kunming 650032, China; 2. The State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology and Kunming Primate Research Center, the Chinese Academy of Sciences, Kunming 650223, China; 3. Graduate School of the Chinese Academy of Sciences, Beijing 100101, China; 4. The Second People's Hospital of Yuxi City, Yuxi 653101, China; 5. Yunnan Mental Health Hospital, Kunming 650224, China)
Association ofRELNpromoter SNPs with schizophrenia in the Chinese population
CHANG Lü-Hua1, LI Ming2,3, LUO Xiong-Jian2,3, LIU Xing-Yan4, YIN Li-De4, YANG Shun-Ying4, DIAO Hong-Bo5, SU Bing2,*, PU Xing-Fu4,*
(1. The First Affiliated Hospital of Kunming Medical College, Kunming 650032, China; 2. The State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology and Kunming Primate Research Center, the Chinese Academy of Sciences, Kunming 650223, China; 3. Graduate School of the Chinese Academy of Sciences, Beijing 100101, China; 4. The Second People's Hospital of Yuxi City, Yuxi 653101, China; 5. Yunnan Mental Health Hospital, Kunming 650224, China)
Previous research on gene expression analysis and association tests have suggested thatRELNis a risk gene for schizophrenia in world populations. Based on the reported down-regulation of RELN in schizophrenia patients compared with normal subjects, we speculated that variants in theRELNpromoter region may confer risk for schizophrenia. In this study, we investigated the associations of three SNPs in the promoter region ofRELNwith schizophrenia in a case-control sample from southwestern China (940 cases and 1 369 controls). The results suggested that none of the SNPs showed significant associations in our sample, indicating the risk variants for schizophrenia inRELNmay not be located in the promoter region. We also performed meta-analysis by combining our data with previously reported data on the Chinese population with a total sample size of 2 843 individuals, and the result remained nonsignificant. Collectively, our results suggested variants in theRELNpromoter may not harbor risk SNPs associated with schizophrenia in the Chinese population.
RELN; Schizophrenia; Promoter; SNP; Chinese population
Schizophrenia is a severe psychiatric disorder with high heritability. Many susceptibility genes have been proposed by linkage analyses, candidate gene studies, and genome wide association (GWA) analyses, including DISC1, NRG1, TCF4, NRGN, and ZNF804A (Thomson et al, 2007; Williams et al, 2004; O'Donovan et al, 2008). However, many cannot be successfully replicated among different populations. A recent GWA study on Ashkenazi Jews identified that a common variant (rs7341475) ofRELNwas significantly associated withschizophrenia in females (Shifman et al, 2008). This association was further supported by replications in a UK study and another Ashkenazi Jew study (Shifman et al, 2008; Liu et al, 2010), but was unable to be replicated in USA and Chinese populations (Shifman et al, 2008; Need et al, 2009), indicating potential genetic heterogeneity inRELNfor schizophrenia between different world populations.
We previously identified severalRELNSNPs and haplotypes associated with schizophrenia in the Chinese population (Li et al, 2011), which is consistent with reported associations in European populations and suggestsRELNis likely a common risk gene for schizophrenia in populations worldwide, though the risk variants differ between reported associations (Kahler et al, 2008; Wedenoja et al, 2008, 2009).
Down-regulation ofRELNmRNA and protein in the brain of schizophrenia patients has been reported previously, but the underlying mechanism remains unknown (Impagnatiello et al, 1998). Hypermethylation ofRELNpromoter in schizophrenic patients is a likely cause; however, negative findings have also been reported (Grayson et al, 2005; Tochigi et al, 2008). One reasonable explanation is that mutations in theRELNpromoter may affect gene expression and function and influence neurodevelopment, leading to schizophrenia susceptibility. Interestingly, short tandem repeats in the promoter region influenceRELNpromoter activity, but the association tests in schizophrenia show negative results (Akahane et al, 2002). Additionally, an investigatedRELNpromoter SNP has shown marginal significance with schizophrenia in a small case-control Chinese sample (P=0.08) (Chen et al, 2002), suggesting theRELNpromoter may harbor risk variants for schizophrenia. In this study, we examined the association between the RELN promoter SNPs and schizophrenia in a Chinese case-control sample.
1 Methods and Materials
1.1 Samples
We recruited 940 unrelated schizophrenia patients (481 females and 459 males, mean age=37.4 a,SD=9.1) and 1 369 unrelated normal controls (732 females and 637 males, mean age=36.7 a,SD=6.8) from southwestern China. The patients were all from Yunnan Mental Health Hospital and The Second People's Hospital of Yuxi City and were diagnosed with having schizophrenia according to DSM-IV and ICD-10 criterion. Patients who had history of alcoholism, substance induced psychotic disorders, epilepsy, neurological diseases, or other symptomatic psychoses were excluded from this study. Control subjects were recruited from the local general populations. All individuals were asked to provide detailed information about medical and family psychiatric histories, and those who had a history of mental disorders, drug abuse, or alcohol dependence were excluded. All patient and control subjects were of Han Chinese origin from the Yunnan province of southwestern China. All individuals were provided with written informed consents for participation, and the research protocol was approved by the internal review board of the Kunming Institute of Zoology, Chinese Academy of Sciences.
1.2 SNP selection
As no SNPs were shown in the -2 kb region upstream from the transcription start site (TSS) ofRELNin Han Chinese from Beijing (CHB) data obtained from HapMap database, we used bi-directional sequencing to search for potential SNPs in the promoter region ofRELNon 100 schizophrenic and 100 healthy randomly selected subjects. Primers used for PCR amplification of the RELN promoter were 5′-AGCCCAGAAGCAATGAA TAAC-3′ (forward) and 5′-TCCCAACTTGTGACTCCATT C-3′(reverse). The PCR program started with an initial incubation at 95oC for 5 min, followed by 40 cycles of 95oC for 30 s, 60oC for 40 sec and 72oC for 1 min, and then held at 72oC for 10 min. Two SNPs were identified (g.-847G>A and g.-888G>C), with g.-888G>C having been investigated previously (Chen et al, 2002). We further carried out case-control association analysis on these SNPs (plus anotherRELNpromoter SNP rs6951875, shown in HapMap data) in our samples.
1.3 SNP genotyping
Venous blood was collected from all participants, and genomic DNA was extracted from the blood sample using the phenol-chloroform method. The DNA samples of the cases and controls were randomly distributed in the case-control DNA plates.
All selected SNPs were genotyped by SNaPShot as described in our previous study (Luo et al. 2008). In brief, genomic fragments which contained selected SNPs were amplified by PCR with a total volume of 25 μL (including 10 ng of genomic DNA) in 96-well plates. Amplified fragments were purified and specific genotyping primers were used to amplify the target site. After one base extension, the reaction was terminated and the products were loaded on an ABI 3130 automatic sequencer (Applied Biosystems). Primer sequences for SnaPShot analysis were 5′-TTTTTATGAGGTATTCTGA CACTGGATGAAGAATAATTAT-3′(rs6951875), 5′-TTTT TTTTTTTTTTTGCGGGAGGGACAGGGGGCCTGGGT GGGAAGGGAGC-3′ (g.-847G>A) and 5′-TTTTTTTTTT TTTTTTTTTTATGAGGCTCTGTCGCTGCCGCGAGGG GCCGGGCGG-3′(g.-888G>C). The SNP genotype callings were automatically performed using ABI GeneMapper 4.0 and verified manually. To ensure accuracy of genotyping, we used bi-directional sequencing on 100 randomly selected individuals. No genotyping errors were found.
1.4 Statistical analysis
The Haploview program was applied to test the genotypic distribution of SNPs for Hardy-Weinberg equilibrium (HWE) between paired SNPs, and to define haplotype blocks (Barrett et al, 2005). Allelic and genotypic associations were accessed with PLINK (Purcell et al, 2007). To detect significant differences in association in female or male samples separately, we conducted statistical analysis with sex as a covariate using PLINK (Purcell et al, 2007). The 95% confidence intervals (CI) of odds ratio were calculated with the online tool (http://faculty.vassar.edu/lowry/odds2x2. html). Haplotype frequency estimation and association tests were performed using PLINK, and only those haplotypes with a frequency of >0.01 in cases and control subjects were considered (Purcell et al, 2007). Power analysis was performed using G*power program (Erdfelder et al, 1996). For meta-analysis, we used the Mantel-Haenszel method with a fixed-effects model. Analysis was conducted by RevMan manager (The Cochrane Collaboration, 2002).
2 Results
Due to genotyping failure of some samples, analyses were based on 2 230 samples for SNP g.-847G>A (861 cases and 1 369 controls), 2 307 samples for SNP g.-888G>C (940 cases and 1 367 controls), and 2 296 samples for SNP rs6951875 (940 cases and 1 356 controls). The overall genotype calling rate was 98.6%.
Genotype distributions of the three SNPs in both cases and controls were in HWE (P>0.05). The LD map of the tested SNPs in cases and control samples are shown in Fig. 1.

Fig. 1 The LD map of the tested SNPs in case and control samples
None of the tested SNPs were significantly associated with schizophrenia (Tab.1 and Tab.2). We also investigated the association of these SNPs with schizophrenia in females and males separately, and nosignificant results were observed (Tab. 1 and Tab. 2). Finally, we performed a meta-analysis by combining a previous study with a total sample size of 2 843 (Chen et al, 2002), but still failed to find a significant association between g.-888G>C and schizophrenia (OR=0.86, 95%CI=0.71–1.04,P=0.12) (Tab. 3).

Tab. 1 Allele frequencies and single SNP association (P-value) analysis

Tab. 2 Genotype counts and p-values of the tested SNPs
We further performed a haplotype-based analysis of the three SNPs with schizophrenia in all samples, female samples and male samples. No haplotypes were significantly associated with schizophrenia (Tab. 4). Notably, the globalp-value of haplotype analyses in the female samples reached significance (P=0.007), which is likely due to a false positive effect caused by the low frequency haplotypes.

Tab. 3 Meta-analysis of association between g.-888G>C and schizophrenia
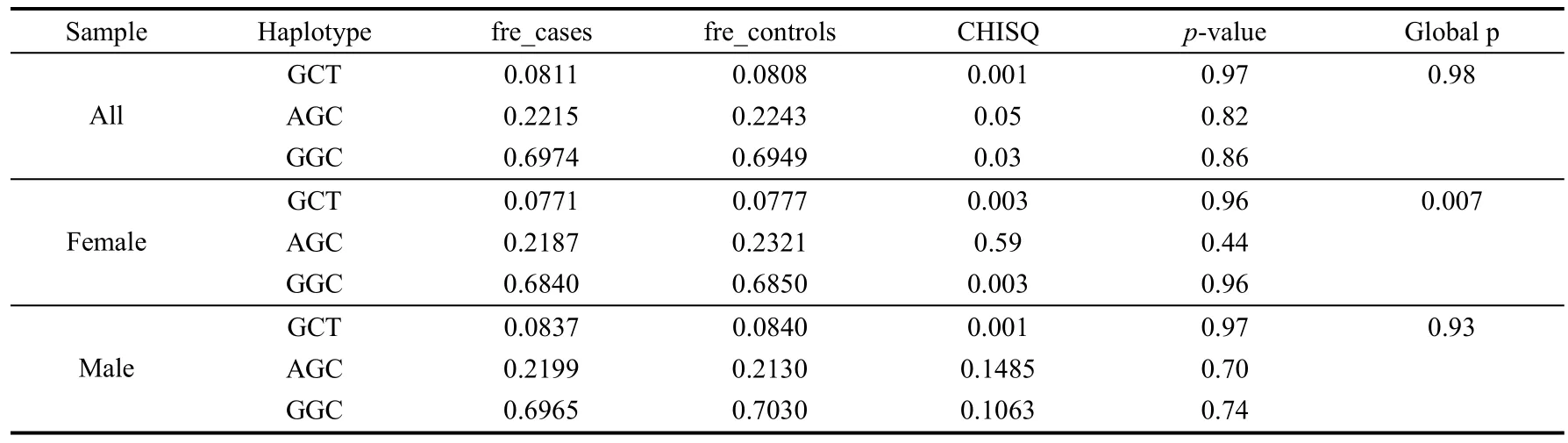
Tab. 4 Analysis of the 3 SNPs for haplotypic association with schizophrenia
We performed a power calculation using the G*power program. The present sample size revealed >99% power to detect a significant association (α<0.05) with an effect size index of 0.1 (corresponding to a ‘weak’ gene effect).
3 Discussion
It has been reported that SNP g.-888G>C shows an association tendency with schizophrenia (P=0.08) (Chen et al, 2002). Our current study, however, showed no significant associations, suggesting SNPs in the promoter region ofRELNconfer no risk for schizophrenia or that there is a potential genetic divergence among regional Han Chinese populations.
In our previous study, we observed multiple SNPs withinRELNsignificantly associated with schizophrenia in Han Chinese. These SNPs were all located in the intron region and not causal SNPs (Li et al, 2011), however, suggesting the possibility of finding risk SNPs in the promoter region which had not been systematically screened before. We failed to find significant association betweenRELNpromoter SNPs and schizophrenia, implying that the causal SNP was not located in the promoter region. However, there are several possibilities that could explain the negative results in this study. Firstly, since we only sequenced 2 kb of theRELNpromoter region, we could not detect other potential genetic variants risks not located in the -2 kb promoter region ofRELN. Secondly, though our sample size was relatively large, it was smaller than the GWA study sample sizes and was unlikely to identify the risk variant with weak effect. Taken together, our findings indicate that the SNPs in the core promoter region (-2 kb upstream of TSS) ofRELNwere not likely associated with schizophrenia in the Chinese population. Further studies should focus on other regions, and a replication study with large sample size is needed.
Akahane A, Kunugi H, Tanaka H, Nanko S. 2002. Association analysis of polymorphic CGG repeat in 5' UTR of the reelin and VLDLR genes with schizophrenia[J].Schizophr Res, 58, 37-41.
Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps[J].Bioinformatics, 21: 263-265.
Chen ML, Chen SY, Huang CH, Chen CH. 2002. Identification of a single nucleotide polymorphism at the 5' promoter region of human reelin gene and association study with schizophrenia[J].Mol Psychiatry, 7: 447-448.
The Cochrane Collaboration. 2002. Review Manager (RevMan) [Computer program][CP]. Version 4.2 for Windows. Oxford: England[s.n.].
O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR. 2008. Identification of loci associated with schizophrenia by genome-wide association and follow-up[J].Nat Genet, 40, 1053-1055
Erdfelder E, Faul F, Buchner A. 1996. G*Power: a general power analysis program[J].Behav Res Meth,Instrum Comput, 28:1–11.
Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, Costa E. 2005. Reelin promoter hypermethylation in schizophrenia[J].Proc Natl Acad Sci USA, 102, 9341-9346.
Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, Uzunov DP, Smalheiser NR, Davis JM, Pandey GN, Pappas GD, Tueting P, Sharma RP, Costa E. 1998. A decrease of reelin expression as a putative vulnerability factor in schizophrenia[J].Proc Natl Acad Sci USA, 95, 15718-15723.
Kahler AK, Djurovic S, Kulle B, Jonsson EG, Agartz I, Hall H, Opjordsmoen S, Jakobsen KD, Hansen T, Melle I, Werge T, Steen VM, Andreassen OA. 2008. Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene[J].Am J Med Genet B: Neuropsychiatr Genet, 147B, 1089-1100.
Li M, Luo XJ, Xiao X, Shi L, Liu XY, Yin LD, Ma XY, Yang SY, Pu XF, Yu J, Diao HB, Shi H, Su B. 2011. Analysis of common genetic variants identifies RELN as a risk gene for schizophrenia in Chinese population[J].World J Biol Psychiatry,2011 Jul 11 [Epub ahead of print].
Liu Y, Chen PL, McGrath J, Wolyniec P, Fallin D, Nestadt G, Liang KY, Pulver A, Valle D, Avramopoulos D. 2010. Replication of an association of a common variant in the Reelin gene (RELN) with schizophrenia in Ashkenazi Jewish women[J].Psychiatr Genet, 20.184-186.
Luo XJ, Diao HB, Wang JK, Zhang H, Zhao ZM, Su B. 2008. Association of haplotypes spanning PDZ-GEF2, LOC728637 and ACSL6 with schizophrenia in Han Chinese[J].J Med Genet, 45(12): 818-826.
Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, Shianna KV, Yoon W, Kasperaviciūte D, Gennarelli M, Strittmatter WJ, Bonvicini C, Rossi G, Jayathilake K, Cola PA, McEvoy JP, Keefe RS, Fisher EM, St Jean PL, Giegling I, Hartmann AM, Möller HJ, Ruppert A, Fraser G, Crombie C, Middleton LT, St Clair D, Roses AD, Muglia P, Francks C, Rujescu D, Meltzer HY, Goldstein DB. 2009. A genome-wide investigation of SNPs and CNVs in schizophrenia[J].PLoS Genet, 5, e1000373.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses[J].Am J Hum Genet, 81, 559-575.
Shifman S, Johannesson M, Bronstein M, Chen SX, Collier DA, Craddock NJ, Kendler KS, Li T, O'Donovan M, O'Neill FA, Owen MJ, Walsh D, Weinberger DR, Sun C, Flint J, Darvasi A. 2008. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women[J].PLoS Genet, 4, e28.
Thomson PA, Christoforou A, Morris SW, Adie E, Pickard BS, Porteous DJ, Muir WJ, Blackwood DH, Evans KL. 2007. Association of Neuregulin 1 with schizophrenia and bipolar disorder in a second cohort from the Scottish population[J].Mol Psychiatry, 12, 94-104.
Tochigi M, Iwamoto K, Bundo M, Komori A, Sasaki T, Kato N, Kato T. 2008. Methylation status of the reelin promoter region in the brain of schizophrenic patients[J].Biol Psychiatry, 63, 530-533.
Wedenoja J, Loukola A, Tuulio-Henriksson A, Paunio T, Ekelund J, Silander K, Varilo T, Heikkila K, Suvisaari J, Partonen T, Lonnqvist J, Peltonen L. 2008. Replication of linkage on chromosome 7q22 and association of the regional Reelin gene with working memory in schizophrenia families[J].Mol Psychiatry, 13, 673-684.
Wedenoja J, Tuulio-Henriksson A, Suvisaari J, Loukola A, Paunio T, Partonen T, Varilo T, Lonnqvist J, Peltonen L. 2009. Replication of association between working memory and Reelin, a potential modifier gene in schizophrenia[J].Biol Psychiatry, 67, 983-991.
Williams NM, Preece A, Morris DW, Spurlock G, Bray NJ, Stephens M, Norton N, Williams H, Clement M, Dwyer S, Curran C, Wilkinson J, Moskvina V, Waddington JL, Gill M, Corvin AP, Zammit S, Kirov G, Owen MJ, O'Donovan MC. 2004. Identification in 2 independent samples of a novel schizophrenia risk haplotype of the dystrobrevin binding protein gene (DTNBP1) [J].Arch Gen Psychiatry, 61, 336-344.
RELN基因启动子区SNP位点与精神分裂症的相关性分析
常履华1, 李 明2,3, 罗雄剑2,3,刘兴彦4, 尹利德4, 杨顺英4, 刁红波5, 宿 兵2,*, 普兴富4,*
(1.昆明医学院第一附属医院 神经内科,云南 昆明650032; 2.中国科学院昆明动物研究所 遗传资源与进化重点实验室,云南 昆明650223; 3.中国科学院研究生院,北京100101; 4.玉溪市第二人民医院,云南 玉溪653101; 5.云南省精神病院,云南 昆明650224)
目前有很多证据证明RELN基因在世界人群中是一个精神分裂症的致病基因。基于之前报道过的RELN基因在精神分裂症患者中表达下降的事实, 可以推测在RELN基因启动子区可能包含影响精神分裂症发生的多态位点。该研究分析了中国西南地区病例——对照人群中(940位患者和1 369位正常人)RELN基因启动子区的3个单核苷酸多态性位点与精神分裂症的相关性。研究结果显示, 这些多态位点都不与精神分裂症相关, 表明RELN基因的致病位点并不在其启动子区。将前人研究结果与该研究结果进行综合分析(共2 843个样本), 结果仍不显著。因此, 该研究表明,RELN基因启动子区的单核苷酸多态性位点在中国人群中并不与精神分裂症相关。
RELN; 精神分裂症; 启动子; 单核苷酸多态性位点; 中国人群
Q987.2; Q593.2; R749.3
A
0254-5853-(2011)05-0504-05
2011-04-25;接受日期:2011-07-22
10.3724/SP.J.1141.2011.05504
date: 2011-04-25; Accepted date: 2011-07-22
*Corresponding authors (通信作者), E-mail: sub@mail.kiz.ac.cn; yxpuxingfu@gmail.com
猜你喜欢
杂志排行
Zoological Research的其它文章
- 白眉山鹧鸪冬季觅食地选择
- Decreased contrast sensitivity of visual cortical cells to visual stimuli accompanies a reduction of intracortical inhibition in old cats
- Complete mitochondrial genome of the laced fritillary Argyreus hyperbius (Lepidoptera: Nymphalidae)
- Possible genetic reproductive isolation between two tilapiine genera and species: Oreochromis niloticus and Sarotherodonmelanotheron
- A new blind loach species, Triplophysa huanjiangensis (Teleostei: Balitoridae), from Guangxi, China
- Seasonal dynamics of wintering waterbirds in two shallow lakes along Yangtze River in Anhui Province
