镍离子与酵母乙醇脱氢酶的相互作用
2010-12-12尹国维李芝芬王保怀杜为红
尹国维 尉 薇 徐 佳 李芝芬 王保怀 杜为红,*
(1中国人民大学化学系,北京 100872;2北京大学化学与分子工程学院,物理化学研究所,北京 100871)
Metal ions are indispensable in kinds of biomacromolecules, especially in metalloproteins.They acts as cofactor or inhibitor in various proteins and in even engineered proteins[1-3].Yeast alcohol dehydrogenase(YADH)is a metalloenzyme which catalyze the fermentation reaction of alcohol to acetaldehyde[4].This fermentation has been widely studied for its implications in wine and beer production[5-6],and an increasing interest in its application for biotechnological processes of bioconversion of different organic wastes into ethanol to be used as solvent or fuel[7-8]. YADH is a tetramer(150 kDa)with four subunits held together[9]. Each subunit contains two zinc ions with one zinc ion located at the active site(catalytic zinc)and bound to two cysteines,one histidine and a water molecule.The other zinc ion bound to four cysteines and maintains the tertiary structure of the enzymes (structuralzinc).TheZn(II)sitesareconservedamongADHsfrom different species[10-14].
The structure-activity relationships of ADHs are widely concerned in recent years.Evaluation of Hofmeister effects on ADH and other proteins indicated that the protein kinetics stability could be influenced by salt species and their concentration,and the thermodynamic parameters were also affected by some small molecules[15-16].To improve the activity of ADH,some works focused on the transition metal substitution for zinc ions,and possible substrates and inhibitors were studied[12,17-18].Moreover,the quantum mechanics method was used to show the kinetic isotope effects and enzyme motion[19].The interaction of mithramycin and chromomycin with ADH were performed to check the binding affinity of the two anticancer antibiotics to bivalent cations,i.e.,zinc ions in structural site or catalytic site[20-21].This means that the inhibitors of ADH are widely distributed.They include not only the classical reagent 4-methyl-pyrazole[12]and the anticancer compounds,but also different metal ions,such as copper[22]and bismuth[23].
Among the bivalent cations,Ni(II)is reported to inhibit ADH in a mode of mixed type mechanism in previous study[22],but little is known about the detailed information on Ni(II)-YADH interaction.In this paper,we have characterized the interaction and inhibition of Ni(II)to YADH.UV-Vis spectroscopy and fluorenscence spectroscopy were used to investigate the binding process of Ni(II)and YADH.Ellman method[24]was carried out to determine the thiolate group binding to Ni(II).And the inhibition mechanism was studied by enzymatic reaction.Furthermore,the differential scanning calorimetry(DSC)and fast protein liquid chromatography(FPLC)were performed to evaluate the thermodynamic stability of protein.
1 Materials and methods
1.1 Samples
YADH and nicotinamide adenine dinucleotid(NAD)were purchased from Sigma-Aldrich Co.(USA).The enzyme was used without further purification.Nickelous acetate tetrahydrate,trihydroxymethyl aminomethane(Tris),and 5,5′-dithiobis(2-nitrobenzoic acid)(DTNB)were purchased from BBI company (USA).All other reagents were of analytical grade.
1.2 UV-Vis spectroscopy
UV-Vis spectroscopy was used to study the binding process of Ni(II)and YADH.The lyophilized powder of YADH was dissolved in 2.0×10-2mol·L-1Tris-HCl buffer at pH 8.0.Enzyme concentration was determined from the UV absorbance at 280 nm with an absorption coefficient(ε280)of 1.89×105mol-1· L·cm-1[25].The spectrum width was from 300 to 500 nm.40 folds of Ni(II)(2.4×10-4mol·L-1)was added to 6.0×10-6mol·L-1YADH in 2.0×10-2mol·L-1Tris-HCl buffer at pH 8.0,298.2 K. The course of the reaction was monitored up to 300 min.All UVVis spectra were recorded on a Cary 50 spectrometer(Varian, USA)with thermostat holders at 298.2 K.
1.3 Fluorescence spectroscopy
The experiment was carried out on a Perkin-Elmer LS55 fluorescence spectrometer.A solution of 6.0×10-7mol·L-1YADH reacted with 40 folds of Ni(II)in 2.0×10-2mol·L-1Tris-HCl buffer at pH 8.0,298.2 K.The excitation wavelength was at 295 nm and the exit slit was set to 4 nm.The changes in emission intensities were obtained at regular time intervals.Each spectrum was corrected by blank subtraction using 2.0×10-2mol·L-1Tris-HCl buffer at pH 8.0.
1.4 Enzyme catalysis reaction
The concentrations of NAD+and NADH were determined using the extinction coefficients of 6.22×103mol-1·L·cm-1at 340 nm[26].Enzyme activity was determined by the changes of initial rate of absorbance at 340 nm corresponding to the reduction of NAD+to NADH as previously reported[27].The solution of YADH was incubated in the presence of excess of Ni(II)for 5 min.An aliquot was withdrawn and added to a solution containing 1.5×10-3mol·L-1NAD+and 0.2 mol·L-1EtOH.The final enzyme concentration was 2.5×10-9mol·L-1in solution.The initial rates(r0)of reaction were recorded at different concentrations of ethanol ranging from 5.0×10-3to 50.0×10-3mol·L-1.The Michaelis constant(Km)and the maximal reaction velocity(rmax)for inhibited reactions and control were obtained from Lineweaver-Burk plots[28].

1.5 Thiolate group analysis of YADH
Ellman′s method[24]was utilized to determine the free thiolate content(SH)of YADH before and after reacting with Ni(II). YADH was incubated with 100 folds of Ni(II)at 298.2 K,in 2.0× 10-2mol·L-1Tris-HCl buffer,pH 8.0.A solution of DTNB(1.0× 10-2mol·L-1)was added to the mixture.The final solution contained 2.0×10-6mol·L-1YADH,2.0×10-4mol·L-1Ni(II)and 5.0× 10-4mol·L-1DTNB.The reaction solution was incubated for 4 h until the absorbance at 412 nm did not change.The amount of generated p-nitrothiolate was determined using the extinction coefficient(ε412)of 1.42×104mol-1·L·cm-1[29].
1.6 Differential scanning calorimetry
Differential scanning calo rimetry experiments were performed with a Setaram(Lyons,France)Micro DSC III calorimeter.The mixture of 40 folds of 2.4×10-4mol·L-1Ni(II)and 6.0× 10-6mol·L-1YADH was incubated for 12 h in 2.0×10-2mol·L-1Tris-HCl buffer at pH 8.0,298.2 K.Then it was measured using the scanning rate of 1.0 K·min-1.The experimental temperature range was from 298.2 to 383.2 K.Temperature correction and baseline correction had been done before proceeding with the experiment.The sample volume was 0.8 mL.Tris-HCl buffer was used as the reference in all the three repeat experiments.
1.7 Fast protein liquid chromatography
A solution of 10 μmol·L-1YADH was incubated with 40 foldsofNi(II)in 100 mmol·L-1Tris-HCl at pH 8.0,298.2 K.After 120 min,the mixture was injected to fast protein liquid chromatography(FPLC)system.The Tris-HCl buffer(100 mmol·L-1, pH 8.0)was used as an elution solvent.Chromatograms were recorded by monitoring the absorbance at 280 nm with a UV detector.Control experiments were performed in the absence of Ni(II).The molecular mass calibration curve for the column was obtained using bovine serum albumin(65 kDa)and cytochrome C(12 kDa)as standards.
2 Results
2.1 Binding of Ni(II)and YADH
The interaction of Ni(II)with YADH leads to a new UV-Vis absorption band(Fig.1A).With the mixture of 40 folds of Ni(II) to YADH solution,the absorbance centered at 320 nm increased gradually.This band was assigned as S--Ni(II)ligand-to-metal charge transfer(LMCT)transitions due to Ni(II)binding to the thiolate ligand.It could be used to monitor the progress of the reaction between Ni(II)and YADH.Kinetics of the reaction was described by the dependence of absorption spectrum on time (Fig.1B).It was characterized by an initial rapid increase in absorbance,then a progressive increase for the duration.The twokinetic steps could be resolved,which obey first-order kinetics and fit to a bi-exponential growth using the non-linear least square method:

where k1and k2are the rate constants of the two kinetic phases, A1and A2are the corresponding amplitudes that show the contribution of the individual kinetic phases to the observed change in absorbance.The rate constant k1,was measured to be 0.091 min-1, and contributed 28%to the whole reaction[23].And the rate constant k2,had a value of 6.9×10-3min-1representing the rest of the reaction.
2.2 Conformational change in YADH due to the binding of Ni(II)
Fluorescence spectroscopy is widely used in protein conformational investigation since the tryptophan and tyrosine residues can produce intrinsic fluorescence[30].YADH has five tryptophan residues in each subunit.These residues produce an intrinsic fluorescence for YADH at 340 nm.With the mixture of 40 folds of Ni(II)to YADH solution,the fluorescence emission intensity decreased obviously(Fig.2A).It revealed that conformational changesoccurredinYADHuponNi(II)binding.The decrease ofYADH intensity versus time was also in a biphasic process and could be fitted by a two-exponential function as used in UV data processing(Fig.2B).The rate constant k1for the fast step was measuredto be 0.31 min-1,which contributed to 25%of the reaction,while the rate constant k2for the slow step had a value of 2.4×10-2min-1that represented the rest of the reaction.The rates are slightly higher than the corresponding values obtained from UV-Vis spectroscopy.

Fig.1 (A)Time scale of absorption spectrum,(B)kinetics of the reaction of Ni(II)to YADH at 320 nmsolution containing YADH(6.0×10-6mol·L-1)and 40 folds of Ni(II)in 2.0×10-2 mol·L-1Tris-HCl at pH 8.0,298.2 K;the broad band centered at 320 nm in Fig.1A indicating formation of Ni(II)-S(thiolate)bonds,reaction time from bottom to top:0,2,4,6,8,10,15,20,50,80,145,300 min
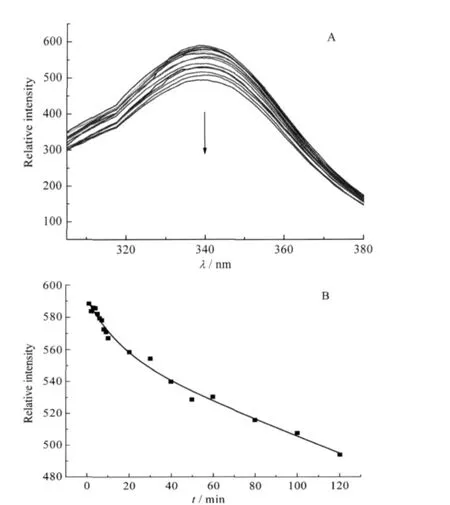
Fig.2 (A)Time scale of fluorescence emission spectra with an excitation wavelength of 295 nm,(B)kinetics of the reaction of Ni(II)to YADH at 340 nm emission intensitysolution containing YADH(6.0×10-7mol·L-1)and 40 folds of Ni(II)in 2.0×10-2 mol·L-1Tris-HCl at pH 8.0,298.2 K;reaction time from top to bottom: 0,1,2,3,4,5,6,7,8,9,10,20,30,40,50,60,80,100,120 min

Fig.3 Lineweaver-Burk plots of the enzyme catalysis reactionThe solution is composed of 2.5×10-9mol·L-1YADH and 1.5×10-3mol·L-1NAD+ at 298.2 K,2×10-2mol·L-1Tris-HCl,pH 8.0.Kmfor control reaction(◆)was found to be 8.37×10-3mol·L-1.The value is in agreement with that of control in the presence of 60(■)and 240(▲)folds of Ni(II)
2.3 Inhibition of Ni(II)to YADH activity
Based on the interaction study of YADH and Ni(II),we measured the rate of ethanol oxidation catalyzed by YADH at different substrate concentrations in the presence of Ni(II).The kinetics of enzyme catalysis reaction can be described by a Michaelis-Menten model.In the present work,the Kmand rmaxwere calculated to be 8.3×10-3mol·L-1and 41.48 OD·s-1(OD:optical density)respectively for the uninhibited control reaction,which were acceptable for further enzymatic inhibition analysis[31](Fig. 3).And the Kmvalues obtained in the presence of 60 folds and 240 folds of Ni(II)were almost the same as that in the absence of Ni(II).However,the rmaxdecreased significantly due to the increase of Ni(II).The Lineweaver-Burk plots showed a typical mode of noncompetitive inhibition[28].
2.4 Thiolate group analysis of YADH

Fig.4 DSC curves for pure YADH in the presence and absence of Ni(II)In the absence of Ni(II),the onset temperature is measured at(330.0±0.2)K (curve a),the corresponding value in the presence of 40 folds of Ni(II)is measured at(335.8±0.3)K(curve b)
The free thiolate contents were determined using DTNB by Ellman′s method so as to investigate whether Ni(II)binds to free Cys residues of YADH.The amount of generated p-nitrothiolate was determined using the absorbance at 412 nm as described in the experimental section[29].Totally,36 free Cys residues are found in each YADH subunit after treatment with dithiolthreitol (DTT)[32].19 free SH groups in the native enzyme were determined in the present work.After incubation with 100 folds of Ni(II)for 4 h at pH 8.0 Tris-HCl buffer,298.2 K,the number of free thiolate groups was determined to be 15.Therefore,there is one free thiol group loss in each subunit of YADH compared to its intact form.
2.5 Thermal denaturation of YADH upon binding of Ni(II)
The DSC method provided significant information about the thermodynamic properties of protein molecules,and the influence of molecular interactions on the stability of proteins and nucleicacids[33-34].The denaturation experimentofYADH byDSC started from 298.2 to 383.2 K and returned from 383.2 to 298.2 K.YADH showed an irreversible denaturation process(Fig.4). There was an exothermic peak at(330.0±0.2)K(onset point), which was very similar as reported[16].The molar enthalpy change of denaturation of(-7.6±0.5)×104kJ·mol-1was too large for conformational change and possibly due to protein sedimenta-tion.Addition of Ni(II)resulted in the increase of molar enthalpy change and denaturation temperature to(-8.9±0.3)×104kJ·mol-1and(335.8±0.3)K(onset point),respectively.

Fig.5 FPLC profiles of YADH after incubation with Ni(II)(A)native YADH,(B)YADH with 40 folds of Ni(II)incubation after120 min;a solution of ca 10 μmol·L-1YADH was incubated with 40 folds of Ni(II)in 100 mmol·L-1Tris-HCl,pH 8.0
2.6 Fast protein liquid chromatography study
Metal ions are known to affect signal transduction and protein-protein interaction.We carried out this experiment in order to demonstrate whether Ni(II)interferes with the quaternary structure of the native YADH.After loading YADH solution into the column,the relative retention volume value was observed at 62.9 and 74.0,respectively.Peak a in Fig.5A corresponds to a 150 kDa species based on the mass calibration curve,while peak b corresponds to a component with a molecular mass about 75 kDa.Therefore,they can be assigned to the tetramer and dimer of YADH,respectively.The existence of dimer might be from conformational equilibrium of ADH in solution.After incubation with Ni(II),the peak at 62.9 decreased in its relative intensity obviously(Fig.5B).While the peak at 74.0 increased in its relative intensity gradually.This suggests that part of tetrameric YADH dissociates into a dimer,presumably due to the binding of Ni(II)to the enzyme.
3 Discussion
YADH is a classical enzyme which contains two zinc(II)ions in each subunit,one in its active site and another in auxiliary site.The enzymatic activity has been reported to be inhibited by some metal ions at various conditions[18,22-23].Ni(II)is an important transition metal that takes part in many biological processes[34]. In the present work,we reported the interaction of Ni(II)with YADH.The results show that Ni(II)can bind to YADH and change the conformation of YADH.
UV-Vis spectroscopy reveals that the binding of Ni(II)leads to the appearance of 320 nm absorbance band.The time scale shows two kinetics steps for Ni(II)binding.The rate constants are less than those obtained from fluorescence spectrum,which indicate that the conformational change is prior to the binding of Ni(II)to thiolate group of ADH.
Although the inhibition of Ni(II)on recombinant ADH exhibits a mixed type mechanism[22],our data show a noncompetitive inhibition in the enzyme catalysis reaction at the beginning of Ni(II)binding.Since the binding process is time dependent and the enzymatic conformation changes gradually,the conclusive mechanism of inhibition is inenarrable.The complexity might be induced by the anion effect compared with previous result[22],since anion plays a key role in enzymatic activity and protein stability[12,15,35].The used sample in this case was nickelous acetate tetrahydrate,and further work is needed to compare the binding mechanism by different Ni(II)compounds in order to make it clear.
Ni(II)binding could lead to the dissociation of YADH from tetramer to dimmer,which is verified by FPLC experiments.The relative stability of dimer indicates that YADH could be described as a“Etetramer of dimers”with two identical interfaces. Hence in the DSC process,the binding of Ni(II)induces YADH in a higher denaturation temperature and molar enthalpy change. And we might conclude that conformational change arised from Ni(II)binding influences the path of YADH thermal denaturation.
Metal ions inhibition of YADH reveals various but exciting results.The investigation on interaction of Ni(II)and YADH makes a whole profile of the metal binding than ever.And it provides more information to understand metal-protein interaction.
1 Lu,Y.;Berry,S.M.;Pfister,T.D.Chem.Rev.,2001,101:3047
2 Xu,K.;Yang,X.D.;Wang,K.Chem.J.Chin.Univ.,2008,29: 2525 [徐 崑,杨晓达,王 夔.高等学校化学学报,2008,29: 2525]
3 Huang,Z.X.Prog.Chem.,2002,14:318 [黄仲贤.化学进展, 2002,14:318]
4 Ramaswamy,S.;Kratzer,D.A.;Hershey,A.D.;Rogers,P.H.; Arnone,A.;Eklund,H.;Plapp,B.V.J.Mol.Biol.,1994,235:777
5 Blandino,A.;Caro,I.;Cantero,D.Biotechnol.Lett.,1997,19:651
6 Onnela,M.L.;Suihko,M.L.;Penttil,M.;Keraen,S. J.Biotechnol.,1996,49:101
7 Fernandez,M.R.;Biosca,J.A.;Martinez,M.C.;Achkor,H.; Farres,J.;Pares,X.Adv.Exp.Med.Biol.,1997,414:373
8 Lortie,R.;Fassouane,A.;Laval,J.M.;Bourdillon,C.Biotechnol. Bioeng.,1992,39:157
9 Vanni,A.;Pessione,E.;Anfossi,L.;Baggiani,C.;Cavaletto,M.; Gulmini,M.;Giunta,C.J.Mol.Catal.B-Enzym.,2000,9:283
10 Magonet,E.;Hayen,P.;Delforge,D.;Delaive,E.;Remacle,J. Biochem.J.,1992,287:361
11 Meijers,R.;Adolph,H.W.;Dauter,Z.;Wilson,K.S.;Lamzin,V. S.;Cedergren-Zeppezauer,E.S.Biochemistry,2007,46:5446
12 Reimers,M.J.;Hahn,M.E.;Tanguay,R.L.J.Biol.Chem.,2004, 279:38303
13 Rubach,J.K.;Plapp,B.V.Biochemistry,2002,41:15770
14 Winberg,J.O.;Brendskag,M.K.;Sylte,I.;Lindstad,R.I.; McKinley-McKee,J.S.J.Mol.Biol.,1999,294:601
15 Broering,J.M.;Bommarius,A.S.J.Phys.Chem.B,2005,109: 20612
16 Nath,S.;Satpathy,G.R.;Mantri,R.;Deep,S.;Ahluwalia,J.C. J.Chem.Soc.Faraday Trans.,1997,93:3351
17 Kleifeld,O.;Rulisek,L.;Bogin,O.;Frenkel,A.;Havlas,Z.; Burstein,Y.;Sagi,I.Biochemistry,2004,43:7151
18 Vanni,A.;Anfossi,L.;Pessione,E.;Giovannoli,C.Int.J.Biol. Macromol.,2002,30:41
19 Billeter,S.R.;Webb,S.P.;Agarwal,P.K.;Iordanov,T.;Hammes-Schiffer,S.J.Am.Chem.Soc.,2001,123:11262
20 Das,S.;Devi,P.G.;Pal,S.;Dasgupta,D.J.Bio.Inorg.Chem., 2005,10:25
21 Devi,P.G.;Chakraborty,P.K.;Dasgupta,D.J.Biol.Inorg.Chem., 2009,14:347
22 Cavaletto,M.;Pessione,E.;Vanni,A.;Giunta,C.J.Biotechnol., 2001,84:87
23 Jin,L.;Szeto,K.Y.;Zhang,L.;Du,W.H.;Sun,H.Z.J.Inorg. Biochem.,2004,98:1331
24 Ellman,G.L.Arch.Biochem.Biophys.,1959,82:70
25 Buhner,M.;Sund,H.Eur.J.Biochem.,1969,11:73
26 Tkachenko,A.G.;Winston,G.W.Arch.Biochem.Biophys.,2000, 380:165
27 Vallee,B.L.;Hoch,F.L.Proc.Natl.Acad.Sci.U.S.A.,1955,41: 327
28 Wang,J.Y.;Zhu,S.G.;Xu,C.F.Biochemistry.Beijing:Higher Education Press,2002:351-383 [王镜岩,朱圣庚,徐长法.生物化学.北京:高等教育出版社,2002:351-383]
29 Riddles,P.W.;Blakeley,R.L.;Zerner,B.Meth.Enzymol.,1983, 91:49
30 Jornvall,H.;Eklund,H.;Branden,C.I.J.Biol.Chem.,1978,253: 8414
31 Dickinson,F.M.;Monger,G.P.Biochem.J.,1973,131:261
32 Harris,I.Nature,1964,203:30
33 Du,W.H.;Han,W.;Li,Z.F.;Wang,B.H.Thermochim.Acta, 2000,359:55
34 Du,W.H.;Wang,L.;Li,J.;Wang,B.H.;Li,Z.F.;Fang,W.H. Thermochim.Acta,2007,452:31
35 Buhler,R.;Von Wartburg,J.P.FEBS Lett.,1984,178:249
