Effects of Triptolide on Histone Acetylation and HDAC8 Expression in Multiple Myeloma in vitro
2010-07-18FeiZhaoLinglanZengYanChenRuiLiYuanLiuLuWenYiquanChengChunZhang
Fei Zhao, Ling-lan Zeng, Yan Chen, Rui Li, Yuan Liu, Lu Wen, Yi-quan Cheng Chun Zhang
Department of Hematology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, China
Effects of Triptolide on Histone Acetylation and HDAC8 Expression in Multiple Myeloma in vitro
Fei Zhao, Ling-lan Zeng*, Yan Chen, Rui Li, Yuan Liu, Lu Wen, Yi-quan Cheng Chun Zhang**
Department of Hematology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, China
Objective:Multiple myeloma is a kind of malignant plasma cell disease that originated from B lymphocyte and secrete great amount of monoclonal immunoglobulin. It is still one of the refractory diseases at present. Numerous studies show that there is an intensive relationship between the disequilibriumof histone acetylation and the occurance of multiple myeloma. Here we investigated the effect of triptolide(TPL) on the proliferation, apoptosis, histone H3 and H4 acetylation and expression of histone deacetylase 8 (HDAC8)in vitro, to explore its antimyeloma mechanism.
Methods:The effect of triptolide on the growth of RPMI8226 was studied by 3-(4,5-Dimethyl-2-thiazolyl) -2,5-diphenyl-2H-tetrazolium(MTT) assay. Apoptosis was detected by Hoechst 33258 staining. The protein expressions of acetyl-histone H3 and H4 were determined by Western blot, and the expression of HDAC8 was assessed by RT-PCR, Western blot and confocal microscopy.
Results:Triptolide inhibited the proliferation of RPMI8226 and induced apoptosis in a time- and dosedependent manner. The 36h IC50value was (105.370 ± 0.189)nmol/L. Triptolide increased the acetylation of histone H3 and H4 greatly. Furthermore, triptolide significantly down-regulated the mRNA and protein expression of HDAC8.
Conclusion:Triptolide can inhibit proliferation and induce apoptosis of RPMI8226 significantly. Triptolide reduces the expression of HDAC8 in order to increase the histone H3 and H4 acetylation, which is possibly the anti-myeloma mechanism of triptolide.
Triptolide, Histone acetylation, HDAC8, Multiple myeloma
INTRODUCTION
As a diterpene triepoxide, triptolide(TPL), the principal active ingredient of extracts from the Chinese herb Tripterygium wilfordii Hook.F (TwHF), is a member of the Celastraceae family[1](The structure shown in Figure 1). Triptolide has extensive pharmacological activities including immunodepression, anti-inflammation[2,3]and anti-tumor properties[4]. In diverse hematological tumorstriptolide shows anti-tumor activity and many studies are carried out to elucidate the potential anti-tumor mechanisms[5-8]. Multiple myeloma is a kind of malignant plasma cell dyscrasia originated from B lymphocytes, and a great amount of monoclonal immunoglobulins are secreted by the tumor cells. Nowadays multiple myeloma is still a refractory disorder. Numerous studies show that there is an intensive relationship between the disequilibriumof histone acetylation and the initiation and progression of multiple myeloma[9]. Multiple histone deacetylase inhibitors can inhibit the proliferation of myeloma cells, and induce apoptosis of the myeloma cells[10,11]. Histone acetylation is a reversible process that is regulated by the opposing activities of histone actyltransferases(HATs) and histone deacetylases (HDACs). Generally, hyperacetylation of histones results in transcriptional activation, whereas deacetylation correlates with transcriptional silencing. Correspondingly, transcriptional activators are often associated with HAT activity whereas HDACs frequently form complexes with transcriptional repressors. There are 11 known isoforms of the HDAC family, denoted HDAC 1–11[12]. The human HDACs are divided into three different classes based on their similarity to yeast HDAC proteins. Class I enzymes are ubiquitously expressed and include HDAC1, -2, -3, and -8[13]. HDAC8 is different from the typical class I enzyme (HDAC1, -2, and -3) in several respects, e.g. it lies near the boundary of the class I and class II enzymes[14], binds various metals including Fe(II) and K+ to its active site, and is phosphorylated by cyclic AMP-dependent protein kinase A (PKA) instead of protein kinase CK2[15,16]. HDAC8 causes histone H3 and H4 deacetylation[17]. All of these observations intrigered us to investigate the relationship between HDAC8 and multiple myeloma, and to explore the effect of triptolide on HDAC8 expression and histone acetylation status in multiple myelomain vitro.

Figure 1. Chemical structure of triptolide from Tripterygium wilfordii Hook F.
In this report, we describe for the first time the HDAC8 inhibitory effects of triptolide in human multiple myeloma cells and the corresponding changes in cell growth, apoptosis, and histone acetylation, trying to explore the anti-myeloma mechanism thereby.111111
MATERIALS AND METHODS
Cell Line and Reagents
Human multiple myeloma cell line RPMI8226 was obtained from Bio-Mart Company (Shanghai, China) and cultured in RPMI-1640 medium (Gibco BRL, Grand Island, NY, USA) containing 10% fetal bovine serum (Sijiqing Co. Ltd, Hangzhou, China) in 5% CO2incubator at 37°C. Triptolide with 98% purity was purchased from Sigma (St. Louis, MO, USA), initially dissolved in dimethylsulfoxide (DMSO), stored at -20°C, and then thawed before use. 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazo lium (MTT), DMSO and Hoechst33258 were purchased from Sigma. RNA trizol was purchased from Invitrogen (CA, USA). Reverse transcriptionpolymerase chain reaction (RT-PCR) kit was purchased from TOYOBO(Japan). Anti-acetyl histone H3 (acetyl-histone H3), anti-acetyl H4 (acetyl-histone H4), anti-HDAC8, and anti-β-actin antibodies were purchased from Santa Cruz (California, USA), Tetrametrylrhodarnine isothiocyante (TRITC)-labeled secondary antibodies, horse radish peroxidase (HRP)-labeled secondary antibodies and chemiluminescence (ECL) reagent kits were purchased from Pierce Biotechnology(Rockford, IL, USA).
MTT Assay
The antiproliferative effect of triptolide on RPMI 8226 cells was determined using the MTT dye uptake method. In brief, the cells (50,000 per well) were incubated in quintuplicate in 96-well plates. Different doses of triptolide were added, with the final concentrations of 40, 60, 80, 100 and 120nmol/L for 24, 36 and 72 h. Thereafter, 20 μl MTT solution (5mg/ml in phosphate-buffered saline) was added to each well. After 4h at 37°C, the supernatant was removed and 150μl DMSO was added. When the blue crystal was dissolved, the optical density(OD) was detected at 570 nm wavelength by a 96-well multiscanner autoreader (Bio-Rad M450,USA). The following formula was used∶ cell proliferation inhibited(%)=[1-(OD of the experimental samples/ OD of the control)] ×100%. The IC50was taken as the concentration that caused 50% inhibition of cell proliferation.
Hoechst 33258 Staining
Nuclear fragmentation was visualized by Hoechst 33258 of apoptosis nuclei. Apoptosis cells were collected by centrifugation, washed with phosphatebuffered saline (PBS), and fixed in 4% paraformaldehyde for 20 min at room temperature. Subsequently the cells were washed and resuspended in 20μl PBS before being deposited on poly lysine-coated coverslips and left to adhere to the cover slips for 30 min at room temperature, after which the cover slips were washed twice with PBS. The adhered cells were then incubated with0.1%Triton X-100 for 5 min at room temperature and rinsed with PBS three times. The cover slips were treated with Hoechst 33258 at 37°C for 30min, rinsed with PBS, and mounted on slides with glycerol-PBS. The cells were viewed with an Olympus fluorescence microscope (Japan).
Western Blot Analysis
After treatment, cells were harvested and lysed in 100 μL of lysis buffer (10mmol/L Tris-HCL [pH7.5], 1mmol/L EDTA, 1% Triton X-100, 150mmol/L NaCl, 1mmol/L dithiothreitol, 10% glycerol, 0.2mmol/L phenylmethylsulfonyl fluoride and protease inhibitors) by incubation on ice for 30 min, and then the extracts were centrifuged at 18,000×g for 15 min to remove cell debris. After the addition of 5× loading buffer, protein samples were electrophoresed on a 12% sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto nitrocellulose membranes. The membranes were then immunoblotted with primary antibodies directed against acetyl-histone H3, acetyl-histone H4, and HDAC8 (dilution of 1∶200). After incubation overnight at 4°C, the blots were exposed to the HRP-labeled sencondary antibody at a final dilution of 1∶2000 in Western washing solution. Finally, the blots were visualized by ECL and autoradiography system. Quantification of the bands was carried out using the Quantity One densitometric analysis software (Bio-Rad, CA, USA).
Reverse Transcription-polymerase Chain Reaction
Total cellular RNA was extracted using Trizol reagent. Reverse transcription-polymerase chain reaction(RT-PCR) was performed with the appropriate primers, following the protocol of TOYOBO kit. PCR reaction mixture 20μl was initially amplified. Primer pairs were all designed from human cDNA sequences available in GenBank and synthesized by Shanghai Bioengineering Company (Shanghai, China). The sequences of the primers and product sizes for HDAC8 and β-actin were as follows∶ For hdac8, the forward primer used was 5'-TGG GCA GTC GCT GGT-3'; reverse primer, 5'-GTG GCT GGG CAG TCA TAA-3' (product size∶ 285bp). Polymerase chain reaction was carried out at 94°C for 5 min, followed by 34 cycles of 94°C for 30s, and 55.5°C for 30s, then 72°C for 30s, finally with an extension at 72°C for 7 min. For β-actin, the forward primer used was 5'-GAG CTA CGA GCT GCC TGA CG-3'; reverse primer, 5'-CCT AGA AGC ATT TGC GGT GG-3' (product size∶ 416bp). Polymerase chain reaction was carried out at 94°C for 5 min, followed by 34 cycles of 94°C for 30s, and 60°C for 30s, then 72°C for 30s, finally with an extension at 72°C for 7 min. The amplified PCR products were separated by electrophoresis on a 2% agarose gel and quantitated by relative intensities of the bands, as compared to those of β-actin using Smart View Bio-Electrophoresis Image Analysis System (Furi, Shanghai, China).
Immunofluorescence with Confocal Microscope
After incubation with 40μmol/L of triptolide for 24 h, cells were collected and fixed in 4% paraformaldehyde for 10 min. The suspensions were permeabilized with 0.25% Triton X-100 for 10 min, and blocked with 3% bovine serum albumin for 30 min. After that, the primary antibody against HDAC8 (diluted at 1∶100) was used overnight at 4°C. Then, the samples were exposed to TRITC-labeled secondary antibody (diluted at 1∶100) for 1 h and stained with Hoechst 33258 (10μg/ml) to visualize the DNA. Images were captured using an FV-500 confocal microscope (Olympus,Japan).
Statistical Analysis
Results were expressed as ¯x±sand analyzed using SPSS 13.0. Student’st-test was used to compare quantitative data populations with normal distribution and equal vriance. A value ofP<0.05 was considered statistically significant. At least three independent experiments were performed.
RESULTS
Effects of Triptolide on the Proliferation of RPMI8226 Cells by MTT
The effect of triptolide on the proliferation of RPMI8226 cells was determined by MTT assay. RPMI8226 cells were treated with different concentrations of triptolide (0, 40, 60, 80, 100 nmol/L) for 0h, 24h, 36h, 48h and 72h respectively, which resulted in the inhibition of cell proliferation in a dose- and time- dependent manner. As shown in Figure 2, The OD value of the triptolide-treated group was significantly lower than that of the untreated group. The IC50value for 36 h of the RPMI8226 was 105.37 ± 0.19 nmol/L. As the exposure time of triptolide increased, the IC50value decreased dramatically.
Effects of Triptolide on Apoptosis of RPMI8226 Cells by Hoechst 33258 Staining
In order to study the effect of triptolide on cell apoptosis, we used Hoechest 33258 staining to investigate the change in the nuclei of RPMI8226 cells, as shown in Figure 3. Apoptosis bodies containing nuclear fragments were found in the triptolide-treated cells, where the chromatin became condensed and marginalized, the nuclear envelope appeared lytic and the cytoplasm were shrinked (Figure 3B), while none of these was found in untreated cells (Figure 3A). When treated with 80 and 160 nmol/L triptolide for 24h, there were more shrinked and lytic nuclei as shown in Figure 3C and 3D.

Figure 2. Time-effect and dose-effect relationship of proliferation inhibiition by triptolide on RPMI8226 cells.
Expressions of Acetyl-Histone H3, H4 and HDAC8 in RPMI8226 Cells by Western Blot
To investigate the effects of triptolide on H3 and H4 actylation and the action mechanism, we examined the protein expressions of acetyl-histone H3, acetylhistone H4, and HDAC8. RPMI8226 cells were treated with triptolide (40, 80 and 160nmol/L) for 48 h. After treatment, histones were extracted from the cells, and the protein expression of acetyl-histone H3, H4 and HDAC8 were measured by western blot analysis. Exposure to triptolide (40, 80 and 160 nmol/L) for 48 h could increase the global levels of acetyl-histone H3 and H4 while the protein expression of HDAC8 declined significantly, which is shown in Figure 4.
Effects of Triptolide on the mRNA Level of HDAC8 in RPMI8226 Cells
We explored the effects of triptolide on the mRNA level of HDAC8 as well. RPMI8226 cells were treated with triptolide (40, 80 and 160nmol/L) for 24 h. After that, the mRNA level of HDAC8 decreased significantly in a dose-dependent manner by RT-PCR which was shown in Figure 5. So it is concluded that triptolide induces a dose-dependent decline of HDAC8 both at transcriptional level and translational level in RPMI8226 cells.
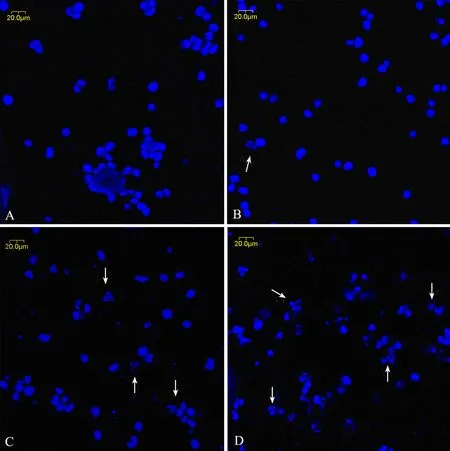
Figure 3. The morphological changes of RPMI8226 cells treated with triptolide by Hoechst33258 staining (×400). A∶ Untreated RPMI8226 cells; B∶ Cells treated with 40 nmol/L triptolide for 24 h; C∶ Cells treated with 80 nmol/L triptolide for 24 h; D∶ Cells treated with 160 nmol/L triptolide for 24 h.
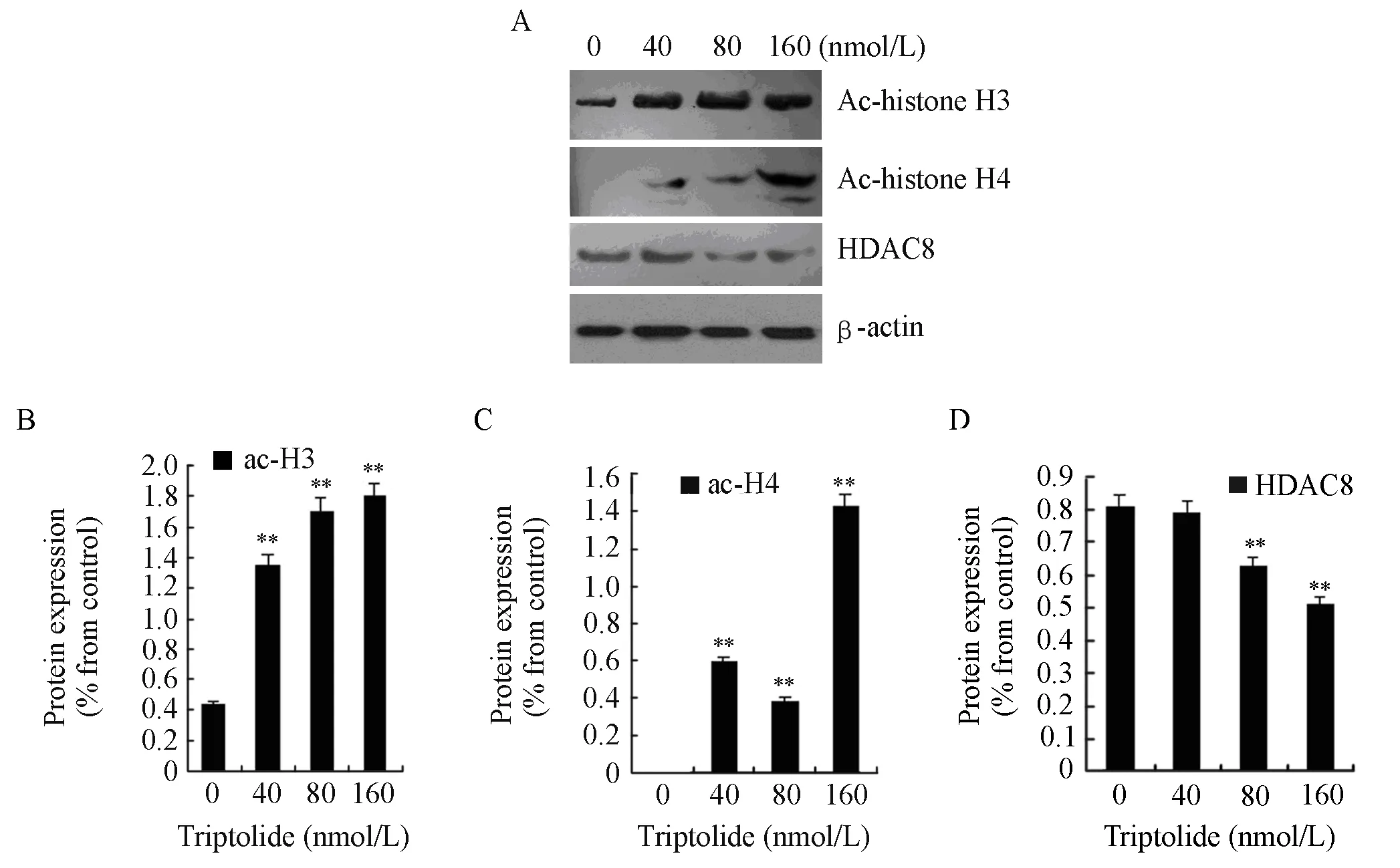
Figure 4. The protein expressions of acetyl-histone H3, H4 and HDAC8 in RPMI8226 cells when treated for 48 h by triptolide of different concentrtion (**P<0.01). A∶ RPMI8226 cells were treated with 0, 40, 80 or 160 nmol/L of triptolide and then cell lysates were subjected to Western blot with anti-acetyl-histone H3, anti-acetyl-histone H4, anti-HDAC8 and anti-β-actin; B∶The expression of actyl-histone H3 was quantified by densitometry; C∶ The expression of acetyl-histone H4 was quantified by densitometry; D∶ The expression of HDAC8 was quantified by densitometry.
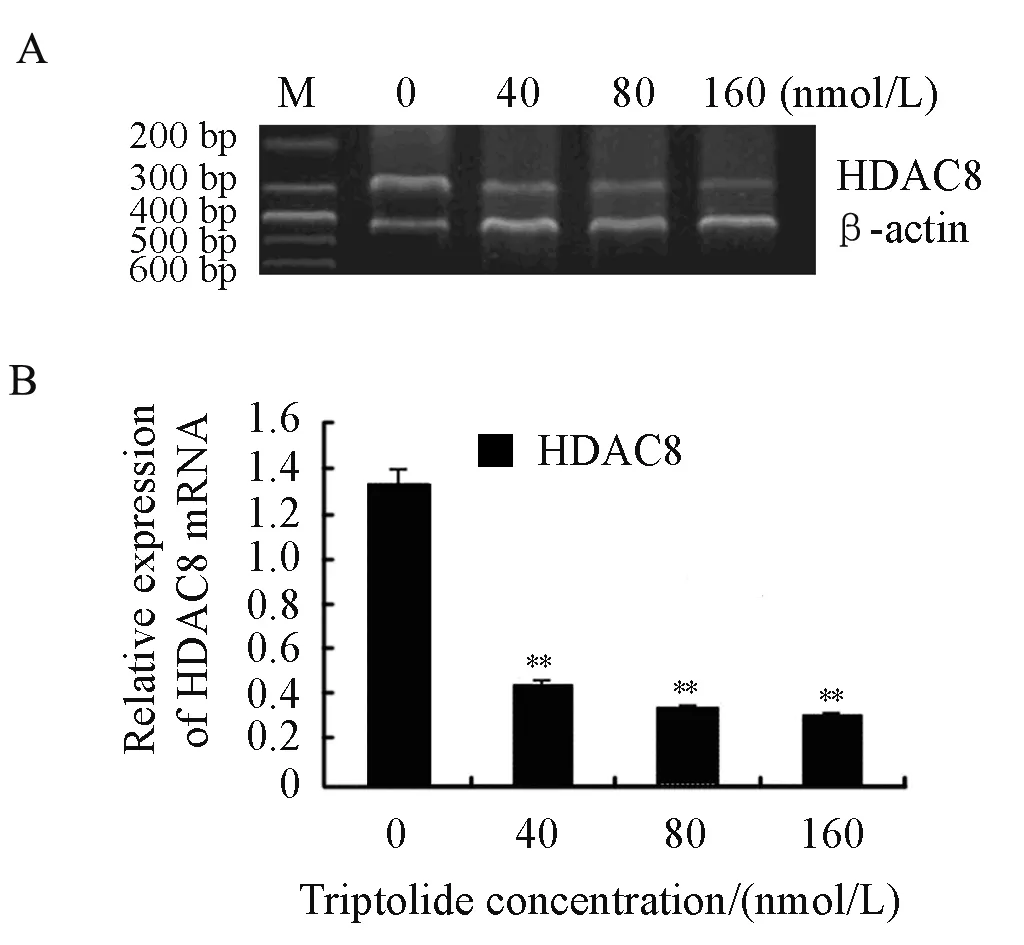
Figure 5. Effects on the mRNA of HDAC8 when treated by triptolide for 24 h in RPMI8226 cells (**P<0.01) A∶ RT-PCR was performed with the primers of HDAC8 and β-actin. B∶The relative expression of HDAC8 mRNA was shown above.
Location and Protein Expression of HDAC8 in RPMI8226 Cells with Confocal Microscope
To visualize the subcellular location and expression of HDAC8 in RPMI8226 cells, confocal microscope was used. As shown in Figure 6, HDAC8 was localized in cell nucleus. The mean fluorescence density value of the control was 39.86 ± 0.47 (Figure 6A). After treatment with triptolide (40nmol/L) for 24h,the fluorescence density decreased dramatically, as the mean fluorescence density value was 21.96 ± 0.34 (Figure 6D). Our results revealed that compared with the control, the expression of HDAC8 in RPMI8226 cells was remarkably inhibited by the addition of triptolide, while the subcelluar localization was still in the nucleus after treatment.

Figure 6. Effect of triptolide on subcellular localization and expression of HDAC8 in RPMI8226 cells (×400). A∶ Control, HDAC8 stained with TRITC; B∶ Control, DNA stained with Hoechst 33258; C∶ Merging figure A with Figure B; D∶ Cells treated by triptolide, HDAC8 stained with TRITC; E∶ Cells treated by triptolide, DNA stained with Hoechst 33258; F∶Merging Figure D with Figure E.
DISCUSSION
Triptolide is a kind of natural compound that possesses multiple biological activities such as inhibiting proliferation, inducing apoptosis and controlling the metastasis of tumor cells. Triptolide interferes with the cell cycle, improves the activity of p21waf1/cip1[18,19], inhibits cell proliferation[20,21], activates caspase8, 9 and 3, lyses the DNA repair enzyme PARP[22,23], and induces cell apoptosis directly or by means of inhibiting nuclear factor κB (NFκB) through upregulating nuclear factor κB inhibiting protein (IκBα)[24]. Triptolide is associated with up-regulation of the Bax gene[25], and causes down-regulation of the Bcl-2 gene which results in control of cell growth in tumors[26]. Recent studies suggests that triptolide overcomes dexamethasone resistance and enhances PS-341-induced apoptosis via PI3k/Akt/NF-κB pathways in human multiple myeloma cells[27]. Our experiments showed that triptolide could inhibit the proliferation of RPMI8226 cells significantly in a time- and dose-dependent manner. Hoechst 33258 staining showed typical cellular apoptosis morphologic changes when treated with triptolide. With the increase of drug concentration, the apoptotic rate of RPMI8226 cells increased significantly.
Histone acetyltransferase(HAT) and HDAC control the addition and removal of acetyl groups on proteins and maintain a dynamic balance of steady-state acetylation. The role of histone deacetylase in oncogenesis may be a consequence of abnormal transcription factors usurping the normal mechanisms regulating gene transcription. Transcription silence of the tumor suppressor genes in malignant tumor cells has a close relationship with the decrease of histone acetylation of the tumor suppressor genes. HDAC8 is a HDAC subtype which has a molecular weight of 49kDa and belongs to the Class I HDAC enzymes. HDAC8 has the deacetylase activity in many kinds of tissues and cells[28], and removes the acetyl groups from the N-terminal of histone H3 and H4 thereby deregulating histone acetylation[17,29]. Histone deacetylase inhibitors upregulate histone acetylation through inhibition of the HDAC activity. Histone acetylation and deacetylation change the superior structures through affecting the interaction between DNA and histone[30,31]. Histone deacetylation is closely related to cell proliferation, while histone acetylation is connected with cell cycle inhibition, cell differentiation and apoptosis[32]. Research showed that when K562 chronic myelogenous leukemia cells was induced differentiation artificially, histone H3 acetylation increased in the gene regulatory region of Rho family GTPase Rac2 which closely connected with myeloid cell differentiation[33]. In a comprehensive panel of normal tissues, cancer cell lines and primary tumors, a global aberrant pattern of histone H4 modification, characterized by a loss of mono-acetylation at Lys16 and trimethylation at Lys20, predominantly located at DNA repetitive sequences, was seen only in cancer cells and was associated with early tumorigenesis[34]. Histone hypo-acetylation and overexpression of HDAC was also observed in human cancer tissues from a range of organs such as stomach, colon and breast. The level of histone hypo-acetylation correlates with tumor initiation[35,36]. We examined the regulation of acetyl histone H3 and H4 with triptolide in RPMI8226 cells using Western blot, and found that after treatement with triptolide for 48h, the level of acetyl histone H3 and H4 rose strikingly in a dose dependent manner. We subsequently examined the expression of HDAC8 in RPMI8226 cells by RT-PCR and Western blot, and found that after triptolide treatment, the mRNA and protein levels of HDAC8 decreased significantly in a dose dependent manner. The detection with confocal microscope showed that HDAC8 localized in the cellular nucleus. When treated with triptolide, there was no subcellular localization difference, but the expression of HDAC8 was remarkably declined.
Abnormal transcription factors disrupting the normal gene transcription may be the reason why histone deacetylation induces tumors. Studies showed that in patients with multiple myeloma and myeloma cell lines the overexpression of c-myc could be detected[37]. As the disease developed, the expression of c-myc rose significantly, while the inhibition of HDAC can suppress the expression of c-myc[38,39]. Further studies indicated that in patients with multiple myeloma and myeloma cell lines which all treated with melphalan, the upregulation of histone acetylation could be detected at the H3K9 loci of promoters of c-myc and CCND1, while the protein expression levels of Myc and CyclinD1 declined correspondingly[28]. We speculate that triptolide declines the protein expression of Myc through inhibiting the deacetylation of c-myc and subsequently increasing the histone acetylation of c-myc, which makes it possible that triptolide specifically inhibits multiple myeloma-causing genes so that decreases or even reverses the tumor.
In all, our experiments showed that triptolide inhibited the proliferation of RPMI8226 cells in a time- and dose-dependent manner and induced dosedependent apoptosis. Triptolide strikingly inhibited HDAC8 accompanied by accumulation of acetylated H3 and H4 histones in RPMI8226 cells. The regulation of histone acetylation is probably theanti-multiple myeloma mechanism of triptolide, which provides important theoretical foundation for the clinical application of triptolide.
Acknowledgement
The authors would like to thank the Department of Central Laboratory, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China, for offering relevant experimental facilities and technical support.
REFERENCES
[1] Carter BZ, Mak DH, Schober WD, et al. Triptolide induces caspase-dependent cell death mediated via the mitochondrial pathway in leukemic cells[J]. Blood 2006; 108∶ 630-7.
[2] Kiviharju TM, Lecane PS, Sellers RG, et al. Antiproliferative and proapoptotic activities of triptolide (PG490), a natural product entering clinical trials, on primary cultures of human prostatic epithelial cells[J]. Clin Cancer Res 2002; 8∶ 2666-74.
[3] Xiang M, Zhang C. Advances in studies on immunosuppression of Tripterygium wilfordii[J]. Chin Trad Herb Dru(in Chinese) 2005; 36∶ 458-61.
[4] Yang S, Chen J, Guo Z, et al. Triptolide inhibits the growth and metastasis of solid tumors[J]. Mol Cancer Ther 2003; 2∶ 65-72.
[5] Yang M, Shen JK, Huang J, Du HP, Ma QL, Jin J. Interleukin-6-independent expression of glucocorticoid receptor is upregulated by triptolide in multiple myeloma[J]. Leuk Lymphoma 2009; 50∶802-8.
[6] Carter BZ, Mak DH, Schober WD, et al. Triptolide sensitizes AML cells to TRAIL-induced apoptosis via decrease of XIAP and p53-mediated increase of DR5[J]. Blood 2008; 111∶ 3742-50.
[7] Zhang C, Cui GH, Liu F, et al. Inhibitory effect of triptolide on lymph node metastasis in patients with non-Hodgkin lymphoma by regulating SDF-1/CXCR4 axis in vitro. Acta Pharmacol Sin. 2006; 27∶1438-46.
[8] Borja-Cacho D, Yokoyama Y, Chugh RK, et al. TRA2L and triptolide∶ an effective combination that induces apoptosis in pancreatic cancer cells[J]. Gastroinbtest Surg. 2010; 14∶ 252-60.
[9] Krejcí J, Harničarová A, Streitová D, et al. Epigenetics of multiple myeloma after treatment with cytostatics and gamma radiation[J]. Leukemia Res 2009; 33∶ 1490-8.
[10] Feng R, Ma H, Hassig CA, et al. KD5170, a novel mercaptoketone-based histone deacetylase inhibitor, exerts antimyeloma effects by DNA damage and mitochondrial signaling[J]. Mol Cancer Ther 2008; 7∶1494-505.
[11] Dong XF, Song Q, Li LZ, et al. Histone deacetylase inhibitor valproic acid inhibits proliferation and induces apoptosis in KM3 cells via downregulating VEGF receptor[J]. Neuro Endocrinol Lett 2007; 28∶ 775-80.
[12] de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, et al. Histone deacetylases (HDACs)∶ characterization of the classical HDAC family[J]. Biochem J 2003; 370∶737-49.
[13] Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression[J]. Curr Opin Genet Dev 2003; 13∶143-53.
[14] Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family∶functional implications of phylogenetic analysis[J]. J Mol Biol 2004; 338∶ 17-31.
[15] Waltregny D, Glénisson W, Tran SL, et al. Histone deacetylase HDAC8 associates with smooth muscle alpha-actin and is essential for smooth muscle cell contractility[J]. FASEB J 2005; 19∶ 966-8.
[16] Lee H, Sengupta N, Villagra A, et al. Histone deacetylase 8 safeguards the human ever-shorter telomeres 1B (hEST1B) protein from ubiquitinmediated degradation[J]. Mol Cell Biol 2006; 26∶5259-69.
[17] Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A[J]. Mol Cell Biol 2004; 24∶ 765-73.
[18] Lavelle D, Chen YH, Hankewych M, et al. Histone deacetylase inhibitors increase p21(WAF1) and induce apoptosis of human myeloma cell lines independent of decreased IL-6 receptor expression[J]. Am J Hematol 2001; 68∶ 170-8.
[19] Nian H, Delage B, Pinto JT, et al. Allyl mercaptan, a garlic-derived organosulfur compound, inhibits histone deacetylase and enhances Sp3 binding on the P21WAF1 promoter[J]. Carcinogenesis 2008; 29∶1816-24.
[20] Ficner R. Novel structural insights into class I and II histone deacetylases[J]. Curr Top Med Chem 2009; 9∶235-40.
[21] Oehme I, Deubzer HE, Wegener D, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis[J]. Clin Cancer Res 2009; 15∶ 91-9.
[22] Choi YJ, Kim TG, Kim YH, et al. Immunosuppressant PG490 (triptolide) induces apoptosis through the activation of caspase-3 and downregulation of XIAP in U937 cells[J]. Biochem Pharmacol 2003; 66∶ 273-80.
[23] Balasubramanian S, Ramos J, Luo W, et al. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas[J]. Leukemia 2008; 22∶ 1026-34.
[24] Yinjun L, Jie J, Yungui W. Triptolide inhibits transcription factor NF-kappaB and induces apoptosis of multiple myeloma cells[J]. Leuk Res 2005; 29∶99-105.
[25] Zhou GX, Ding XL, Huang JF, et al. Apoptosis of human pancreatic cancer cells induced by Triptolide [J]. World J Gastroenterol 2008; 14∶ 1504-9.
[26] Lin J, Chen LY, Lin ZX, et al. The effect of triptolide on apoptosis of glioblastoma multiforme (GBM)cells[J]. J Int Med Res 2007; 35∶ 637-43.
[27] Yang M, Huang J, Pan HZ, et al. Triptolide overcomes dexamethasone resistance and enhanced PS-341-induced apoptosis via PI3k/Akt/NF-κB pathways in human multiple myeloma cells[J]. Int J Mol Med 2008; 22∶ 489-96.
[28] Buggy JJ, Sideris ML, Mak P, et al. Cloning and characterization of a novel human histone deacetylase, HDAC8[J]. Biochem J 2000; 350∶199-205.
[29] Gurard-Levin ZA, Mrksich M. The Activity of HDAC8 Depends on Local and Distal Sequences of Its Peptide Substrates[J]. Biochemistry 2008; 47∶6242-50.
[30] Lee DY, Hayes JJ, Pruss D, et al. A positive role for histone acetylation in transcription factor access to nucleosomal DNA[J]. Cell 1993; 72∶ 73-84.
[31] Wolffe AP, Guschin D. Chromatin structural features and targets that regulate transcription[J]. J Struct Biol 2000; 129∶102-22.
[32] Marks PA, Richon VM, Breslow R, et al. Histone deacetylase inhibitors as new cancer drugs[J]. Curr Opin Oncol 2001; 13∶ 477-83.
[33] Muthukrishnan R, Skalnik DG. Identification of a minimal cis-element and cognate trans-factor(s) required for induction of Rac2 gene expression during K562 cell differentiation[J]. Gene 2009; 440∶ 63-72.
[34] Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at lys16 and trimethylation at lys20 of histone H4 is a common hallmark of human cancer[J]. Nat Genet 2005; 37∶ 391-400.
[35] Yasui W, Oue N, Ono S, et al. Histone acetylation and gastrointestinal carcinogenesis[J]. Ann N Y Acad Sci 2003; 983∶ 220-31.
[36] Song J, Noh JH, Lee JH, et al. Increased expression of histone deacetylase 2 is found in human gastric cancer[J]. APMIS 2005; 113∶ 264-8.
[37] Chiecchio L, Dagrada GP, Protheroe RK, et al. Loss of 1p and rearrangement of MYC are associated with progression of smouldering myeloma to myeloma∶sequential analysis of a single case[J]. Haematologica 2009; 94∶1024-8.
[38] Avet-Loiseau H, Gerson F, Magrangeas F, et al. Rearrangements of the c-myc oncogene are present in 15% of primary human multiple myeloma tumors[J]. Blood 2001; 98∶ 3082-6.
[39] Shou Y, Martelli ML, Gabrea A, et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma[J]. Proc Natl Acad Sci USA 2000; 97∶ 228-33.
R733.3 Document code: A Article ID: 1000-9604(2010)02-0148-08
10.1007/s11670-010-0148-y
2009−10−27; Accepted 2010−02−23
This work was supported by the National Natural Science Foundation of China(No.30700882)
*Contributed equally to this study.
**Corresponding author.
E-mail∶ Zhangchun23@yahoo.com.cn
© Chinese Anti-Cancer Association and Springer-Verlag Berlin Heidelberg 2010
杂志排行
Chinese Journal of Cancer Research的其它文章
- Triptolide Inhibits Cell Growth and Induces G0- G1 Arrest by Regulating P21wap1/cip1 and P27 kip1 in Human Multiple Myeloma RPMI-8226 Cells
- Different Outcome of Myeloid Sarcoma with Spinal Cord Compression Preceding Acute Myeloid Leukemia: Report of Two Cases and Review of Literatures
- CGI-100 Specific shRNA Inhibits Proliferation and Induces Differentiation in Leukemia K562 Cells
- Expression of Embryonic Stem Cell Marker Oct-4 and Its Prognostic Significance in Rectal Adenocarcinoma
- Differential Diagnosis of Warthin's Tumor Complicated with Lung Adenocarcinoma by 18F- FDG PET/CT Imaging and Radioisotope Scanning with Tc-99m Pertechnetate: A Case Report and Literature Review
- Synergistic Action of fMLP-boanmycin Combination on the Growth of Mouse Colon Carcinoma and Its Action Mechanisms
