CGI-100 Specific shRNA Inhibits Proliferation and Induces Differentiation in Leukemia K562 Cells
2010-07-18YongqiangWangTingmeiChenYanLeiHongleiFengKeWangYanZhang
Yong-qiang Wang, Ting-mei Chen, Yan Lei, Hong-lei Feng, Ke Wang, Yan Zhang
The Key Laboratory of Laboratory Medical Diagnostics of Ministry of Education,Department of Clinical Biochemistry, Chongqing Medical University, Chongqing 400016, China
CGI-100 Specific shRNA Inhibits Proliferation and Induces Differentiation in Leukemia K562 Cells
Yong-qiang Wang, Ting-mei Chen*, Yan Lei, Hong-lei Feng, Ke Wang, Yan Zhang**
The Key Laboratory of Laboratory Medical Diagnostics of Ministry of Education,Department of Clinical Biochemistry, Chongqing Medical University, Chongqing 400016, China
Objective:To investigate the characteristics of CGI-100- knockdown K562 cells and the effect of CGI-100 RNA interference (RNAi) on matrine-treated K562 cells.
Methods:Three oligonucleotides targeting CGI-100 gene and a pair of negative control containing the same nucleotide composition with a different sequence were devised and chemically synthesized. The inhibition efficiency of CGI-100 expression by shRNA-CGI-100 in K562 cells was determined using semiquantitative RT-PCR and dot blot hybridization. The effect of CGI-100 RNAi on the growth of K562 cells was examined using MTT assay and cell differentiation was measured by distinct approaches including flow cytometry, benzidine staining and electron microscope. After CGI-100-konckdown K562 cells were incubated with 0.2 mg/ml of matrine or 30 μmol/L of hemin for 48 h, the expression levels of Glycophorin A(GPA)(CD235a) and Growth factor independence-1B mRNA(Gfi-1B mRNA) were measured by RT-PCR and the protein levels of GPA, CD14 and CD15 were detected by flow cytometry.
Results:The eukaryotic expression vectors of CGI-100 RNAi were successfully constructed. The K562/shRNA-CGI-100 cell line was established in which the inhibition efficiency of CGI-100 gene expression by shRNA-CGI-100 was 54%. CGI-100-knockdown inhibited the proliferation and induced erythroid differentiation in K562 cells. Compared with the control K562 cells, the K562/shRNA-CGI-100 cells showed decreased absorbance value detected by MTT assay, decreased enchromation, increased heterochromation, increased percentage of G0/G1phase cells, decreased population of S phase cells, decreased PI (proliferation index of cells), and elevated percentage of benzidine-positive cells. Moreover, the sensitivity of K562/shRNA-CGI-100 cells to either matrine or hemin was enhanced and the sensitivity of these cells to matrine was higher than that to hemin. Compared with the control K562 cells, matrine treatment in K562/shRNA-CGI-100 cells resulted in increased inhibitory rate of proliferation, elevated percentage of benzidine-positive cells, obviously up-regulated mRNA expressions of GPA and Gfi-1B, and increased mean fluorescence intensity (MFI) of GPA. No CD14 expression was detected and no statistical significance was found for the detected CD15. Finally, the MFI of GPA increased in K562/shRNA-CGI-100 cells treated with hemin and was 1.7 times less than that in cells exposed to matrine.
Conclusion:These results suggest that the function of CGI-100 gene is correlated with the deregulated proliferation and the block of erythroid differentiation in K562 cells and may also be involved in matrine-induced erythroid differentiation in K562 cells.
CGI-100; RNA interference; Leukemia; Erythroid differentiation; Matrine
INTRODUCTION
The function of CGI-100 gene which is located on chromosome 1p22.1 is largely unknown. Its full-length sequence is 3636 bp and the region coding protein is located between 113 bp and 802 bp. TheCGI-100 protein lies in the fifth transport region of human transmembrane protein which is called as TMED5 (Homo sapiens transmembrane emp24 protein transport domain containing 5), an important member of P24 protein family[1].
Our previous studies indicated that CGI-100 gene was highly expressed in K562 cells. Its overexpression might accelerate the proliferation of K562 cells and increase the percentage of S phase cells and degree of malignancy of these cells. Besides these, the expression of CGI-100 mRNA decreased rapidly in matrine-induced erythroid differentiation and apoptosis in K562 cells and maintained at a low level after a prolonged time of matrine exposure[2]. Therefore, we hypothesized that CGI-100 gene may be involved in malignant proliferation of leukemia cells and the pathway of matrine-mediated differentiation and apoptosis in K562 cells.
To prove this hypothesis, we successfully constructed the eukaryotic expression vectors of CGI-100 RNAi and established the K562/shRNACGI-100 cell line using CGI-100 specific shRNA. We found that CGI-100 specific shRNA inhibited cell growth and induced erythroid differentiation in K562 cells. In addition, CGI-100 gene knockdown increased the sensitivity of K562 cells to matrine and hemin, respectively. The sensitivity of the K562/shRNACGI-100 cells to matrine was higher than that to hemin. These results suggest that the function of CGI-100 gene is correlated with the deregulated proliferation and the block of erythroid differentiation in K562 cells and may also be involved in matrine-induced erythroid differentiation in K562 cells.
MATERIALS AND METHODS
Short-hairpin RNA Design
Three pairs of shRNA were designed according to CGI-100 gene sequence in the Genbank following the rules of shRNA interference[3]. Each pair containing a unique19-nt double-stranded human CGI-100 sequence is presented as an inverted complementary repeat and separated by a loop of -nt spacer plus BamHI restriction site in the upstream sequence and plus HindIII restriction site in downstream. In order to exclude the possibility that other genes were interfered, the interfere sequence were compared with the sequence from GeneBank. The same base composition with a different sequence was regarded as a negative control. Then the shRNA interference sequences were synthesised by Invitrigen Company. Annealed oligonucleotides were subcloned into the pGenesil-1 expression vectors (Jingsai, Wuhan) and sequence verified, as shown in Table 1.
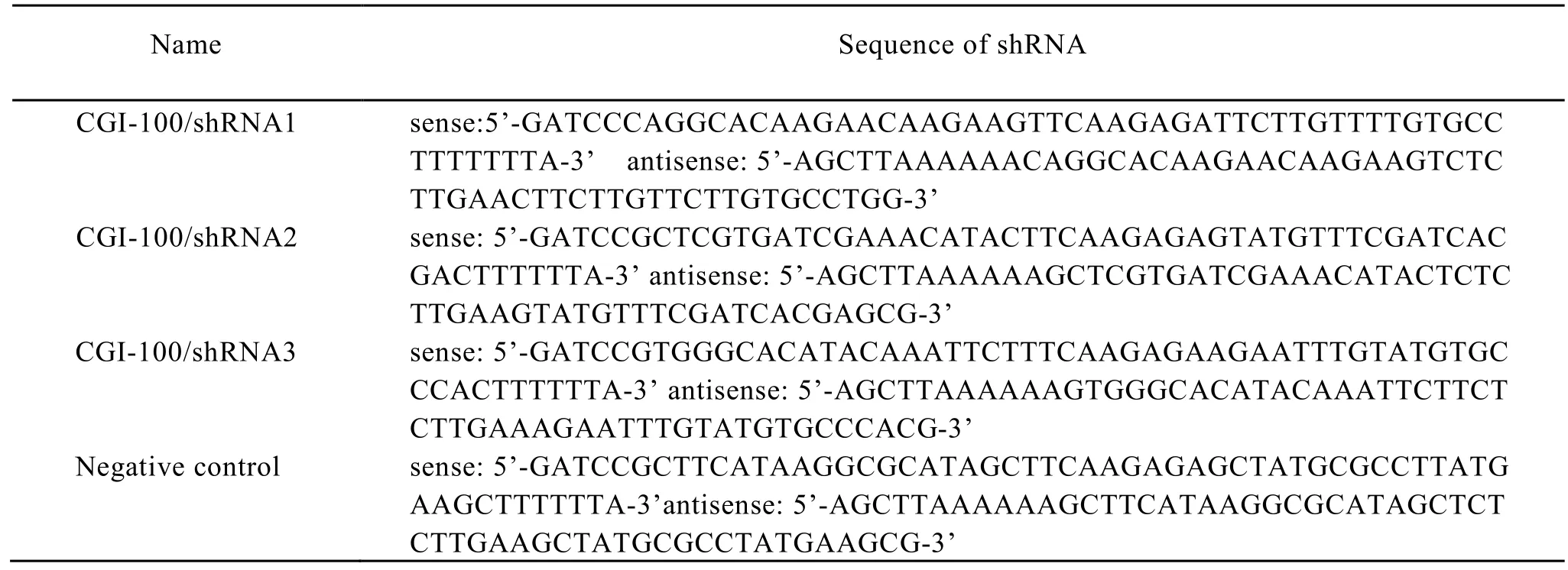
Table 1. Oligonucleotide sequences of GCI-100 specific CGI-100 shRNA and negative control shRNA
Cell Culture and Transfection
K562 cells (leukemia cell line, from Shanghai Institute for Biological Sciences, Chinese Academy of Sciences) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Sijiqing, Hangzhou), at 37°C with 5% CO2.According to manufacturer’s recommendation, K562 cells were transfected by LipofectamineTM2000 (Invitrogen, USA) with empty pGenesil, negative control/shRNA, CGI-100/shRNA1, CGI-100/ shRNA2 and CGI-100/ shRNA3 respectively. Cells were seeded in a 24-wellplate at a concentration of 1×106cells/well and allowed overnight growth to reach 80% confluency. Cells were then transfected with the mixture of 0.8 µg plasmid DNA and 2.4 µl of LipofectaminTM 2000. Following transfection for 48 h, G418 were added to the culture medium for resistance screening.
RNA Isolation and Semiquantitative RT-PCR
Total RNA was extracted from cells of each group. First-strand cDNA was synthesized from 1 µg of total RNA using Prime Script Kit (TAKARA, Dalian). The gene expressions of β-actin, CGI-100, GPA and Gfi-1B (growth factor independent 1B transcription repressor) were quantified by semiquantitative RT-PCR. The sequences of primers (Invitrogen, USA) used in the semi-quantitative RTPCR assays were listed in Table 2. β-actin was used as an endogenous control. Gene expression analyze was performed with the Quantity One software (BIO-RAD, USA). All experiments were repeated for at least three times.
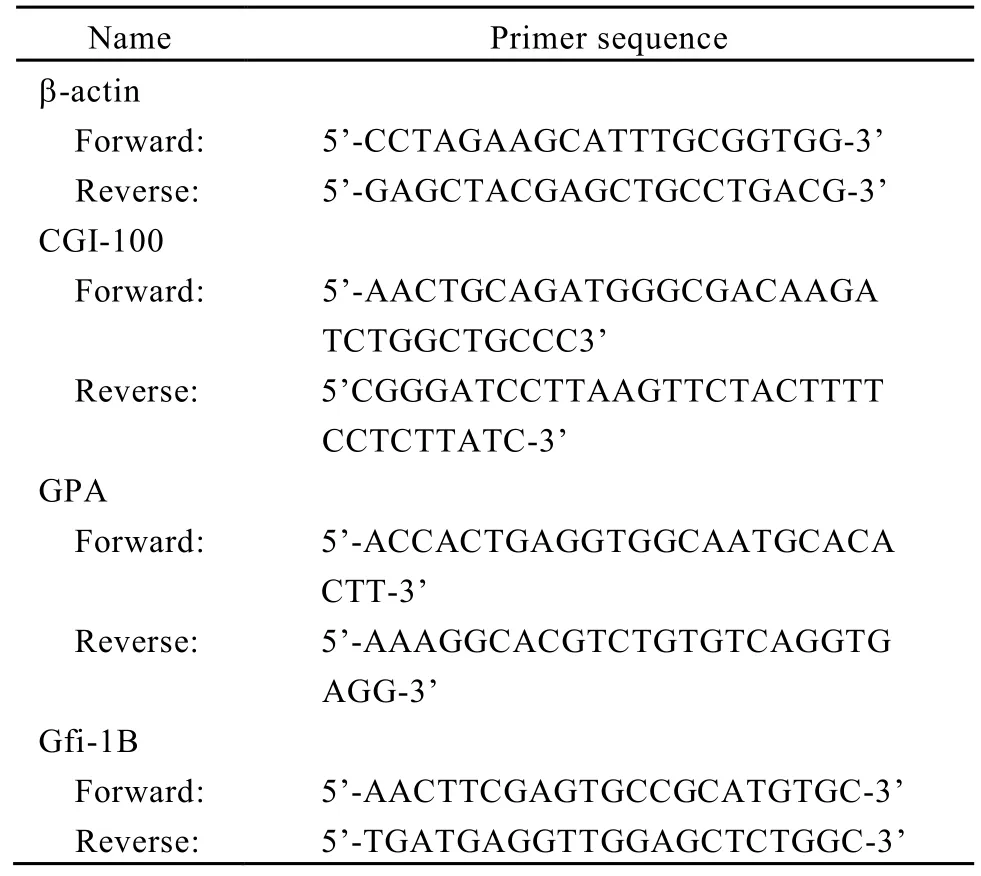
Table 2. Primers for semi-quantitative RT-PCR
CGI-100 mRNA Detection by Dot Blot Hybridization
CGI-100 and Vimentin probe were labeled with DIG random primer strictively according to the instructions provided by the Company(Roch). Labeling efficiency was analyzed with immune coloration. Total RNA was extracted from cells of each group and blotted on the membrance by specific device (BIO-RAD, USA). At the same time, internal controls (Vimentin gene) were set up for each sample. Hybridization and immune coloration were carried out according to the statement by the company(Roch). All experiments were repeated for at least three times.
Assessment of Cell Proliferation with MTT
A total of 5×103cells in 100 µl of the medium were plated respectively in 96-well plates. And 24 h later, 20 µl of the MTT reagent (Promega, Madison, WI) was added and incubated for 2 h. After the incubation, absorbance was measured at 490 nm using a microplate reader. All experiments were repeated for at least four times. The absorbance value was measured and proliferation inhibition rate(IR) was calculated as∶ Inhibition rate (IR)=(control group A-experimental group A)/control group A × 100%.
Cell Cycle Analysis by Flow Cytomety
A total of 1×104cells at logarithmic phase were seeded into a 6-well culture plate. Then the cells were harvested by centrifugation and washed twice with ice-cold PBS (pH 7.4). The cells were fixed in icecold 70% ethanol at least for 24 h at 4°C. Next, the cells were washed twice with PBS and resuspended in 1 ml DNA staining solution (50 μg/ml propidium iodide and 100 μg/ml RNase A in PBS) for 30 min. The analysis of cell cycle distribution was performed by Flow Cytometer and analyzed by CellQuest software package. Each experiment was repeated three times. Cell proliferation index (PI) was Calculated using the cell proliferation index formula∶PI (%)=(S + G2/M)/(G1 + S + G2/M) × 100%.
Transmission Electron Microscopy
Cells were collected, adjusted to 2×106/ml, treated with 4% glutaraldehyde, fixed by 1% osmium tetroxide, dehydrated with ethanol and acetone gradually, embedded by resin, and finally, observated with transmission electron microscopy after citrate staining.
Benzidine Staining
According to the method described in previous report[4], cells were seeded into a 24-well culture plate and cultured at 37°C with 5% CO2for 48 h. Then, 30μl of fresh benzidine staining were added at room temperature for 5 min. Blue cells observed by microscope were the positive cells and the percentage of positive cells was calculated from a total of 200 cells.
Detection of GPA, CD14 and CD15 Protein
A total of 1×105cells were seeded into a 6-well culture plate. After treatment the cells were harvested and monoclonal antibody (PE-tag, eBiosciecce Company) was added. The differentiation antigens of GPA, CD14 and CD15 were analyzed by flow cytometry. Moreover, logarithmic phase cells of each group were incubated with 30 μmol/L hemin (Sigma Company) for 24 h, the GPA protein tested.
Statistical Analysis
Statistical analysis was carried out using SPSS 11.5 software package for Windows. Data were showed as ¯x±s. The analysis of variance was used to assess the difference in expression level of multi-group comparison. LSD method was used for inter-group comparison. APvalue less than 0.05 was considered statistically significant.
RESULTS
CGI-100-knockdown by shRNA
shRNA recombinant vectors including CGI-100/ shRNA1, CGI-100/shRNA2, CGI-100/shRNA3 and negative control/shRNA were successfully established and correctly sequence-verified. Compared with other groups,inhibitory efficiencyof CGI-100 gene expression in CGI-100/shRNA1 group reached 54%. Statistical analysis showed no significant difference among the K562 control group, empty vector group and negative control /shRNA group (Figure 1). Hence, K562/shRNA-CGI-100-1 named as K562/shRNACGI-100 cells with the highest inhibition of CGI-100 expression was selected for the following experiments.
Effects of CGI-100-knockdown on K562 Cell Proliferation and Ultrastructures
The growth curve of each cell group indicated that the cell proliferation rate was much lower in K562/shRNA-CGI-100 cells compared with the untransfected K562 cells and K562/shRNA-pGenesil cells(P<0.05). The untransfected K562 and K562/ shRNA-pGenesil cells amplified very quickly, but no statistical significance was found for the difference (P>0.05) (Figure 2A). Additionally, we analyzed the effect of CGI-100-knockdown on ultrastructure in K562 cells. As shown in Figure 2B, electron microscopy indicated that most euchromatin, endocytoplasmic reticulum and bioblast were prone to be fond in K562/shRNA-CGI-100 cells. However, those organelles were much small in K562 cells. Additionally, phenomenona of chromatospherite excursion and nucleus introcession were obviously observed in K562 cell.
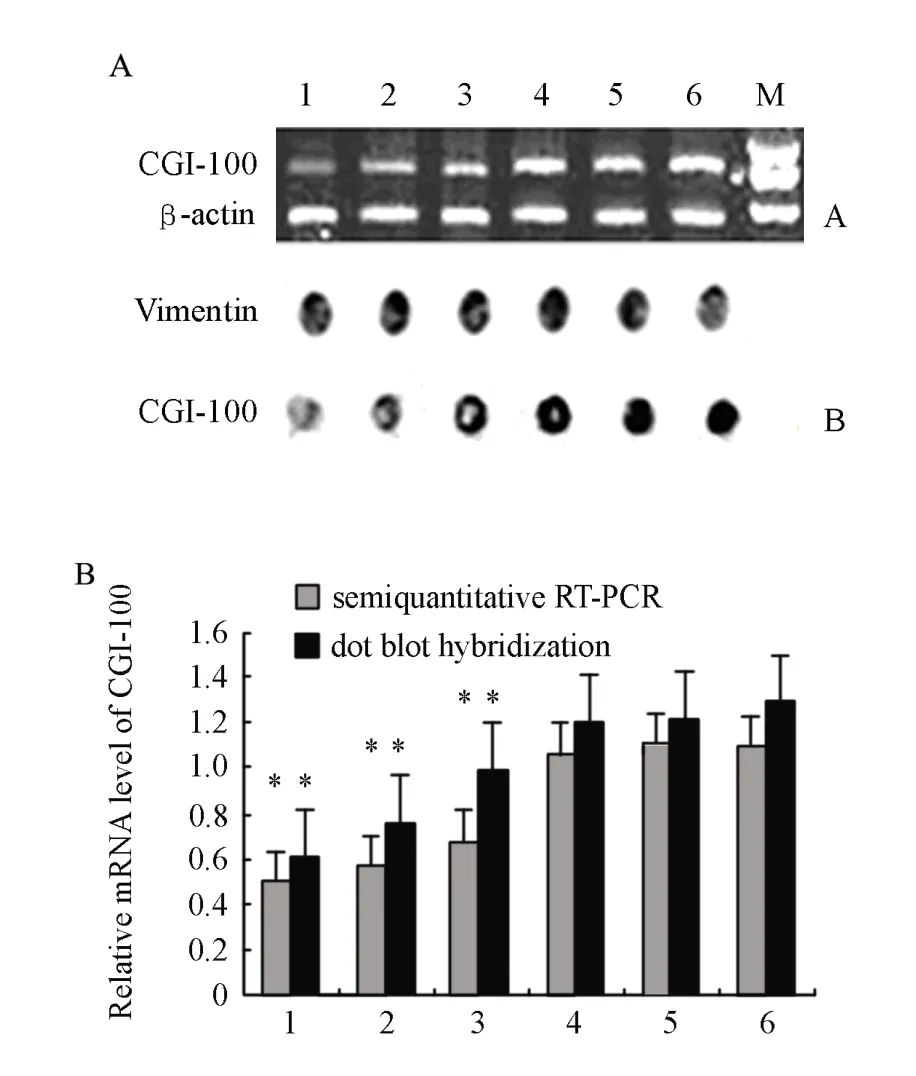
Figure 1. RNA interference mediated gene silencing of CGI-100 in K562 cells 2 weeks post-transfection. (A) CGI-100 mRNA transcripts detected by semi- quantitative RT-PCR and dot blot hybridization. M. DNA 2,000 marker; Lane 1. K562/shRNA-CGI-100-1; Lane 2. K562/shRNACGI-100-2; Lane 3. K562/ shRNA-CGI-100-3; Lane 4. K562/shRNA- pGenesil; Lane 5. K562/shRNA-Negative; Lane 6. K562. (B) Diagram presentation of OD ratios obtained for each corresponding lane or dot. Obviously, K562 cells transfected with CGI-100/shRNA1 exhibit the least transcription of CGI-100.*∶P<0.05vsK562/ shRNA-pGenesil and K562/shRNA-Negative and K562.
Effects of CGI-100-knockdown on Benzidine-Positive Staining
Compared with control K562 cells, the percentage of benzidine-positive staining cells of K562/shRNA-CGI-100 increased from 3% to 7% (P<0.05). After both groups were treated with 0.2mg/ml of matrine, the percentage of benzidinepositive staining cells increased from 16% to 23% (P<0.05). However, the percentage of benzidinepositive staining cells between untransfected K562 and K562/shRNA-pGenesil cells was almost the same and no statistical significance was found (P>0.05) (Figure 4A).
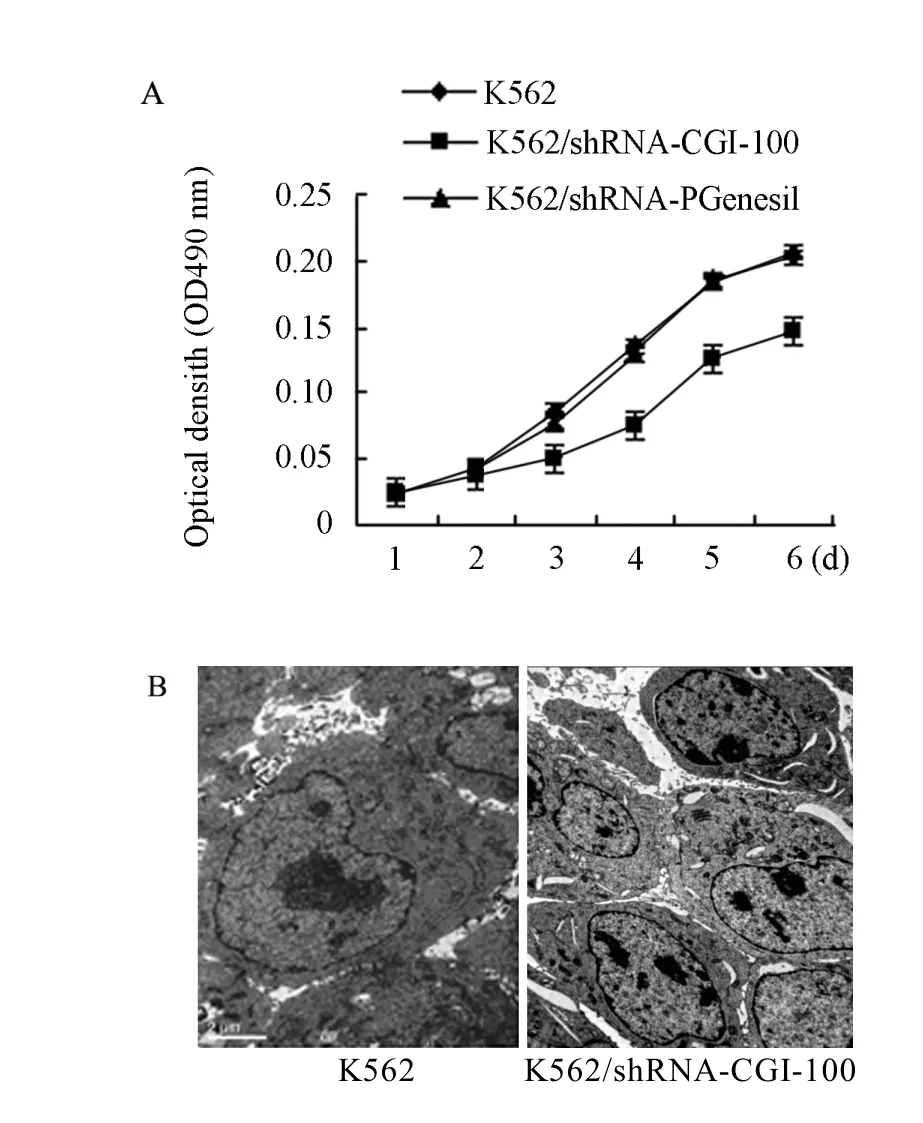
Figure 2. CGI-100 specific shRNA inhibites the proliferation of K562 cells (A) Proliferation curve of cells. (B) Electron microscopy of K562 and K562/ shRNA-CGI-100 cells; more euchromatin and endocytoplasmic reticulun and bioblast were found in K562/shRNA-CGI-100 cells, and chromatospherite exursion and nucleus introcession were obvious in K562 cells.*∶P<0.05 vs control group.
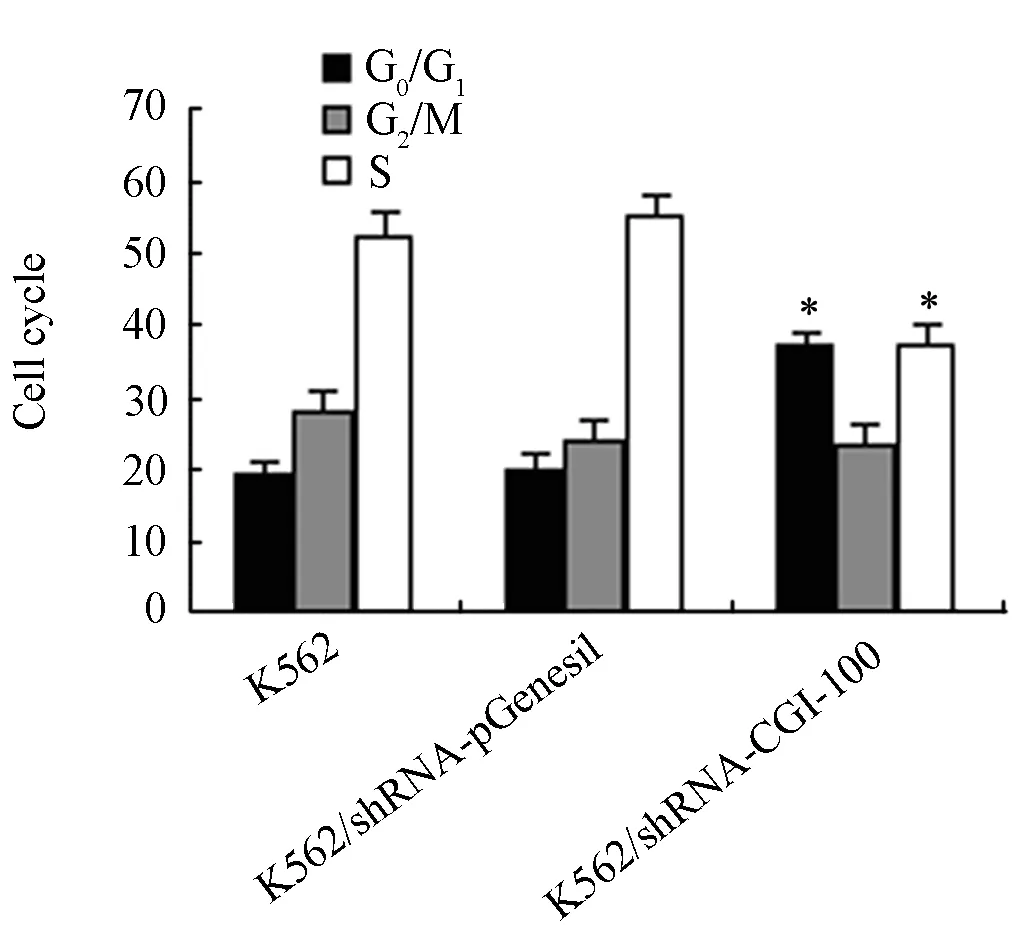
Figure 3. The effects of CGI-100 specific shRNAs on cell cycle.*∶P<0.05 vs control group.
Effects of CGI-100-konckdown on Cell Cycle
After inhibition of CGI-100 expression in K562 cells, Cell cycle was examined. Figure 3 showed the mean values of triplicate experiments. In comparison with in K562/shRNA-pGenesil and untransfected K562 cells, the percentage of G0/G1stage cells in K562/shRNA-CGI-100 cells increased from 21% to 35%. Cell population at S phase decreased from 56% to 38% (P<0.05). However, there was no statistically significant difference between K562/shRNA-pGenesil and untransfected K562 cells.
Effects of CGI-100-konckdown on the Inhibition Effect of Matrine
It was reported that matrine which were extracted fromkuhsenginhibited proliferation of K562 cells. Here we investigated whether sensitivity of K562 to matrine can be enhanced after knockdown of CGI-100 gene. Compared with control K562 cells, the inhibitory rate of proliferation of K562/shRNA-CGI-100 cells increased from 41% to 62% (P<0.05) after exposure to 0.2mg/ml of matrine (Figure 4B).
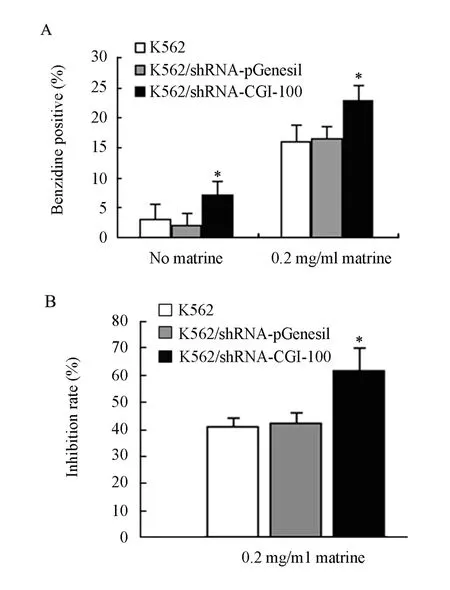
Figure 4. The effect of CGI-100 specific shRNAs on the percentage of benzidine-positive cells (A) and inhibition rate (B)*∶P<0.05vscontrol group.
Effects of CGI-100-knockdown on the Expressions of GPA and Gfi-1B mRNA
After K562/shRNA-CGI-100 and control K562 cells were treated with 0.2 mg/ml matrine, GPA mRNA expressions in both samples were increased. But the increase of GPA mRNA expression in K562/shRNA-CGI-100 was higher than in control K562 cells (P<0.05). Likewise, the mRNA expression of Gfi-1B in K562/shRNA-CGI-100 was obviously increased (P<0.05) (Figure 5).
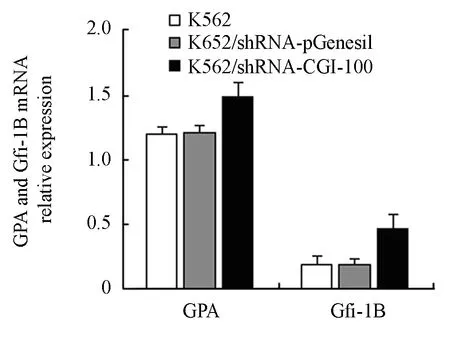
Figure 5. The effect of CGI-100 specific shRNAs on GPA and Gfi-1B mRNA expression measured by RT-PCR.*∶P<0.05 vs control group.
Effects of CGI-100-konckdown on Protein Levels of CD14, CD15 and GPA
To determine the effect of CGI-100-konckdown on matrine-induced differentiation in K562 cells, we investigated the expression levels of GPA, CD14 and CD15 in K562/shRNA-CGI-100 cells as well as control K562 cells treated with 0.2 mg/ml of matrine respectively. No expression of CD14 was tested. Although CD15 was found to be expressed in all the cells, no statistical significance was obtained (P>0.05). However, compared to control K562 cells, the mean fluorescence intensity (MFI) of GPA in K562/shRNA-CGI-100 cells increased by 31% (P<0.05) (Figure 6).
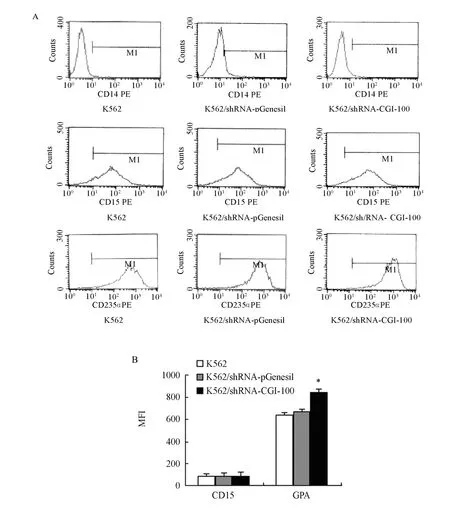
Figure 6. MFI of CD14, CD15 and GPA (MFI∶ mean fluorescence intensity). (A) MFI of CD14, CD15 and GPA in each group by flow cytometry; (B) MFI of GPA and CD15 expression in K562 cells, K562/shRNA-pGenesil cells and K562/shRNA-CGI-100 cells (n=4, MFI, ¯x±s).*P<0.05 vs control group.
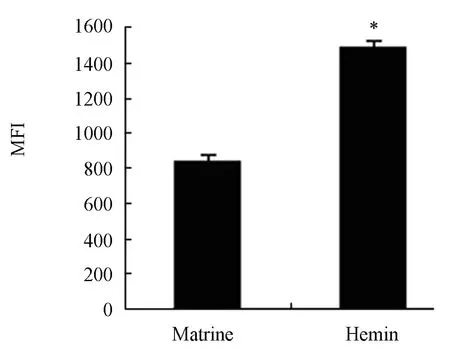
Figure 7. MFI values of GPA expression in K562/ shRNACGI-100 cells (n=4, MFI,¯x±s).*P<0.05 vs control group.
To evaluate the efficiency of matrine as an inducer of erythroid differentiation in K562 cells, K562/shRNA-CGI-100 cells and control K562 cells were treated with 30μmol/L hemin, which is generally accepted as a powerful erythroid differentiation inducer. Compared with control K562 cells, the MFI of GPA in K562/shRNACGI-100 cells increased by 20% (P<0.05), which was 1.7 times less than that in cells incubated with 0.2 mg/ml of matrine (P<0.05) (Figure 7 and Figure 8). K562 cells with CGI-100 knockdown were sensitive to matrine than to Hemin (Figure 9).
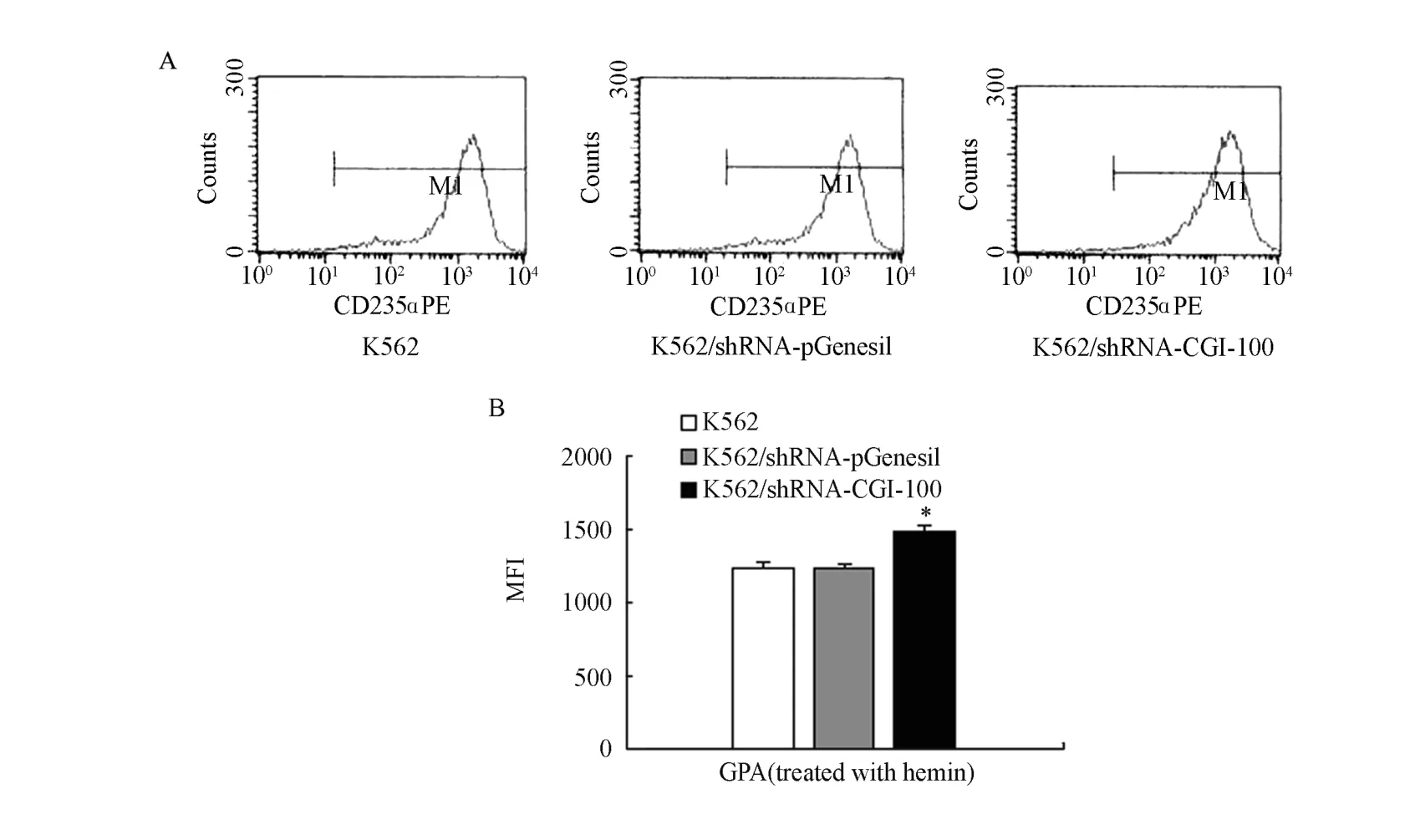
Figure 8. MFI of GPA expression (MFI∶ Mean fiuorescence intensity). (A) MFI of GPA in K562 cells, K562/shRNA-pGenesil cels and K562/shRNA-CGI- 100 cells deteced by flow cytometry; (B) MFI values of GPA expression in K562cell, K562/ shRNA-pGenesil cells and K562/shRNA-CGI-100 cells (n=4, MFI, ¯x±s).*P<0.05 vs control group.
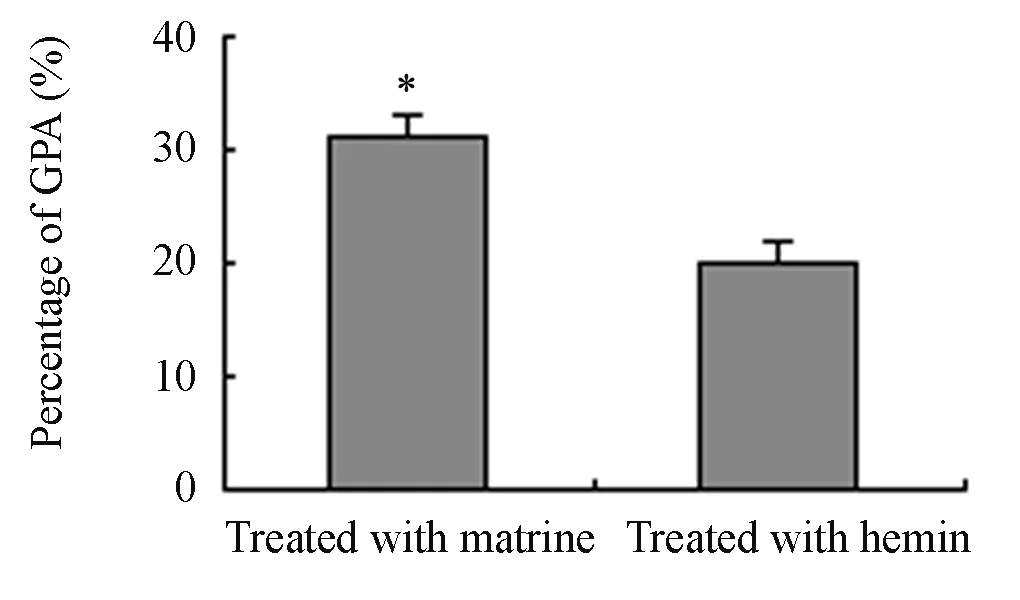
Figure 9. The MFI values of GPA expression in K562/ shRNA-CGI-100 and control K562 cells which were treated with matrine and hemin (n=4, %, ¯x±s).*P<0.05vsK562/shRNA-CGI-100 (GPA, Hemin treated) (MFI∶ Mean fluorescence intensity).
DISCUSSION
In this study, we established the K562/shRNACGI-100 cell line using CGI-100 specific shRNA and found that CGI-100 specfic shRNA inhibited cell growth and induced erythroid differentiation in K562 cells. Moreover, CGI-100 gene knockdown increased the sensitivity of K562 cells to matrine and hemin. The sensitivity of the K562/shRNA-CGI-100 cells to matrine was higher than that to hemin.
RNA interference technology which was discovered and named by Frie has been successfully used to research signal transduction and gene functionas well as gene therapy due to its much convenient and valid blockade of the gene expression[5,6]. At present, many methods creating siRNA were candidated such as chemosynthesis and transcription mediated by lentivirus and plasmid respectively. Nevertheless, the last one is considered to be the best method because of its simpleness, convenience and economy and has been widely used to study gene function especially for higher expression gene.
P24 protein family plays an important role in mediating transport of secreting proteins from endocytoplasmic reticulum to Golgi apparatus. Previous studies show that P24 protein interacted with Gas1p (glycosyl phosphatidyl inositol and GPI fixed protein) which is proved to be involved in the P53-mediated inhibition of growth or apoptosis of different tumor cells[7,8]. Our previous studies indicated that CGI-100 gene was highly expressed in K562 cells and the expression of CGI-100mRNA decreased rapidly in the course of erythroid differentiation and apoptosis induction of K562 cells treated by matrine.
To evaluate the effect of down-regulation of CGI-100 gene expression on K562 cells, we conductedin vitrotranscription of a eukaryotic expression plasmid-pGenesil-1 containing transcription promoter and three pairs of shRNA targeting CGI-100. As a result, three sets of recombinant vector named CGI-100/shRNA were successfully constructed and transfected into K562 cells. After identification and selection by RT-PCR and dot blot hybridization, one of the modified K562 cells, named as K562/shRNA-CGI-100 cells, which exhibited the highest inhibition efficiency of CGI-100 mRNA level by 54% was used. The negative control vector and empty plasmid did not decrease CGI-100 gene expression. The results suggest that the obtained shRNA-CGI-100 was specific.
Most of studies have shown that leukemia cells exhibit more powerful metabolism than normal blood cell, which primarily reflects in the nucleic acid metabolism, protein metabolism and glucose metabolism. The result of the metabolic changes affects not only the nucleus and cell organelles such as mitochondria, endoplasmic reticulum and lysosome, but also the structure and function of cells. One of the important biological features is that leukemia cells can continue to divide and proliferate unlimitedly, so we examined the proliferation of K562/shRNACGI-100 cells. As shown in Figure 1, the proliferation of K562/shRNA-CGI-100 cells decreased dramatically on day 3 and the tendency continued to day 6 compared with that of the control K562 cells(P<0.05). The K562/shRNA-CGI-100 cells also showed an arrest in G0/G1 phase, while cells in S phase increased. Besides this, the results observed by microelectron microscopy indicated that abundant euchromatin, endoplasmic reticulum, mitochondria and other organelles appeared in K562/shRNACGI-100 cells, while the phenomenona of nucleolar shift and nuclear convolution were obvious in control K562 cells. Meantime, the percentage of benzidinepositive cells increased. These results suggest that the down-regulated expression of CGI-100 can inhibit the proliferation and reduce the immaturation degree of K562 cells through DNA replication and DNA synthesis. Simultaneously, the erythroid differentiation potential of K562/shRNA-CGI-100 cells was much stronger than that of control K562 cells.
Differentiation of tumor refers to that malignant cells can develop to mature or nearly mature cells in exposure to inducersin vivoandin vitro. In recent years, tumor-induced differentiation has been widely studied and utilized as a treatment for cancers. Among erythroid differentiation inducers of leukemia cells, hemin is considered to be a strong inducer which is generally accepted as an erythroid differentiation inducer. Hemin was reported to activate the ferritin H gene transcription mediated by Ref-1, to inhibit PKC signal pathway and to activate the expression of EDAG gene related to erythroid differentiation through activation of NF[9,10].
In recent years, it was frequently reported that matrine which is extracted fromkuhsenghas anti-tumor activities. Matrine at low concentration was found to induce cell differentiation and promote cell apoptosis. But matrine caused tumor cells necrosis directly at high concentration. The reported mechanism underlying its apoptosis induction was that matrine induces the release of cytochrome C in K562 cells, activates caspase cascades, and inhibits the activity of tyrosinase[11]. The mechanism responsible for matrine-induced erythroid differentiation is related to up-regulation of expression of N-ras and P53 mRNA and reduction of c-myc gene expression[12,13]. Our early studies have shown that matrine with a low concentration changed the morphologic characteristic of K562 cells into late red blood cells[14]. Despite these findings, the mechanism underlying its effect of erythroid differentiation on K562 cells remains unclear. In this study, we observed the increased sensitivity to matrine in K562 cells with down-regulation of CGI-100 gene. MTT results indicated that compared to control K562 cells, matrine increased the inhibitory rate of proliferation of K562/shRNA-CGI-100 cells. Collectively, the results suggest that K562 cells with down-regulated CGI-100 gene are the good model forevulating both the function of CGI-100 and mechanism underlying the effect of matrine-induced erythroid differentiation.
In this study, several differentiation-related proteins were examined during the erythroid differentiation of K562 cells. Both CD14 and CD15 are the markers of monocytes and macrophages. Glycoprotein A, known as CD235a, is a sialoglycoprotein. All of these proteins are expressed on the surface of erythroblastic precursor cells, reticulocytes and mature red blood cells. They are often used as hallmarkers to distinguish category of blood cells. Compared with control cells, the percentage of benzidine-positive cells increased in K562/shRNACGI-100 cells after incubation with matrine, the expressions of GPA mRNA and the MFI of GPA protein were increased. Expression of CD14 was not detected. CD15 was tested, but no statistical significance was obtained. The increase of the MFI of GPA in K562/shRNA-CGI-100 cells treated with hemin was found but was less than that in cells incubated by marine. Although the potential of hemin-induced erythroid differentiation exceeded that of matrine, CGI-100 knockdown in K562 cells resulted in cells with higher sensitivity to matrine than that to hemin. These results suggest that CGI-100 gene may be associated with the pathway involved in matrine-indcued erythroid differentiation in K562 cells.
Regulation of cell differentiation was a process of multi-factor and multi-step complex. Gfi-1B is a zinc finger protein which is found to be able to bind to hematopoietic transcription factor GATA protein and contribute to the expression of erythroidspecific genes such as EKLF, NF-E2, α and β globin and EPO genes[15]. Our results indicated that Gfi-1B gene expression in K562/shRNA-CGI-100 cells elevated significantly. The results suggest that Gfi-1B gene may be involved in the process of erythroid differentiation induced by matrine
In summary, we found that CGI-100-knockdown inhibited the proliferation and induced erythroid differentiation in K562 cells. This down-regulation also enhanced the sensitivity of cells to matrine and hemin. Thus further study is worth to explore whether CGI-100 gene can serve as a potential target for leukemia treatment.
REFERENCES
[1] Muňiz M, Nuoffer C, Hauri HP, et al. The EMP24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles[J]. Cell Biol 2000; 148∶ 925-30.
[2] Huang FX, Zhang Y, Wang WJ. Effects of Matrine on CGI-100 Gene Expression and Proliferation in K562 Cells[J]. J Exp Hematol(in Chinese) 2008; 16∶525-30.
[3] Elbashir MS, Elbashir, Harborth J, Weber K, et al. Analysis of gene function in somatic mammalian cells using small interfering RNAs [J]. Methods 2002; 26∶199-213.
[4] Wang WJ, Zhang Y, Huang FX.The expression of IER3IP1 gene in K562 cells treated by matrine and its effect on the cell growth[J]. Zhonghua Xue Ye Xue Za Zhi(in Chinese) 2007; 28∶ 823-7.
[5] Hu X, Su F, Qin L, et al. Stable RNA interference of ErbB-2 gene synergistic with epirubicin suppresses breast cancer growthin vitroandin vivo[J]. Biochem Biophys Res Commun 2006; 346∶ 778-85.
[6] Crnković-Mertens I, Muley T, Meister M, et al. The anti-apoptotic livin gene is an important determinant for the apoptotic resistance of non-small cell lung cancer cells[J]. Lung Cancer 2006; 54∶135-42.
[7] Schimmőller F, Singer-Krüger B, Schroder S, et al. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi[J]. EMBO J 1995; 14∶1329-39.
[8] Peng R, De Antoni A, Gallwitz D. Evidence for overlapping and distinct functions in protein transport of coat protein sec24p family members[J]. J Biol Chen 2000; 275∶11521-8.
[9] Jasek E, Mirecka J, Litwin JA. Effect of differentiating agents (all-trans retinoic acid and phorbol 12-myristate 13-acetate) on drug sensitivity of HL60 and NB4 cellsin vitro[J].Folia Histochem Cytobiol 2008; 46∶ 323-30.
[10] Iwasaki K,Mackenzie EL,Hailemariam K, et al. Hemin-mediated regulation of an antioxidantresponsive element of the human ferritin H gene and role of Ref-1 during erythroid differentiation of K562 cells.,Mol Cell Biol 2006;26∶2845-2856.
[11] Li CY,Zhan YQ,Xu CW, et al. EDAG regulates the proliferation and differentiation of hematopoietic cells and resists cell apoptosis through the activation of nuclear factor-kappa B[J].Cell Death Differ2004; 11∶1299-308.
[12] Liu XS, Jiang J. Molecular mechanism of matrineinduced apoptosis in leukemia K562 cells[J]. Am J Chin Med 2006; 34∶ 1095-103.
[13] Jiang H,Hou C,Zhang S, et al. Matrine upregulates the cell cycle protein E2F-1 and triggers apoptosis via the mitochondrial pathway in K562 cells[J].Eur J Pharmacol 2007; 559∶ 98-108.
[14] Zhang Y, Ma LD, He YJ, et al.Gene expression profile in early differentiation of K562 cells induced by matrine[J]. Zhonghua Xue Ye Xue Za Zhi(in Chince) 2004; 25∶ 342-5.
[15] Saleque S, Kim J, Rooke HM, et al. Epigenetic Regulation of Hematopoietic Differentiation by Gfi-1 and Gfi-1b Is Mediated by the Cofactors CoREST and LSD1[J]. Molecular Cell 2007; 27∶ 562-72.
R733.7 Document code: A Article ID: 1000-9604(2010)02-0126-09
10.1007/s11670-010-0126-4
2009−10−28; Accepted 2010−02−25
This work was supported by the National Natural Science Foundation of China(No.30171150);
*Contributed equally to this study
**Corresponding author.
E-mail∶ zhangyan2753@yahoo. cn
© Chinese Anti-Cancer Association and Springer-Verlag Berlin Heidelberg 2010
杂志排行
Chinese Journal of Cancer Research的其它文章
- Triptolide Inhibits Cell Growth and Induces G0- G1 Arrest by Regulating P21wap1/cip1 and P27 kip1 in Human Multiple Myeloma RPMI-8226 Cells
- Different Outcome of Myeloid Sarcoma with Spinal Cord Compression Preceding Acute Myeloid Leukemia: Report of Two Cases and Review of Literatures
- Effects of Triptolide on Histone Acetylation and HDAC8 Expression in Multiple Myeloma in vitro
- Expression of Embryonic Stem Cell Marker Oct-4 and Its Prognostic Significance in Rectal Adenocarcinoma
- Differential Diagnosis of Warthin's Tumor Complicated with Lung Adenocarcinoma by 18F- FDG PET/CT Imaging and Radioisotope Scanning with Tc-99m Pertechnetate: A Case Report and Literature Review
- Synergistic Action of fMLP-boanmycin Combination on the Growth of Mouse Colon Carcinoma and Its Action Mechanisms
