A mutation in the promoter of the yellow stripe-like transporter gene in cucumber results in a yellow cotyledon phenotype
2024-03-12JiaweiPanJiaSongRahatSharifXuewenXuShutongLiXuehaoChen
Jiawei Pan ,Jia Song ,Rahat Sharif ,Xuewen Xu ,Shutong Li ,Xuehao Chen#
1 School of Horticulture and Landscape Architecture, Yangzhou University, Yangzhou 225009, China
2 Institute of Vegetables, Tianjin Academy of Agricultural Sciences, Tianjin 300384, China
Abstract Leaf color mutants in higher plants are considered to be ideal materials for studying the chlorophyll biosynthesis,photosynthesis mechanism and chloroplast development. Herein,we identified a spontaneous mutant,yc412,in cultivated cucumber that exhibited yellow cotyledons. The yellow-lethal mutant was diagnosed with an abnormal chloroplast ultrastructure,and reduced photosynthetic capacity and pigment content. Through bulked segregant analysis-based whole-genome sequencing and fine genetic mapping,we narrowed the yellow cotyledons (yc) locus to a 96.8 kb interval on chromosome 3. By resequencing and molecular cloning,we showed that Csyc is a potential candidate gene,which encodes a yellow stripe-like (YSL) transporter. The T to C mutation in the promoter region of Csyc caused the yellow cotyledon phenotype in yc412. Compared to YZU027A (WT),the expression of Csyc was significantly downregulated in the cotyledons of yc412. Silencing of Csyc in cucumber via virus-induced gene silencing resulted in chlorotic leaves,mainly by suppressing the chlorophyll content. Furthermore,a comparative transcriptome analysis revealed that chloroplast-related genes and chlorophyll biosynthesis genes were significantly downregulated in yc412 cotyledons. Our results provide new insights into the molecular function of the YSL transporter in plant chloroplast development and chlorophyll synthesis.
Keywords: cucumber,yellow-lethal cotyledons,chloroplast development,yellow stripe-like transporter
1.Introduction
Plant leaves are an important organ that decides the fate of the photosynthetic mechanism under normal and adverse environmental conditions (Wanget al.2016;Fuet al.2019). The frequency of leaf color variation is relatively high in the plant kingdom,and the sources of mutation are many,so color variation is an easy-toidentify genetic phenomenon (Parket al.2007;Sugimotoet al.2007;Fenget al.2019;Zhenget al.2019). Plant leaves are predominantly green,resulting from the combined effect of light energy absorption by chloroplast pigments (Frommeet al.2003;Yamataniet al.2022).Differences in the plant growth environment and genotype often lead to changes in leaf color (Estebanet al.2008;Luet al.2012). Mutations in leaf color often compromise photosynthesis,which can seriously affect crop yield and economic benefits (Chenet al.2013;Huanget al.2016).Yellow leaf mutants have been identified in various crops,including rice (Yooet al.2009),maize (Xinget al.2014),soybean (Zhuet al.2020),cotton (Maoet al.2019),Arabidopsis(Nagataet al.2005),cabbage (Zhanget al.2017),tomato (Chenget al.2022),and pepper (Shiet al.2022). These mutants can be used not only to study chlorophyll biosynthesis and catabolism,but also as markers for identifying seed purity (Sakowskaet al.2018;Donget al.2020;Panget al.2022).
Many chlorophyll and chloroplast development related mutations affecting leaf coloration have been reported.For instance,Wuet al.(2007) identified a rice Chldeficient mutant,yellow-greenleaf1(ygl1),with yellowgreen leaves in young plants due to disrupted chlorophyll synthesis. In addition,for a new type of guanylate kinase gene,pt/mtGK,rice plants with loss-of-function mutations develop chlorotic leaves with inhibited chloroplast differentiation and disrupted chloroplast translation machinery during early leaf development (Sugimotoet al.2007). The yellow stripe-like (YSL) transporter family is also involved in leaf color change. This family is mainly responsible for the transport of heavy metal ions in gramineous plants,particularly Fe3+(Curieet al.2009).Fe is one of the essential trace elements for plant growth and development,and it is a key in plant photosynthesis,respiration,and enzymatic reactions (Kobayashi and Nishizawa 2012). Fe deficiency can lead to impaired chlorophyll synthesis and the promotion of leaf chlorosis,and in the process affect crop productivity (Gaoet al.2022;Zhaiet al.2022). Maize (Zeamays)YellowStripe1is the founding member of theYSLfamily,which mediates Fe uptake from the soil (Curieet al.2001).TheArabidopsisysl1ysl3double mutant plants can grow normally,but they display Fe deficiency symptoms,including strong interveinal chlorosis (Waterset al.2006).
In cucumber (CucumissativusL.,2n=2x=14),several leaf color mutants have been identified. Miaoet al.(2016) found a virescent leaf mutant showing yellow cotyledons and true leaves,which gradually turn green during leaf development. The phenotype is caused by a single nucleotide mutation in the promoter region ofCsaCNGCs,which encodes a cyclic-nucleotidegated ion channel protein. The chlorophyll-deficient golden cotyledon and true leaf mutation is due to a nonsynonymous mutation in the third exon ofCsChlI,which encodes a subunit of magnesium chelatase (Gaoet al.2016). Songet al.(2018) identified a virescentyellow leaf mutant that exhibits reduced pigment contents and delayed chloroplast development.Fine genetic mapping and sequence alignment showed that the mutation was due to a nonsynonymous mutation (C to T)in the first exon ofCsa4G637110,which encodes a DnaJlike zinc finger protein. Dinget al.(2019) found that four missense mutations in four tandem13-Lipoxygenasegenes resulted in the yellow-green cotyledon and true leaf phenotype. A spontaneously mutated virescent leaf mutant,104Y,had yellow cotyledons and the upper five true leaves. With increasing chlorophyll content,the yellow true leaves gradually turned green from top to bottom in 104Y due to a mutation in the auxin F-box protein gene (Zhanget al.2020). An EMS-induced young yellow leaf mutant,C777,presented yellow cotyledons and emerging true leaves. The small dots on these true leaves turned green gradually with leaf growth. The geneCsHDencoding a histidine and aspartic acid (HD)-containing protein was identified as the causal gene(Huet al.2020). Xionget al.(2021) identified an EMSinduced mutant,yl2.1,with yellow cotyledons,while the true leaves did not turn green.CsYL2.1encodes a plastid isoform of triose phosphate isomerase (pdTPI),which catalyzes the reversible conversion of dihydroxyacetone phosphate (DHAP) to glyceraldehyde-3-phosphate(GAP) in chloroplasts. Recent studies have shown that a 7 kb deletion mutation in theCsSRP43gene coding sequence and promoter region leads to abnormal chloroplast development and yellow cotyledons along with true leaves (Zhanget al.2022).CscpFtsYencodes one homolog of the chloroplast signal-recognition particle(cpSRP) receptor,and it controls the cucumber yellow leaf phenotype (Zhaet al.2022). In the present study,a novel yellow-lethal cotyledon mutant,yc412,was identified from the cucumber inbred line YZU027A. Our results showed that the mutation is due to a single nucleotide substitution in the promoter ofCsaV3_3G026880,which encodes a YSL transporter.
2.Materials and methods
2.1.Plant materials and mapping populations
A spontaneous mutant,yc412,possessing yellow-lethal cotyledons was obtained from the cucumber inbred line YZU027A,which was originally collected from Central/West Asia. The mutant seedlings failed to produce true leaves and exhibited reduced growth and lethality at the cotyledon stage. Sinceyc412showed growth arrest and death,normal self-pollination or hybridization could not be performed. Therefore,an F2population was constructed by crossing a heterozygous plant for the mutant allele fromycwith the YZU055A (an inbred line with green leaves) in this study. All the plant materials were grown in a greenhouse.
2.2.Measurement of chlorophyll contents and photosynthetic characteristics
The cotyledons of YZU027A andyc412plants were selected when they were fully expanded after germination.The contents of chlorophylla(Chla),chlorophyllb(Chlb)and total chlorophyll were determined. Leaves were ground and mixed in even proportions. A 0.2 g sample was weighed into a 50 mL centrifuge tube,20 mL of extract liquid(75% ethanol solution in an 8:1 volume ratio) was added in a fume hood,and the sample was placed in the dark for two days until the leaves turned white. The supernatant was collected by centrifugation,and the absorbance values at 663,645 and 470 nm were measured using a UV spectrophotometer (MAPADA,UV-1800PC,Shanghai,China). Wild type (WT) andyc412plants were selected,and three replicates were set. We used the following equations to calculate the chlorophyll contents:
Chla(mg g–1)=(12.7OD663–2.69OD645)×(20/0.2×1,000)
Chlb(mg g–1)=(22.9OD645–4.68OD663)×(20/0.2×1,000)
Chlorophyll (mg g–1)=Chla(mg g–1)+Chlb(mg g–1)
Carotenoid (mg g–1)=[OD470×(1/100)–3.27×Chla–104×Chlb]/198
The uniform cucumber cotyledons of YZU027A andyc412plants were used to measure photosynthetic rate,stomatal conductance,intercellular CO2concentration,and transpiration rate with a portable chlorophyll fluorescence spectrometer (LI-COR,LI-600,Beijing,China). Each independent biological replication included three individuals,and three biological replications were repeated thrice. Values presented are the mean±SD.
2.3.Transmission electron microscopy
For transmission electron microscopy,cotyledons from one-week-old seedlings were cut into 1 m3pieces,fixed in 2.5% glutaraldehyde in 0.1 mol L–1phosphate buffer (pH=7.4) at 4°C for 4 h,rinsed,and further fixed overnight in 1% OsO4solution at 4°C. The samples were subsequently dehydrated,embedded,sectioned,and stained,as described by Longet al.(2022). The chloroplast structure of cotyledon cells was observed with a JEM-1400 Plus transmission electron microscope(JEOL,Tokyo,Japan).
2.4.DNA isolation and sequencing
We randomly selected 25 normal green and 25 yellow leaf individuals from the YZU055A×YZU027A F2population to construct two DNA pools. Genomic DNA was extracted by the CTAB method (Murray and Thompson 1980). BSA was performed for the initial mapping of theyclocus using the two DNA bulks,the green pool and yellow pool,which were made by bulking equal amounts of tissues. YZU027A,yc412,YZU055A,and the two pools were subject to paired-end sequencing with the DNB seq T7 platform. The five libraries were constructed and sequenced at Anoroad Technologies Corporation(Beijing,China). After filtering raw data,the clean reads were mapped to the cucumber V3 reference genome(http://cucurbitgenomics.org/organism/20) with Burrows-Wheeler-Alignment (Li and Durbin 2009). Variants(SNPs and InDels) were called using Genome Analysis Toolkit (GATK) (McKennaet al.2010). We used the welldocumented Euclidean distance (ED) method,followed by loess regression analysis,to calculate the genotype frequency of each SNP between the two pools (Hillet al.2013). The ED values were calculated as follows:
where Ay,Cy,Gy,and Ty represent the depths of bases A,C,G,and T on an SNP site in the yellow pool,respectively,and Ag,Cg,Gg,and Tg represent the depths of bases A,C,G,and T on the same SNP site in the green pool,respectively. Only the peak region with the loess-fitted values of the markers above the threshold of the 99% confidence interval was considered.
2.5.Molecular cloning and phylogenetic analysis
To identify the candidate gene associated withyc,the DNA sequences of the candidate gene were cloned fromyc412,YZU027A and YZU055A. Oligo synthesis and Sanger sequencing of amplicons were performed by Tsingke Inc.(Beijing,China). The DNAMAN was used for carrying out multiple sequence alignment. A phylogenetic tree was constructed with the MEGA X software based on the amino acid sequences of Csyc and its homologs in other species (Saitou and Nei 1987).The amino acid sequences used in the multiple sequence alignment and phylogenetic tree construction were from the following species:Cucumissativus(accession no.XP_004148009.1),Cucumismelo(accession no.XP_008450132.1),Cucurbitamaxima(accession no.XP_022989604.1),Momordicacharantia(accession no.XP_022156489.1),Benincasahispida(accession no.XP_038894292.1),Cucurbitamoschata(accession no.XP_022928209.1),Brassicanapus(accession no.XP_009111552.1),Arabidopsisthaliana(accession no.NP_566806.1),Gossypiumhirsutum(accession no.XP_016678190.2),Capsicumannuum(accession no.XP_016561741.1),Ziziphusjujubavar.spinosa(accession no.XP_015868704.1),Ricinuscommunis(accession no.XP_015570953.1),Prunusavium(accession no.XP_021823044.1),Oryzasativa(accession no.NP_001389280.1),andZeamays(accession no.NP_001131175.1) (Thompsonet al.1994).
2.6.Histochemical GUS staining assay
TheCsycpromoter sequences were amplified from YZU027A andyc412and cloned into theHindIII andPstI restriction sites of the pCAMBIA1391 vector to generate thepCsyctandpCsyccconstructs. After Sanger sequencing,the recombinant constructs were transformed intoAgrobacteriumtumefaciensGV3101. Tobacco leaves of five-week-old plants were infiltrated withAgrobacteriumcultures carrying thepCsyctandpCsyccconstructs separately. For GUS staining,fresh five-week-old tobacco leaves after two days of inoculation were collected and incubated with X-Gluc solution (Solarbio,Beijing,China). After incubation at 37°C for 12 h,the tissue was washed several times with 70% ethanol until the tissue had cleared before mounting and imaging with a stereo microscope (MZ62,Mshot,Guangzhou,China). Primers for the GUS construct are shown in Appendix A.
2.7.Virus-induced gene silencing
The function ofCsycwas characterized by VIGS following the method of Liuet al.(2020).The pV190 vector was kindly provided by Dr.Qinsheng Gu from the Zhengzhou Fruit Research Institute,Chinese Academy of Agricultural Sciences.TheCsyc(+115 to +414 bp) coding sequences were amplified from YZU027A by PCR to produce 300 bp fragments. After confirmation by Sanger sequencing,the fragments were cloned into theBamHI restriction site of the pV190 vector to generate the pV190-Csycconstruct.The recombinant construct and the empty pV190 vector were transformed intoA.tumefaciensstrain GV3101. Positive monoclonals were transferred to 200 μL LB containing 50 mg mL–1kanamycin and 50 mg mL–1rifampicin,and the cells were incubated overnight at 28°C on a shaker. The bacterial solution was mixed with LB at a ratio of 1:100,incubated overnight at 28°C,centrifuged at 6,000×g for 5 min,and the cultured cells were collected and resuspended in induction buffer containing 0.2 g L–1acetosyringone,0.4 g L–1l-cysteine and 5 mL L–1Tween 20 (Solarbio). The final OD600value was adjusted to 0.8 and the suspension was soaked for 4 h at room temperature. The resuspended bacterial solution was infiltrated into the cotyledons of 7-day-old seedlings with a 1 mL syringe. Finally,the infiltrated plants were placed in a dark incubator at 18°C and transferred to normal growth conditions (28°C/18°C day/night) after 24 h. Two weeks after inoculation,the VIGS efficiency ofCsycin the agroinfiltrated cucumber leaves was evaluated using qRTPCR. Three technical and three biological replicates were used for each sample. Primers for the VIGS construct are shown in Appendix A.
2.8.RNA-sequencing and comparative transcriptome analysis
The cotyledons of YZU027A andyc412were harvested and immediately frozen in liquid nitrogen and stored at–80°C. Total RNA was extracted using the RNA Easy Fast Plant Tissue Kit (Tiangen,Beijing,China) according to the manufacturer’s instructions. Single-stranded cDNA libraries were prepared for each sample using 500 ng of total RNA with a TruSeq RNA Sample Prep Kit V2 (Illumina,San Diego,CA,USA) according to the manufacturer’s instructions. Six libraries were sequenced using the MGI DNBSEQ T7 platform by ANOROAD Technologies(Beijing,China). The clean reads were mapped to the cucumber 9930 version 3 reference genome assembly(http://cucurbitgenomics.org/organism/20) using HISAT2(Sirenet al.2014). After alignment,the number of reads for each cucumber gene model was obtained and normalized by Fragments Per Kilobase of transcript per Million mapped fragments (FPKM) (Trapnellet al.2010). The R package ‘DESeq’ was used to identify the differentially expressed genes (DEGs) between theyc412and YZU027A samples (Loveet al.2014) based on |log2FC(fold change)|≥1 coupled with an adjusted false discovery rate≤0.05. Pathway enrichment analysis was conducted to elucidate the significant pathways of DEGs based on the Gene Ontology (GO) database (http://www.geneontology.org/) and Kyoto Encyclopedia of Gene and Genomes (KEGG) databases (http://www.genome.jp/kegg). The raw RNA-sequencing reads have been deposited in the NCBI Sequence Read Archive (SRA)under accession number PRJNA848016.
2.9.qRT-PCR analysis
Total RNA was extracted from the cotyledons of YZU027A andyc412using the RNA Easy Fast Plant Tissue Kit (Tiangen,Beijing,China) according to the manufacturer’s instructions. First-strand cDNA was synthesized using HiScript®III RT SuperMix for qPCR(+gDNA wiper) (Vazyme,Nanjing,China). Gene expression was analyzed by qRT-PCR using ChamQ SYBR Color qPCR Master Mix (Vazyme,Nanjing,China). The cucumberβ-actin(GenBank AB010922)gene was selected as an internal control to normalize mRNA abundance. All experiments were performed in three biological replicates.Relative gene expression levels were calculated using the 2–ΔΔCtmethod (Livak and Schmittgen 2001). Gene-specific primers for the qRTPCR analysis were designed using the online software Primer 3Plus (https://www.primer3plus.com/index.html)and are listed in Appendix A.
3.Results
3.1.The yc412 mutant displays defective in chloroplast development
The spontaneous mutantyc412exhibits a yellow cotyledon and a lethal phenotype aborted 10 days after germination (Fig.1-A). Chla,Chlb,carotenoid,and total chlorophyll levels were significantly reduced,and respectively accounted for only 2.4,2.2,1.9,and 2.3%of the levels in WT plants (Fig.1-B). The photosynthetic parameters of 8-day-old YZU027A andyc412plants at the cotyledon stage were measured. As expected,the net photosynthetic rate,stomatal conductance and transpiration rate inyc412were significantly reduced compared to WT plants by 67.4,66.7 and 45.7%,respectively,and the intercellular CO2concentration was higher than in WT plants by 31.5% (Fig.1-C–F). The chlorophyll fluorescence parameters of PSI and PSII were further compared to evaluate whether the photosynthetic apparatus was affected inyc412. Apparent reductions in the electron transport rate and maximum photochemical efficiency of photosystem II were observed in theyc412mutant (Fig.1-G and H). These observations suggested that theycmutation had crippled the photosynthesis machinery.
To explore the effect ofycon chloroplast structure development,we compared the ultrastructure of chloroplasts between theyc412and WT plants by transmission electron microscopy. The chloroplasts of WT cotyledons had an intact thylakoid and granum structure with densely arranged grana lamellae (Fig.2-A–C). On the contrary,the chloroplast development was disrupted in theyc412mutants,and the thylakoid membrane structure was abnormal,with loose and disordered grana lamellae. In addition,theyc412chloroplasts accumulated fewer starch granules in the chloroplast stroma than WT(Fig.2-D–F).
3.2.Mapping of the yc locus
To map theyclocus,we crossed YZU027A (which was heterozygous at theyclocus) with YZU055A to construct an F2segregation population (Fig.3-A). All F1plants had normal green coloration,which suggested the recessive nature of the mutation. Among the 1,087 F2plants,959 and 128 had green and yellow leaves,respectively,which was inconsistent with a 3:1 segregation ratio in theχ2test.

Fig. 1 Comparison of the yc412 (mutant) and YZU027A (wild type). A,yc412 and YZU027A seedlings at 5 (left),8 (middle),and 10 (right) days post germination. B,chlorophyll contents of the cotyledons in YZU027A and yc412. Chl,chorophyll;Caro,carotenoid. C–H,photosynthetic parameters in YZU027A and yc412. Values are means±SD (n=9). ****,P<0.0001 and ***,P<0.001 by Student’s t-test.

Fig. 2 Chloroplast structures of YZU027A (wild type;A–C) and yc412 (mutant;D–F). SG,starch granules;TM,thylakoid membranes;GL,grana lamellae. A and D,bar=2 μm;B and E,bar=1 μm;C and F,bar=500 nm.

Fig. 3 Mapping of the yc locus. A,phenotypes of the F2 population derived from the cross between YZU027A (a wild-type plant that is heterozygous for the mutant allele) and YZU055A. B,Euclidean distance (ED) algorithm for mapping genomic regions that possibly harbor the causal mutation. C,linkage analysis with molecular markers on Chr3 placed the yc locus to a 13.5 cM interval flanked by SNP23167277 and SNP23264091. D,sequence alignment of the partial promoter sequence of the Csyc gene between YZU027A,YZU055A,9930 and yc412. There was a base substitution (T to C) in the promoter region of Csyc. E,expression changes of Csyc in cotyledons of YZU027A and yc412. All experiments were performed in triplicate and repeated three times.The expression of YZU027A plants was set to 1. Bars show mean±standard deviation.**,P<0.01 by Student’s t-test.
Among the 1,087 F2,25 individuals with yellow cotyledons and 25 individuals with normal green cotyledons were selected for DNA pools. High-throughput sequencing generated 1,588 million and 152 million paired-end reads in the yellow and green cotyledon pools,respectively. More than 98% of the reads from the two bulked samples were mapped to the cucumber 9930 genome (v3.0). In total,786,134 and 729,855 single nucleotide polymorphisms(SNPs) in the yellow cotyledon pool and the green cotyledon pool,respectively,were identified after their alignments with the reference genome. For each SNP position,the value of the SNP index was obtained and a graph relating SNP positions and SNP index values was generated for all seven cucumber chromosomes (Fig.3-B).Using the top 1% of ED values as a threshold,we mapped theyclocus to the physical position between 22.90 and 23.24 Mb,and 26.56 and 27.23 Mb (above the red dotted line) on Chr3 (Fig.3-B).
To validate the results of the ED method,four polymorphic markers on Chr3,indel21973062,SNP23167277,SNP23264091,and SNP23275525,were used to genotype 850 randomly selected individuals from a total of 1,087 YZU027A×YZU055A F2plants.The linkage analysis using the genotype and phenotype data of the selected F2individuals narrowed theyclocus to a 14.5 cM region flanked by SNP23167277 and SNP23264091 (Fig.3-C). We resequenced five YZU027A (WT) and fiveyc412mutants to identify the causal mutation. An analysis of the variants in YZU027A andyc412detected five SNPs in the 96.8 kb region flanked by SNP23167277 and SNP23264091(Appendix B). Specifically,they are SNP23180528 (T to A),SNP23191522 (A to T),SNP23212419 (A to C),SNP23212420 (C to A),and SNP23218243 (T to C).Among them,only SNP23218243 (T to C) was found to be located in the promoter region ofCsaV3_3G026880,which encodes a YSL transporter. To further confirm the accuracy of the causal mutation,a dCAPs marker was designed to genotype the YZU027A×YZU055A F2plants. The results showed that all F2plants with the mutant phenotype exhibited the same bands withyc412,while all plants with green cotyledons exhibited the same bands with YZU027A or heterozygous bands,suggesting that SNP23218243 is co-segregated with theyclocus. We confirmed the presence of this SNP (T to C) using Sanger sequencing (Fig.3-D). The expression ofCsaV3_3G026880was significantly downregulated in cotyledons ofyc412relative to the WT (Fig.3-E). These results strongly suggested thatCsaV3_3G026880is a potential candidate for theyclocus. We thus namedCsaV3_3G026880asCsycfor future reference.
3.3.A Csyc promoter polymorphism is associated with Csyc expression
Gene expression in eukaryotic cells is typically regulated bycis-acting regulatory elements in the promoter regions(Mikhaylichenkoet al.2018). We cloned the promoter region ofCsycfrom YZU027A andyc412to investigate the cause of the differential expression ofCsycbetween YZU027A andyc412. To characterize thecis-regulatory elements ofCsyc,we used the online PLACE program to search for the putativecis-elements present in the promoter region. This analysis revealed that a T to C mutation led to deletion of the MYC (CATTTG) in the promoter ofCsycinyc412(Fig.4-A).
The 1,575-bp promoter sequences (Chr3: 23,217,201–23,218,775 bp) from YZU027A (pCsyct) and theyc412mutant (pCsycc) were cloned to producepCsyct-GUS andpCsycc-GUS constructs,respectively. Histochemical staining showed that tobacco leaves transformed withpCsyccexhibited weaker GUS expression thanpCsyct(Fig.4-B). This result is consistent with the higher expression level ofCsycin YZU027A than inyc412(Fig.4-B),which also supports the hypothesis thatCsycexpression iscis-regulated.
3.4.Phylogenetic analysis of Csyc
We constructed a phylogenetic tree using putative orthologous proteins from 14 species (Fig.4-C). The results showed thatCsychas the highest sequence identity with XP_008450132.1 fromC.melo. The orthologs from other cucurbitaceous plants (C.maxima,C.moschata,B.hispidaandM.charantia) are located in the same evolutionary branch. We found that the ortholog genes of all dicotyledons,including the cruciferous plants (B.napusandA.thaliana),are clustered together separately,while the ortholog genes of monocotyledons (O.sativaandZ.mays) are located in an outer branch. These results suggested that the amino acid sequences of Csyc and its orthologs are highly conserved in plants (Fig.4-C).
3.5.VlGS assay of Csyc in cucumber
We silencedCsycin cucumber by employing the VIGS technique to confirm its function. The silencing effect of the VIGS system was monitored using cucumber phytoene desaturase (CsPDS,CsGy4G002600) as a visible marker.Agrobacteriumcontaining the pV190-CsPDS vector was infiltrated into the cucumber Superina,which displayed an albino phenotype at 15 days after inoculation,indicating that the VIGS system was effective(Fig.5-B). The silencing effects ofCsycwere confirmed by qRT-PCR. The expression ofCsycwas reduced significantly (54.5%) in cucumber leaves infected with pV190-Csyccompared to those infected with the pV190 empty vector (Fig.5-A). As expected,cucumber plants infected withCsycexhibited chlorotic leaf phenotypes and an approximately 33.7% reduction in chlorophyll content(Fig.5-C). These results indicated thatCsycis required for chlorophyll accumulation in cucumber.
3.6.Transcriptome profiling reveals the regulated network of Csyc

Fig. 4 Effects of mutations in the upstream promoter region of Csyc and phylogenetic analysis of Csyc in cucumber and its homologous proteins in different species. A,cis-element prediction using the Csyc promoter around the T to C mutation. The numbers represent the positions relative to the transcription start site of Csyc. The T to C mutation is marked in yellow. The core sequences of MYC (CATTTG) are highlighted in green. B,qualitative analyses of the activities of the Csyc promoters in tobacco leaves based on GUS histochemical staining. C,CLUSTAL W multi-sequence alignment was performed with amino acid sequences,and an unrooted phylogenetic neighbor-joining tree was constructed using the neighbor-joining method and 1,000 bootstrap replicates built in MEGA X software.
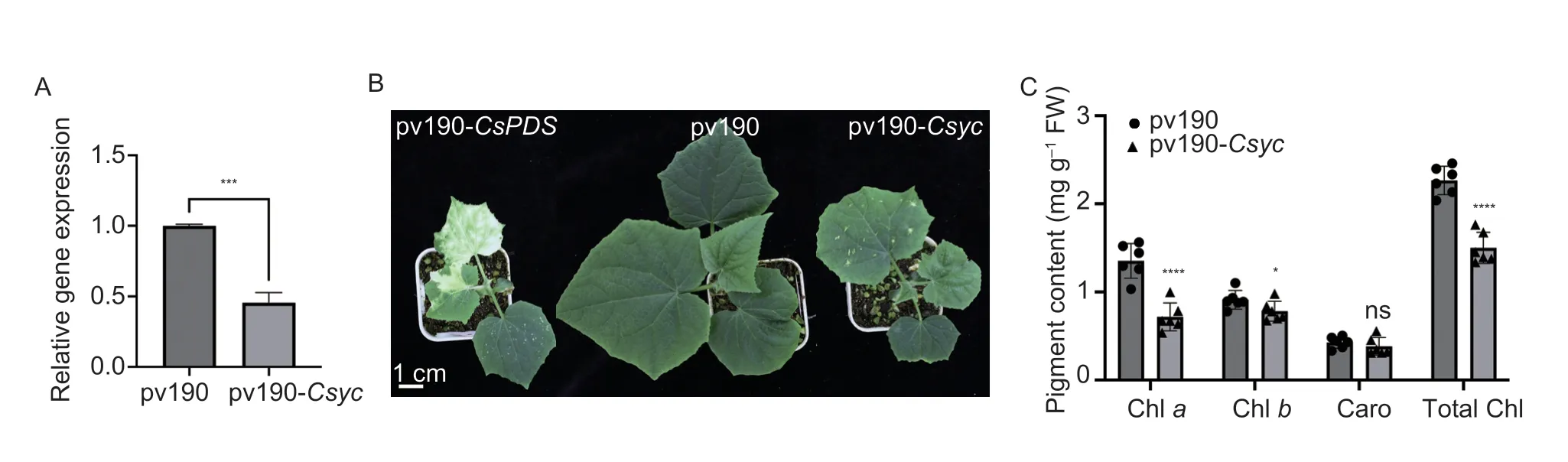
Fig. 5 Functional analysis of Csyc using the virus-induced gene silencing method. A,analysis of Csyc expression levels in cucumber leaves infected with pv190-Csyc or empty pv190 vector. The expression of plants infected with empty pv190 was set to 1. B,treated plants carrying pv190-CsPDS show a photobleaching phenotype (left);control plants carrying pv190 with no silenced phenotypes(middle);treated plants carrying pv190-Csyc show a yellow young leaf phenotype (right). C,chlorophyll content comparison and statistical analysis in control plants and treated pants. Caro,carotenoid. Bars show mean±standard deviation. Student’s t-test,*,***,and **** indicate significance at the P<0.05,P<0.001 and P<0.0001,respectively. ns,not statistically significant.
To elucidate the molecular mechanism ofCsycin cucumber cotyledon color regulation,an RNA-seqbased transcriptome analysis was performed using WT and mutant cotyledons. Groups of 13.6 million and 13.7 million clean paired-end reads were generated for the WT and mutant,respectively (Appendix C). The coefficient values among the three biological replicates ranged from 0.99 to 1 (Fig.6-A),indicating strong correlations.The high-quality clean reads were then aligned to the 9930 genome,with the mapping rate ranging from 96.18 to 97.42% (Appendix C). Using a false discovery rate of 0.05 and fold change 2 as thresholds,a total of 3,205 DEGs were identified,including 1,575 up-and 1,630 downregulated,in the pairwise comparison of mutantvs.WT (Fig.6-B).GO term enrichment analysis was performed to investigate the effect ofCsyc. In cellular components,the DEGs were mainly enriched in the plastid (GO: 0009536),chloroplast (GO: 0009507),and chloroplast stroma (GO: 0009570). Among the biological process category,the DEGs were significantly enriched in chloroplast organization (GO: 0009658). The molecular function category displayed oxidoreductase activity (GO:0016491) as the main component (Fig.6-C;Appendix D). KEGG pathway enrichment analysis was carried out to explore the potential biological functions of the DEGs. Three significant KEGG pathways were identified,including biosynthesis of secondary metabolites,alanine,aspartate and glutamate metabolism,and zeatin biosynthesis (Fig.6-D;Appendix E).
According to the RNA-seq results,the expression levels of plastid-encoded or nucleus-encoded photosynthesisrelated genes were significantly reduced inyc412compared with WT (Fig.7-A–C). These genes included plastid-encoded RNA polymerase (PEP) mediated genes(CsaV3_UNG203670,PsaA;CsaV3_UNG207970,PsbB;CsaV3_7G026830,TAS2R) and nuclear-encoded RNA polymerase (CsaV3_5G024490,rpoA;CsaV3_3G039630,rpoB) co-mediated genes. Fructokinase-like protein(CsaV3_3G034260,fructokinase-like PROTEIN),a component of the cystoid-binding PEP complex,was significantly reduced in the yellow leaf lethal mutants(Fig.7-B and C). The changes in transcript abundances of genes involved in the chlorophyll synthesis pathway were further analyzed (Fig.7-A). The nine key synthetic genes in the chlorophyll biosynthesis pathway,including HEMA(CsaV3_7G007370,encodes glutamyl-tRNA reductase),HEMB (CsaV3_2G033150,encodes 5-aminolevulinate dehydratase),HEMC (CsaV3_3G031800,encodes porphobilinogen deaminase),HEME (CsaV3_4G008050andCsaV3_5G010770,encodes UroporphyrinogenⅢ decarboxylase),GSA (CsaV3_7G007260,encodes Glutamate-1-semialdehyde 2,1-aminomutase),CHLI(CsaV3_6G050640,encodes Magnesium chelatase I subunit),CRD [CsaV3_4G031720,encodes Mgprotoporphynn IX monomethylester (oxidave) cyclases]and PORA (CsaV3_4G033870,encodes NADPH:protochlorophyllide oxidoreductase A) showed significantly down-regulated expression inyc412compared to WT(Fig.7-C). qRT-PCR was performed on six randomly selected photosynthesis-related DEGs to verify the accuracy of the RNA-seq data. The expression trends of the qRT-PCR and RNA-seq analyses are consistent(R2=0.9301),thus further confirming the reliability of the RNA-seq analysis (Fig.7-D).
4.Discussion

Fig. 6 RNA-seq analysis using YZU027A and yc412 cotyledon samples. A,heatmap of sample correlations. B,Volcano map of differentially expressed genes (DEGs) between YZU027A and yc412. The yellow dots represent DEGs with fold change(FC)≥2 and false discovery rate (FDR)<0.01,while black dots represent DEGs with FC≤0.5 and FDR<0.01;the grey dots represent insignificant DEGs with FC between 0.5 and 2 or FDR≥0.01. C,GO enrichment functional classification of DEGs between YZU027A and yc412. The size of each dot indicates the number of genes enriched in the GO term. The green dots indicate cellular component,the yellow dots indicate molecular function,and the blue dots indicate biological process. The yellow line represents the threshold of q/p value=0.05. D,KEGG pathway enrichment of DEGs between the YZU027A and yc412 plants. The size of each dot indicates the number of genes annotated in that pathway. The dot color represents the q-value,with red dots indicating that the pathway was significantly enriched.
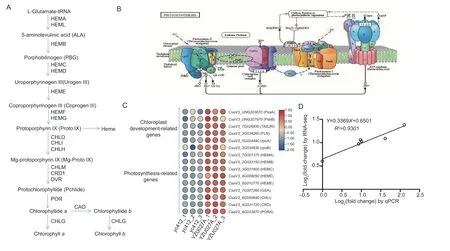
Fig. 7 Differentially expressed genes (DEGs) involved in chlorophyll biosynthesis and chloroplast development. A,chlorophyll biosynthetic pathway. HEMA,glutamyl-tRNA reductase;HEML,glutamate 1-semialdehyde aminotransferase;HEMB,porphobilinogen synthase;HEMC,porphobilinogen deaminase;HEMD,uroporphyrinogen III synthase;HEME,uroporphyrinogen decarboxylase;HEMF,coproporphyrinogen oxidative decarboxylase;HEMG,protoporphyrinogen oxidase;CHLD,magnesium chelatase D subunit;CHLI,magnesium chelatase I subunit;CHLH,magnesium chelatase H subunit;CHLM,magnesium proto IX methyltransferase;CRD1,Mg-protoporphyrin IX monomethylester cyclase;DVR,8-Vinyl reductase;POR,NADPH protochlorophyllide oxidoreductase;CHLG,chlorophyll synthase;CAO,chlorophyllide an oxygenase. B,photosynthesis pathway. The image of the known photosynthesis pathway was obtained from the KEGG database (https://www.kegg.jp/kegg-bin/show_pathway?ko00195).C,expression profile clustering for chloroplast development-related genes and chlorophyll biosynthesis-related genes between YZU027A and yc412. Expression ratios are based on log2FPKM values (Fragments Per Kilobase of transcript per Million mapped fragments),where each vertical column represents a sample (yc412_1,yc412_2,and yc412_3;YZU027A_1,YZU027A_2,and YZU027A_3),and each horizontal row represents a single gene. D,validation of RNA-seq data by qRT-PCR analysis of the six photosynthesis-related genes in (C).
Two yellow cotyledon mutations in cucumber have been reported and identified,includingyc-1andyc-2(Whelanet al.1975). However,the causal genes for these two mutations have not been cloned. Shiet al.(2022)identified a seedling-lethal albino mutant in pepper. In rapeseed,YELLOW COTYLEDON (Bn.yco) mutants were reported by Liuet al.(2021). In the present study,yc412was shown to be a spontaneous mutant in YZU027A with yellow cotyledons and a lethal phenotype (Fig.1-A).The development of the chloroplast was impaired,and the synthesis of chlorophyll was arrested (Fig.1-B;Fig.2). The phenotype ofyc412is distinct from all the previously reported mutants in cucumber. Based on BSA and linkage analysis,we mapped theyclocus to a 96.8 kb interval on Chr3 (Fig.3-B and C). According to the bi-parental resequencing analysis and molecular cloning,theyccandidate gene,Csyc,was shown to encode a YSL transporter (Fig.3-D). We found that a SNP in the promoter ofCsycmay affect the transcription ofCsyc(Fig.3-E),which completely disrupted chloroplast development and obstructed chlorophyll synthesis function(Fig.1-B;Fig.2),thereby reducing the photosynthetic efficiency inyc412(Fig.1).
YSL family members mainly function in iron ion transport,which is highly important for chlorophyll biosynthesis. In line with that,mutants of YSL family genes were shown to be associated with yellowing leaf in rice andA.thaliana(Chuet al.2010;Wuet al.2023).In rice,OsYSL15encodes a functional Fe (III)-DMA transporter involved in the uptake and transport of Fe(III)-DMA from the rhizosphere to the phloem.OsYSL15knockout seedlings exhibited severe germination and early growth stagnation,and were rescued by a high iron supply (Inoueet al.2009). Two insertionalosysl15mutants show a green-deficient phenotype under Fe deficiency. The chlorophyll content inOsYSL15-overexpression plants was higher than that of WT under a 0.1 μm Fe treatment (Leeet al.2009). In addition to the chlorosis phenotype described above,theArabidopsis ysllysl3double mutant exhibits multiple defects during reproduction,with greatly hindered fertility because of defective anther and embryo development (Waterset al.2006). Here,a SNP in the promoter ofyccaused yellowing of the cotyledons. The functions ofCsycwere confirmed by VIGS,in whichCsyc-silenced cucumber plants displayed delayed growth,chlorotic leaves and a reduced chlorophyll content (Fig.5-B). These results suggested the conserved functions ofYSLin plants(Fig.4-C). To our knowledge,this is the first report ofYSLregulating cotyledon color in cucurbits.
The sequence characteristic of theyc412mutant was a T to C mutation in the promoter region ofCsyc,resulting in a downregulation ofCsycexpression (Fig.3-E;4-A and B). The GUS transient assay confirmed that polymorphisms in the promoter region are important forCsycexpression (Fig.4-B). Gene expression variations could arise from polymorphisms incis-acting elements of the promoters (Wanget al.2021). For instance,the copy number variations of a 7 bp indel polymorphism in thecisregulatory sequence in the transcription factorCONSTANSare major determinants of flowering time diversity inArabidopsis(Rosaset al.2014). Here,a T to C mutation led to deletion of the MYC-recognition site (CATTTG) in theCsycpromoter ofyc412(Fig.4-A). The MYC-recognition site (CANNTG) plays an integral role in the regulation of gene transcription (Zhanget al.2016;Yinet al.2020). The T to C mutation resulted in a MYC motif deletion,which may provide an explanation for the downregulation ofCsycmRNA expression levels in theyc412mutant. Further investigations are needed to determine which transcription factor binds directly to the MYC motif.
The regulation of photosynthetic gene expression by the plastid-encoded or nuclear-encoded RNA polymerase(PEP or NEP) is essential for chloroplast development and efficient photosynthesis (Yagi and Shiina 2014;Yuet al.2014). PEP,composed of the four subunits encoded by plastid localized genesrpoA,rpoB,rpoC1andrpoC2,is a multiunit enzyme (Demarsyet al.2006). Loss of function of the proteins associated with the PEP and NEP complexes can give rise to the plant albinic phenotype,eventually leading to seedling death (Yagiet al.2012;Liuet al.2016;Zhenget al.2019). Here,the expression levels of PEP-mediated genes (PsaA,PsbB,andTAS2R)andNEP(rpoAandrpoB) were significantly reduced inyc412(Fig.7-C). Fructokinase-like protein (FLN),a component of the cystoid-binding PEP complex,was also significantly downregulated in the mutants (Fig.7-C).Loss-of-function mutants deficient in FLN develop seedling-lethal albino leaves in pepper (Shiet al.2022).We thus speculate that the SNP in the promoter ofCsycaffects the development of chloroplasts by disturbing the expression trajectory of PEP-mediated transcription. In this study,defects in chloroplast development were also observed in theyc412mutant (Fig.2),indicating that the mutation seriously affected chloroplast development(Fig.7-B and C). Interference in chloroplast development may affect chlorophyll synthesis. The chlorophyll biosynthetic pathway genes (HEMA,HEMB,HEMC,HEME,GSA,CHLI,CRD,andPORA) were suppressed in the mutant (Fig.7-A and C),supporting the notion thatCsycis crucial for chlorophyll biosynthesis and chloroplast development. Notably,four of the nine genes controlling leaf color in cucumber identified by map-based cloning were also differentially expressed in our RNAseq analyses (Miaoet al.2016;Songet al.2018;Dinget al.2019;Zhaet al.2022;Zhanget al.2022). These genes includeCsaV3_2G015690for theyellowleaf2.1(yl2.1) locus (Xionget al.2021),CsaV3_3G042400for theyellowyoungleaf-1(yyl-1) locus (Huet al.2020),CsaV3_3G047850for thevirescent-2(v-2) locus (Zhanget al.2020),andCsaV3_6G050640for theyellowplant(yp) locus (Gaoet al.2016),which are listed in Appendix F.These results indicated thatCsycmay regulate chlorophyll biosynthesis and chloroplast development by making a synergy with these four genes,which provides inroads for future research.
5.Conclusion
The cucumber yellow cotyledon mutation is due to a single nucleotide substitution in the promoter of the yellow stripe-like transporter geneCsYSLthat affects chloroplast development and chlorophyll biosynthesis.
Acknowledgements
This work was supported by the the “JBGS” Project of Seed Industry Revitalization in Jiangsu Province,China (JBGS[2021]018),the Jiangsu Agricultural Innovation of New Cultivars,China (PZCZ201720),and the Open Project Program of Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement,China(K2020030). We thank Dr.Qinsheng Gu from Zhengzhou Fruit Research Institute,Chinese Academy of Agricultural Sciences for kindly providing the pV190 vector.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.11.024
杂志排行
Journal of Integrative Agriculture的其它文章
- Molecular mechanisms of stress resistance in sorghum: lmplications for crop improvement strategies
- Artificial selection of the Green Revolution gene Semidwarf 1 is implicated in upland rice breeding
- Dynamics and genetic regulation of macronutrient concentrations during grain development in maize
- The NAC transcription factor LuNAC61 negatively regulates fiber development in flax (Linum usitatissimum L.)
- The underlying mechanism of variety–water–nitrogen–stubble damage interactions on yield formation in ratoon rice with low stubble height under mechanized harvesting
- Rice canopy temperature is affected by nitrogen fertilizer
