Artificial selection of the Green Revolution gene Semidwarf 1 is implicated in upland rice breeding
2024-03-12ShuliangJiaoQinyanLiFanZhangYonghongTaoYingzhenYuFanYaoQingmaoLiFengyiHuLiyuHuang
Shuliang Jiao ,Qinyan Li ,Fan Zhang ,Yonghong Tao ,Yingzhen Yu ,Fan Yao ,Qingmao Li,Fengyi Hu#,Liyu Huang#
1 Key Laboratory of Biology and Germplasm Innovation of Perennial Rice, Ministry of Agriculture and Rural Affairs/School of Agriculture, Yunnan University, Kunming 650091, China
2 State Key Laboratory of Crop Gene Resources and Breeding, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
3 Wenshan Academy of Agricultural Sciences, Wenshan 663000, China
Abstract Semidwarf breeding has boosted crop production and is a well-known outcome from the first Green Revolution. The Green Revolution gene Semidwarf 1 (SD1),which modulates gibberellic acid (GA) biosynthesis,plays a principal role in determining rice plant height. Mutations in SD1 reduce rice plant height and promote lodging resistance and fertilizer tolerance to increase grain production. The plant height mediated by SD1 also favors grain yield under certain conditions. However,it is not yet known whether the function of SD1 in upland rice promotes adaptation and grain production. In this study,the plant height and grain yield of irrigated and upland rice were comparatively analyzed under paddy and dryland conditions. In response to dryland environments,rice requires a reduction in plant height to cope with water deficits. Upland rice accessions had greater plant heights than their irrigated counterparts under both paddy and dryland conditions,and appropriately reducing plant height could improve adaptability to dryland environments and maintain high grain yield formation. Moreover,upland rice cultivars with thicker stem diameters had stronger lodging resistance,which addresses the lodging problem. Knockout of SD1 in the upland rice cultivar IRAT104 reduced the plant height and grain yield,demonstrating that the adjustment of plant height mediated by SD1 could increase grain production in dryland fields. In addition,an SD1 genetic diversity analysis verified that haplotype variation causes phenotypic variation in plant height. During the breeding history of rice,SD1 allelic mutations were selected from landraces to improve the grain yield of irrigated rice cultivars,and this selection was accompanied by a reduction in plant height. Thus,five known mutant alleles were analyzed to verify that functional SD1 is required for upland rice production. All these results suggest that SD1 might have undergone artificial positive selection in upland rice,which provides further insights concerning greater plant height in upland rice breeding.
Keywords: Green Revolution,Semidwarf 1,high-yield breeding,upland rice,plant height
1.Introduction
The development of high-yielding crop cultivars is essential for meeting the steadily increasing food demands of the global population (Wanget al.2022).Introducing the semidwarf trait into wheat (Triticum aestivumL.) and rice (OryzasativaL.) enabled rapid yield increases because of the associated reductions in lodging and increases in the harvest index (Hedden 2003;Weiet al.2022). Together with the extensive application of chemical fertilizers and pesticides,this development sharply improved crop yields. Therefore,semidwarf crop breeding has been celebrated as launching the Green Revolution (Hargrove and Cabanilla 1979;Khush 1999).
Subsequently,frequent genetic improvements through the introduction of mutant alleles in crops have resulted in rapid increases in grain yields. Mutant alleles,such as riceSemidwarf1(sd1) and wheatRht-B1,Rht-D1,Rth12,andrth18,were selected from mutants of the gibberellic acid (GA) biosynthesis gene and contribute to the semidwarfism trait (Monnaet al.2002;Spielmeyeret al.2002;Hedden 2003). Most modern rice cultivars can be traced back to early Green Revolution short-culmed varieties that have these mutant alleles (Ferrero-Serranoet al.2019;Penget al.2021).
However,high grain yield requires sufficient plant biomass and plant height. Thus,in some cases,Green Revolution gene alleles with less marked phenotypes have been selected to produce semidwarf rather than dwarf traits (Naganoet al.2005).Oryzasativais mainly divided into two primary cultivar lineages,Geng/japonicaandXian/indica. These two lineages feature differentsd1alleles (Muraiet al.2011;Ranaet al.2021). The phenotypes of thesd1mutant alleles used in theGeng/japonicalines are generally less pronounced than those used in theXian/indicavarieties,indicating that the mutant enzymes might have moderate activity (Yuet al.2020).
Long-term natural selection has driven the evolution of variousO.sativaecotype populations,such as upland and irrigated rice,which have evolved their respective characteristic traits under both natural and artificial selection,leading to phenotypic adaptations to their respective environments and simultaneous genetic differentiation between the ecotypes. Therefore,rice provides an ideal model system for studying adaptation to drought and aerobic environments (Xiaet al.2019).Compared with its irrigated counterpart,upland rice has multiple specialized characteristics,such as adaptation to aerobic conditions,greater height,lower tillering potential,and longer and denser roots (Chang 1972;Chang and Vergara 1975). Water deficit and drought stress can severely reduce plant height and consequently affect biomass and grain yield (Vikramet al.2015). We therefore sought to elucidate whether plant height contributes to maintaining the upland rice yield under dryland conditions.In addition,we investigated whether Green Revolution genesd1alleles are present in upland rice.
In this study,we present theSD1haplotypes across various subspecies,ecotypes,and breeding histories.Additionally,we evaluated the plant height,yield,and lodging resistance phenotypes in both the upland rice population and its irrigated counterparts. Artificial mutation ofSD1in upland rice further confirmed that functionalSD1can help maintain the grain yield in upland rice,especially under dryland conditions.
2.Materials and methods
2.1.Plant materials and phenotyping
The 61 irrigated and 72 landrace upland rice accessions chosen for this study were obtained from the Chinese Academy of Agricultural Sciences (CAAS),and their genome sequences are publicly available (Wanget al.2018;Appendix A). Phenotyping was evaluated under both irrigated and dryland conditions during the 1st season of 2021 in Xishuangbanna,Yunnan Province(20°59´N,100°44´E).
For the irrigated conditions,seeds were germinated in a seedbed,and seedlings were then transplanted to a paddy field,where water was pooled on the soil surface throughout the growth and developmental period. For dryland conditions,we conducted direct seeding by dibbling the seeds in dry soil. To fully simulate rainfed conditions,no irrigation was applied in the upland field.When it rained,we drained any excess water to prevent soil submergence,thereby keeping the average soil water content below 20%. The soil water content of the dryland was measured by means of soil moisture meters(TZS-W,Zhejiang Top Instrument Co.Ltd.) at different stages. For each accession,we planted three replicates,and each replicate had 12 individual plants in two rows(six individuals in each row),with a row spacing of 30 cm and a plant spacing of 20 cm. For each cultivar or line,approximately eight individuals were randomly selected and measured. The plant height of the accessions was surveyed at the ripening stage,and the yield per panicle,number of grains per panicle,seed-setting rate,number of productive panicles,and 1,000-grain weight of the accessions were determined. The stem diameter of the basal internode (the first elongated internode of each selected plant) was measured using Vernier callipers at the ripening stage. A spring tension meter was used to measure the tensile force required to break the stem.Data analysis was performed using IBM SPSS Statistics 26 and GraphPad Prism 9 to output the results.
2.2.Construction of the SD1-knockout vector and transgenic rice
The CRISPR/Cas9 binary vector pCAMBIA1300-OsU3-Cas9 (Huanget al.2018) was used to construct theSD1knockout vector. TheSD1target sequence(CGCACGGGTTCTTCCAGGTG),which was inserted upstream of the sgRNA cassette,was designed in the first exon. Next,the target sequence was introduced into the destination vector. A monoclonal colony was selected for Sanger sequencing using OsU3-test-F/OsU3-test-R primers (GCATGGATCTTGGAGGAATCAG/GTGTTGTGTTCACCTGCGAG). TheSD1knockout vector was namedpCRISPR-sd1,transformed into theAgrobacteriumstrainEHA105and then transformed into IRAT104 (aGeng/japonicatype of upland rice) as described by Zhaoet al.(2011).
2.3.Genotyping of transgenic rice and phenotyping of sd1 mutants
Genomic DNA was extracted from the leaves of transgenic rice plants by the cetyltrimethyl ammonium bromide (CTAB) method. Hygromycin (HYG)-resistant rice plants were identified by the HYG-F/R primer (GATGTAGGAGGGCGTGGATATGTCCT/GTATTGACCGATTCCTTGCGGTCCGAA) based on PCR amplification of theHYGfragment. The corresponding specific primersSD1-F/R (CTCAACTCACTCCCGCTCAA/GTCCGCGAAGAACTCCCT) were used to further amplify theSD1target site. The fragments containing the target site were amplified from genomic DNA of the T0and T1plants,and homozygous mutants for each gene were selected by Sanger sequencing.
To evaluate the effects of the mutated alleles,we selected threesd1knockout T1homozygous mutants and wild types from negative T1lines for further phenotyping.Thirty-six T2plants from each line were grown in paddy fields and dryland conditions. Plant height and yieldrelated phenotypes under both paddy and dryland conditions were assessed as described above.
2.4.Genetic analysis of SD1 alleles
The haplotype analysis ofSD1was performed using all single nucleotide polymorphisms (SNPs) within the gene coding sequence region in the 3K-RG,in which synonymous SNPs were ignored (Zhanget al.2021).Phenotypic data for plant height were downloaded from the Rice SNP-Seek Database (Alexandrovet al.2015). One-way ANOVA followed by Duncan’s new multiple range tests was used to compare the phenotypic differences among the five knownsd1major alleles/haplotypes (n≥20 rice accessions) with the agricolae package in R.
3.Results
3.1.Plant height of upland rice is greater and less variable than that of irrigated rice under different conditions
Our previous study indicated that upland rice has a greater height than irrigated rice under both irrigated and dryland conditions (Lyuet al.2014). In the current study,we further evaluated the variations in plant height of rice germplasms under dryland and paddy conditions.The results indicate that almost all rice accessions exhibited reduced height as an adaptation to upland conditions (Fig.1-A). The plant heights of upland rice were approximately 120 and 100 cm in paddy and upland conditions,respectively,while those of irrigated rice were approximately 100 and 75 cm,respectively. Notably,compared with irrigated rice,upland rice not only had greater plant heights under both paddy and upland conditions but also had significantly less variation in height in response to the dryland environment (Table 1).
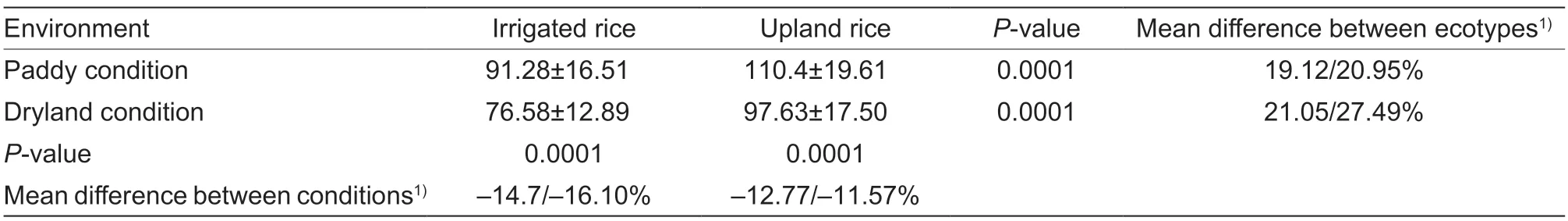
Table 1 Differentiation of plant height between upland and irrigated rice
3.2.Grain yield per panicle is a key trait selected under upland rice domestication,which partially depends on plant height
Because plant height and theSD1locus are positively correlated with grain yield (Zhanget al.2013;Suet al.2021),we sought to determine whether an adequate plant height promotes dryland adaptation and artificial selection in upland rice. Our previous study showed that fewer tillers and higher yield per panicle,as regulated byOsTb2,reflected the domestication for dryland adaptation(Lyuet al.2020).
Based on the findings of our previous study,in which upland rice domestication was reflected in higher grain yield per panicle under dryland conditions,we compared the grain yield per panicle in upland and irrigated rice under both paddy and dryland conditions in the current study. The results showed that the grain yield per panicle in irrigated rice was significantly reduced under dryland conditions when compared with that of irrigated rice grown in paddy fields (Fig.1-B). The grain yield per panicle in upland rice was slightly but not significantly reduced under upland conditions when compared to that of upland rice grown in paddy fields,and was significantly higher than that of irrigated rice (Fig.1-B). The correlation analysis showed that grain yield per panicle was significantly positively correlated with plant height (Fig.1-C).
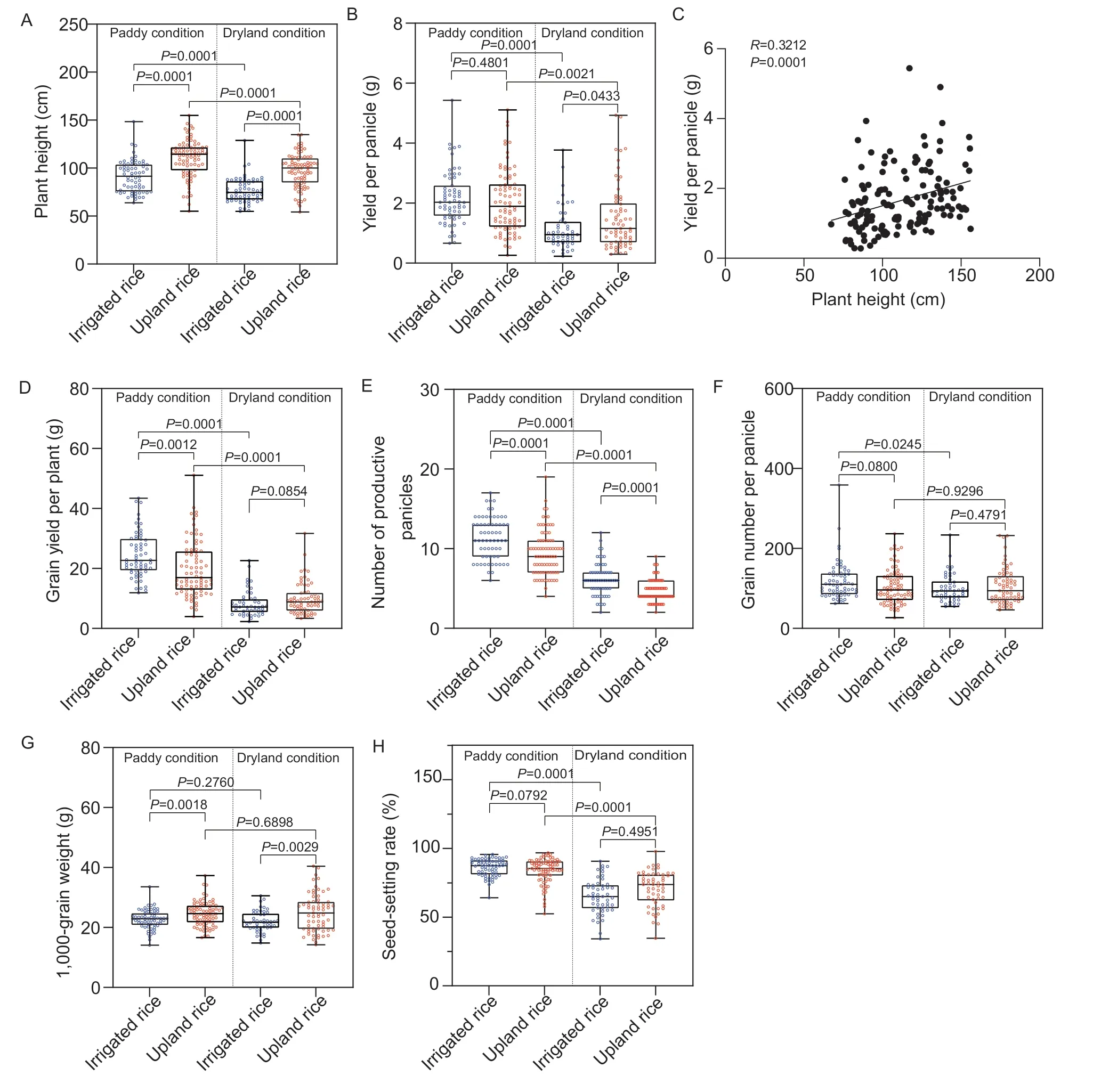
Fig. 1 Phenotypic differentiation between upland and irrigated rice under paddy and dryland conditions. Plant height (A) and yield per panicle (B) were compared between upland and irrigated rice and between paddy and dryland conditions. C,plant height was positively correlated with grain yield per panicle in the 133 cultivars under dryland conditions. Grain yield per plant (D),number of productive panicles (E),grain number per panicle (F),1,000-grain weight (G),and seed-setting rate (H) were compared between upland and irrigated rice and between paddy and dryland conditions. R refers to the Pearson product-moment correlation coefficient;P is the P-value under Student’s t-test analysis. Each dot represents the mean value of approximately eight plants and n>70 cultivars for the statistics.
Studies have also shown that drought stress causes a reduction in productive panicles,which results in reduced grain production (Liuet al.2021). In this study,the upland rice yield per plant was much lower than that of irrigated rice under paddy conditions,mainly due to the intrinsically fewer productive panicles in upland rice,but the variation sharply decreased under upland conditions (Fig.1-D and E). In addition,key yieldrelated traits were measured,including the grain number per panicle and the 1,000-grain weight,under paddy and upland conditions. The grain numbers per panicle under both paddy and upland conditions showed no significant differences,and only that of irrigated rice decreased in response to dryland conditions (Fig.1-F). The 1,000-grain weight of upland rice was greater than that of irrigated rice under both paddy and upland conditions(Fig.1-G). More importantly,the 1,000-grain weights of both upland and irrigated rice were relatively stable under the different conditions (Fig.1-G). However,the seed-setting rates of irrigated and upland rice decreased significantly under dryland conditions,with no significant differences between the two ecotypes (Fig.1-H). All these results suggest that upland rice has better aerobic and dryland adaptability and is domesticated in the trait of grain yield per panicle.
3.3.Upland rice has better lodging resistance than irrigated rice under both paddy and dryland fields
To determine whether the greater plant height of upland accessions could cause lodging problems in rice production,we measured the stem diameter of the basal internodes (the first elongated internodes) of the rice plants (Liuet al.2018). The results reveal that the stems of upland rice are much thicker than those of the irrigated cultivars (Fig.2-A and B),which implies that upland accessions have better lodging resistance than their irrigated counterparts. In addition,we also measured the magnitude of force required to break off the first elongated internode of the rice stem. As shown in Fig.2-C,a greater force is required to break off the stem of upland rice than is required for irrigated rice because of the thicker stem diameter in upland rice.
Plant lodging is significantly associated with plant height. Consequently,the first Green Revolution semidwarf breeding outcome was related to lodging resistance to maintain high yield. The results of this study also demonstrated that plant lodging has a significant positive association with stem diameter (Fig.2-D).Therefore,most upland rice landraces have the trait of lodging resistance,which may not be needed for semidwarf breeding.
3.4.SD1 knockdown mutants in upland rice cultivars can reduce the plant height and grain yield under upland conditions
Although the function ofSD1in regulating plant height by controlling GA synthesis in rice stems is relatively clear,whether thesd1mutation in upland rice reduces yield production remains unknown. The CRISPR/Cas9 system we developed previously (Huanget al.2018) was used to knock outSD1in an upland rice variety (IRAT104)containing wild-type (WT)SD1.
Twenty-four independently transformed rice lines,defined as T0,were regenerated. The disrupted target sites detected by PCR-based Sanger sequencing revealed an 87.5% (21/24) targeting efficiency forSD1. Only two homozygous mutations were identified in the T0generation of the transformed rice plants. In the T1generation grown from the seeds of T0plants,homozygous mutants at the target sites that also lacked the T-DNA segments were screened. Three novel homozygous alleles with different frameshift mutations in T1marker-free plants were obtained for further phenotyping (Fig.3).

Fig. 2 Upland rice has a larger stem diameter and stronger lodging resistance than irrigated rice. A,phenotypes of the cross section at the basal elongated internode in both upland and irrigated rice cultivars under dryland conditions. Scale bars,2 mm.Comparative analysis of the stem diameter (B) and breaking force (C) of the basal elongated internode between upland and irrigated rice. Student’s t-test analysis indicated a significant difference (*,P<0.05;**,P<0.01). D,stem diameter was positively correlated with the breaking force,which represents lodging resistance. R refers to the Pearson product-moment correlation coefficient;P is the P-value under Student’s t-test analysis. Each dot represents the mean value of approximately eight plants per cultivar,and n>15 for the statistics in each ecotype.
Specifically,one allele was an insertion (1 bp),and two alleles were deletions (4 and 1 bp) (Fig.3-A). These new mutant alleles were namedsd1-M1 to M3 (Fig.3).A protein sequence comparison between the WT background and the mutant plants revealed frameshift variations in the WTSd1proteins.
TheSd1mutation alleles (M1 to M3) created by artificial mutagenesis markedly decreased the plant height by almost 30% (Fig.3-B and C). Grain yeilds per panicle were significantly lower in the mutants than in plants of the WT background IRAT104 (Fig.3-D). Grain number of panicle,rather than either seed-setting rate,1,000-grain weight,or productive panicles per plant,was a major contributing factor in the reduced grain yields in dryland conditions (Fig.3-E–H). In addition,the panicle lengths insd1mutants were shorter than those in the WT (Fig.3-I).All these results indicate thatSD1is beneficial for grain yield in upland conditions when the problem of plant lodging is excluded.
3.5.Genetic variations in SD1 and plant height under different breeding histories
TheSD1locus and surrounding region have different degrees of diversity in the rice subpopulations (Wanget al.2018). To determine whetherSD1diversity plays a role in plant height variation among rice germplasms,we performed haplotype analysis ofSD1in the 3K-RG (Huet al.2015;Wanget al.2018). Twenty major haplotypes(n≥20 rice accessions) were identified atSD1using SNPs with minor allele frequency (MAF)>1% in the CDS region in the 3K-RG (Appendices B and C).

Fig. 3 Semidwarf 1 (SD1) mutations in upland rice reduced both plant height and grain yield under dryland conditions. A,gene sequence structure and genotyping of SD1 mutants sd1-M1–M3 in the T1 and T2 generations. Sequences above the peak are SD1 wild-type (WT) (IRAT104) alleles. The peak of each base for the whole sequence was singular and revealed homozygosity.+indicates insertion,– indicates deletion,and the target sequence is shown before the protospacer adjacent motif (PAM) site. B,views of the plant height of WT and sd1 mutants in the mature stage. Scale bars,10 cm. Comparative analysis of plant height (C),yield per panicle (D),grain number per panicle (E),panicle length (F),seed-setting rate (G),1,000-grain weight (H) and number of productive panicles (I) between sd1 mutants and their WT controls. Student’s t-test analysis indicates a significant difference(*,P<0.05;**,P<0.01);ns represents no significant difference.
To verify the relationships between plant height variation andSD1alleles in different ecotypes,we compared rice plant heights under different breeding statuses based on their pedigrees. The results indicated that modern varieties (most of which are irrigated rice),i.e.,improved varieties,were significantly dwarfed compared with the landraces to achieve the goals of the Green Revolution (Fig.4-A). Although a few improved upland rice varieties were included in the 3K-RG and not available for statistical comparisons,some modern typical upland rice varieties showed greater plant height than the modern irrigated rice varieties even under different conditions (Appendix D). Five known mutant alleles ofSD1with different activities in controlling rice plant height were identified (Muraiet al.2011;Penget al.2021),and these alleles have been extensively used in modern cultivars (Fig.4-B;Table 2). Accessions with theSd1_indandSd1_weakalleles (Shaet al.2022) have greater plant heights than accessions with the other alleles (Fig.4-B).For the most part,only these two types of alleles exist in upland rice (Table 2). These results suggest thatSD1might have undergone artificial positive selection in upland rice,which provides further insights concerning the role of a greater plant height in upland rice breeding.
4.Discussion
4.1.Allelic diversity of SD1 contributes the environmental adaptation of rice
Plant height and biomass are known to be positively correlated with crop grain yield. In adapting to various environments,crops have evolved different plant architectures under high-yield selection pressure from humans. The first Green Revolution,which emphasized semidwarf breeding as its theme,serves as a vivid case of adaptive improvement in crop architecture for lodging resistance and fertilizer tolerance. The Green Revolution geneSD1and its allelic variation,specifically expressed in the rice stem,control rice plant height in response to different cultivation conditions. ThisSD1allelic variation is associated with different enzymatic activities and results in different plant architectures with high yields. However,using nonfunctional alleles inXian/indicarice results in poor adaptation to submergence in rainfed production and to widespread water eutrophication due to overfertilization(Mackillet al.1999;Zhang 2007;Kurohaet al.2018).Additionally,studies have shown thatSD1mutation leads to negative effects on the yields ofGeng/japonicarice cultivars (Ogiet al.1993;Muraiet al.2002).
SD1has been involved not only in modern rice breeding,including during the Green Revolution,but also in the early stages of rice domestication (Asanoet al.2011). A recent study suggests that upland rice was directly domesticated fromOryzarufipogon(Luoet al.2022). Dryland environments and even drought stress can cause remarkable reductions in rice plant height and grain yield (Fig.1),and some GA synthesisand metabolism-related genes (such as GA20oxs,GA3oxs,and GA2oxs) regulated by drought stress might be important genetic factors that have also been discovered in other plants (Krugmanet al.2011;Wanget al.2011). Additionally,a functionalSD1encoding GA20ox,the GA synthesis enzyme,can partially resist the restraining influence of dryland environments,which may favor high yields. Upland rice is an ecotype adapted to upland conditions that has been dual-selected by the dryland environment and for high grain yield. Therefore,we can postulate that the functionalSD1allelic variation originating from wild rice became domesticated and contributed to the higher grain yield (Zhanget al.2020).In our study,SD1knockout of upland rice reduced the plant height and grain yield under dryland conditions(Fig.3). In addition,our results further demonstrated that most upland rice has a thicker stem diameter and greater lodging resistance (Fig.2). Although we have no data about the exact differences in nutritional status between upland and paddy fields,typical upland soil may become impoverished by soil erosion. Therefore,it is probably not a coincidence that functional mutations in theGA20oxgene have been selected in the pursuit of higher grain yield in upland rice. A relatively tall plant height is conducive to enhancing grain yield in upland rice. Interestingly,an extreme example of the C9285SD1allele (SD1C9285) that was selected for deepwater rice cultivation and is responsible for submergence-induced internode elongation underlies the adaptation of rice to periodic flooding (Kurohaet al.2018).
4.2.Functional SD1 is beneficial for high-yield upland rice breeding and positively selected during upland rice breeding
Based on the observations of a few upland rice accessions with relatively high plant height and slender stems,reducing the plant height might still be an effective way to improve lodging resistance and enhance grain yield in upland rice breeding. We also found no significant differences in plant height between upland and non-upland rice landrace accessions (Fig.4-A),which was inconsistent with the germplasm population data (Fig.1-A). The reason might be the great genetic difference in their population structures. To reduce genetic background noise,we selected the germplasms to form a balanced population,consisting of halfGeng/japonicaand halfXian/indica(Appendices A and B). In the population filtered from 3K-RG,most accessions wereXian/indica,and only one-third wereGeng/japonica.
Crop breeding has entered the modern era of molecular breeding. Breeders set different breeding targets according to different demands,which are influenced by numerous factors,including water,temperature,fertilizer,and nutritional quality (Kurohaet al.2018). GA signalling pathways (involving biosynthesis and signal transduction)are prime targets for manipulation in the quest for further improvements in cereal yield,but the effects of variousSD1alleles on rice height and yield differ depending on the genetic background (Biswaset al.2020). Taking superindicarice breeding as an example,the weak alleleSD1-EQfromGeng/japonicarice was introduced intoXian/indicarice to achieve the goals related to high plant height and,subsequently,to biomass as well as increased grain production (Yuet al.2020).
In the post-Green Revolution era,targeted genetic modification of crop breeding will accelerate the next Green Revolution (Khush 2001;Pingali 2012). The development of Green Super Rice is an environmentally friendly agricultural effort in pursuit of optimal productivity,and includes aspects such as water efficiency and the reduced use of fertilizer and chemical pesticides (Zhang 2007). To achieve sustainable increases in yield,an ideal plant architecture has been proposed,the main concept of which is to increase plant height and subsequently plant biomass to increase grain production (Yuan 2014).The use of dryland for rice production is one of the most important agricultural means to ensure food security and is known as the Blue Revolution (Sunet al.2022;Xiaet al.2022). Thus,the development of high-yield upland rice cultivars represents a need that demands prompt solutions. Based on this study,we can conclude that the functional or partially functional Green Revolution geneSD1is beneficial for high-yield upland rice breeding and artificial selection in upland rice. Mutations insd1underly the adaptation to the paddy environment in irrigated rice by contributing to lodging resistance and fertilizer tolerance. According to our previous study,upland rice mitigates the trade-off between tillering and adaptation to maintain grain yield per panicle under dryland conditions(Lyuet al.2020). Therefore,a strategy for upland rice breeding was conceived that involves an ideal plant architecture with fewer tillers/panicles,thicker stems,deeper roots,and greater 1,000-grain weight and plant height (Fig.5). In addition to a functionalSD1,the key genes involved in grain weight and grain number,such asGS3(Fanet al.2009),GW2(Songet al.2007),Gn1a(Ashikariet al.2005),DEP1(Sunet al.2014),andIPA1(Miuraet al.2010),might be used for molecular markerassisted selection-and genome editing-based molecular design breeding.
5.Conclusion
We thoroughly analyzed the phenotypic differentiation and variations between irrigated and upland rice under both paddy and dryland conditions. Upland rice was domesticated in plant height,tiller,and 1,000-grain weight under pressure from aerobic adaptation and artificial selection. Artificial selection of the functionalSD1gene results in greater plant height,and ultimately grain yield per panicle,in upland rice under dryland conditions. Hence,our findings not only explain why the Green Revolution genesd1is not present in upland rice but also suggest a strategy for future upland rice breeding.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32272079 and 32060474) and the Yunnan Provincial Science and Technology Department,China (202101AS070001 and 202201BF070001-011).
Declaration of competing interest
The authors declare that they have no conflicts of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.05.010
杂志排行
Journal of Integrative Agriculture的其它文章
- Molecular mechanisms of stress resistance in sorghum: lmplications for crop improvement strategies
- Dynamics and genetic regulation of macronutrient concentrations during grain development in maize
- The NAC transcription factor LuNAC61 negatively regulates fiber development in flax (Linum usitatissimum L.)
- The underlying mechanism of variety–water–nitrogen–stubble damage interactions on yield formation in ratoon rice with low stubble height under mechanized harvesting
- Rice canopy temperature is affected by nitrogen fertilizer
- The first factor affecting dryland winter wheat grain yield under various mulching measures: Spike number
