Three novel umami peptides derived from the alcohol extract of the Pacific oyster (Crassostrea gigas): identification,characterizations and interactions with T1R1/T1R3 taste receptors
2024-02-16BaifngFuDiWuShuzhnChngXianbingXuLingZhangLishuWangHshanmElSiHanxiongLiuMingDu
Baifng Fu,Di Wu,Shuzhn Chng,c,Xianbing Xu,c,Ling Zhang,Lishu Wang,Hshanm R.El-Si,Hanxiong Liu,Ming Du,c,*
a School of Food Science and Technology,Dalian Polytechnic University,Dalian 116034,China.
b Collaborative Innovation Center of Seafood Deep Processing,Dalian Polytechnic University,Dalian 116034,China.
c National Engineering Research Center of Seafood,Dalian Polytechnic University,Dalian 116034,China.
d Division of Hematology and Oncology,Department of Medicine,Medical College of Wisconsin,Milwaukee 53226,USA.
e Division of Pharmacognosy,Department of Medicinal Chemistry,Biomedical Centre,Uppsala University,Uppsala SE-751 23,Sweden
Keywords:Oyster umami peptides iUmami-SCM Taste characterization Molecular docking
ABSTRACT Oyster (Crassostrea gigas),the main ingredient of oyster sauce,has a strong umami taste.In this study,three potential umami peptides,FLNQDEEAR (FR-9),FNKEE (FE-5),and EEFLK (EK-5),were identified and screened from the alcoholic extracts of the oyster using nano-HPLC-MS/MS analysis,iUmami-Scoring Card Method (iUmami-SCM) database and molecular docking (MD).Sensory evaluation and electronic tongue analysis were further used to confirm their tastes.The threshold of the three peptides ranged from 0.38 to 0.55 mg/mL.MD with umami receptors T1R1/T1R3 indicated that the electrostatic interaction and hydrogen bond interaction were the main forces involved.Besides,the Phe592 and Gln853 of T1R3 were the primary docking site for MD and played an important role in umami intensity.Peptides with two Glu residues at the terminus had stronger umami,especially at the C-terminus.These results contribute to the understanding of umami peptides in oysters and the interaction mechanism between umami peptides and umami receptors.
1. Introduction
The oyster,Crassostreagigas,has a long agricultural history and,in recent decades,its global production has exceeded 5 500 000 tons per year[1].Oysters are a great source of complete protein,vitamins,minerals and omega-3 fatty acids,and are essential ingredients for oyster sauce due to their outstanding umami contribution[2].
Umami compounds found in oysters can be perceived via the interaction with umami receptors in the oral cavity[3,4],showing a great potential to improve food acceptance[5,6]. The umami taste of shellfish mainly derives from the confirmed umami compounds such as aspartic acid (Asp),glutamic acid (Glu),succinic acid,succinate,inosinic acid (IMP),guanosine phosphate (GMP)[7],and umami peptides[8,9].Generally,umami peptides were added to seasonings to improve the palatability of food while reducing the amount of sodium chloride added[10].Despite a growing interest in investigating marinederived umami peptides,little has been done to identify umami peptides from oysters[11].
Ultrafiltration,gel chromatography and reversed-phase highperformance liquid chromatography are traditionally used to extract umami peptides[12-14].However,these steps are laborious and cumbersome.It has become increasingly popular to use computer algorithms to predict the activity of peptides[15].Virtual screening with efficiency and rapidity has often been used to assist in the screening of umami peptides.The iUmami-scoring Card Method (iUmami-SCM) (http://camt.pythonanywhere.com/iUmami-SCM) is a computer algorithm used to predict the umami taste of peptides based on the sequence information,such as hydrophobicity,molecular weight and amino acid acidity[15].It has been reported that 18 potential umami peptides are virtually screened fromRuditapes philippinarumandMactra veneriformisclams by iUmami-SCM[16].
The taste receptor T1R1/T1R3 heterodimer is generally considered to be the main umami receptor[17].As the structure and function of the umami receptor are not fully studied,and its crystal structure has not yet been determined,the technology through computer simulation analysis is also very reliable[18].The “Venus fly trap domain” (VFTD),located in T1R3,has a large binding cavity and is regarded as a ligand-binding domain capable of binding umami peptides[19].Homology modeling can be used to establish reliable molecular docking receptor models for T1R1/T1R3.Further,MD techniques are an effective approach for investigating the mechanism of action between umami peptides and umami receptors T1R1/T1R3 heterodimer by analyzing the binding energy and the active sites[20,21].
The objective of this study was to identify potential umami peptides from oysters using a rapid and efficient method of virtual screening to predict and screen their umami contribution.Nano-HPLC-MS/MS was applied to identify the potential peptides.To screen the umami peptide rapidly and efficiently from the identified peptides,the iUmami-SCM website and molecular docking were applied.To verify the umami intensity of the screened peptides,sensory evaluation and electronic tongue (E-tongue) were utilized.And then we analyzed the binding mechanism of umami peptides with the umami receptor T1R1/T1R3 by homology and molecular docking.In this study,we could enrich potential marine food additives with umami flavor and provide insight into the rapid screening of umami peptides.
2. Materials and methods
2.1 Materials and chemicals
Oysters were purchased at Qianhe Market in Dalian,Liaoning Province,China.Their shells were removed and the meat was placed in ice and shipped to the lab within 4 h.Silica C18Packing(40-60 µm) was purchased from Nanomicro Technologies Inc.(Suzhou,China).Acetonitrile (ACN,HPLC grade) was purchased from Sigma-Aldrich (St.Louis,MO,USA).L-Isoleucine (US Pharmacopeia grade) was purchased from Sangon Biotech CO.Ltd.(Shanghai China).Food-grade monosodium glutamate (MSG)(Ajinomoto CO.,INC.,Tokyo,Japan),sucrose (Henan Wanbang Chemical Technology CO.Ltd.,Henan,China),sodium chloride(Henan Tianma Chemical Technology CO.Ltd.Henan,China),and citric acid (Henan Wanbang Chemical Technology CO.Ltd.,Henan,China) were all obtained from the online store.The ethanol used was analytical grade and was purchased from Damao Chemical Reagent Factory (Tianjin,China).All lyophilized components and synthetic peptides were stored at -80 °C for further analysis.
2.2 Preparation of oyster powder (OY-W)
Fresh oyster meat was mixed with distilled water at a ratio of 1:1 (m/V) and boiled for 3 h,followed by filtration with 80-mesh filters (Shanghai SECCO Petrochemical CO.Ltd.,Shanghai,China).Then the filtrate was centrifuged (CF16RXII,Hitachi,Japan) at 3 000 r/min for 5 min under 4 °C.The supernatant was spray-dried at 130 °C using a laboratory low-temperature spray dryer (YC-1800,Shanghai Pilotech Instrument &Equipment Co.Ltd,Shanghai,China)to get oyster power (OY-W).The OY-W was collected and stored for further analysis.
2.3 Purification of peptides
Silica C18Packing was activated with ethanol of 1:3 (m/V) and pre-wetted with distilled water of 1:3 (m/V) before using.The oyster powder dissolved in ultrapure water (20 mg/mL) was loaded on the activated Silica C18Packing.After full mixing adsorption,samples were eluted with the triple volume of the distilled water,20% (V/V)ethanol aqueous solution and pure ethanol,respectively.The three eluted samples were named 0%-E,20%-E,and 100%-E,respectively.
2.4 Identification of peptides
The primary structures and molecular weights of peptides from oysters were identified by nano-HPLC-MS/MS[22].Samples dissolved in 0.1% formic acid in ultrapure water were analyzed by Q Exactive™ coupled to an EASY-nano LC 1200 system (Thermo Fisher Scientific,MA,USA).The mass spectrometer was set to datadependent acquisition (DDA) mode and transitioned between MS and MS/MS mode automatically.
The sample was added to a 25 cm analytical column (75 μm i.d.,1.9 μm resin,Dr.Maisch) and separated with 60 min-gradient starting at 2% buffer B (80% ACN with 0.1% FA) for 3 min followed by a stepwise increase to 35% in 47 min,100% in 1 min and stayed there for 12 min.The column flow rate was maintained at 300 nL/min with a column temperature of 40 °C.The electrospray voltage was set to 2 kV.The Orbitrap was used to acquire a survey of full scan MS spectra (m/z200-1 500) at 70 000 resolutions.The maximum injection time is 60 ms and the automatic gain control (AGC) target was 3e6.The precursor ions were selected into collision cells for fragmentation by higher-energy collision dissociation (HCD),the normalized collection energy was 27.The MS/MS resolution was set to 17 500,while the AGC target was set to 5e4,the maximum injection time of 50 ms,and dynamic exclusion was 20 s.The data collected by mass spectrometry was analyzed by PEAKS Studio version 10.6 and the corresponding species database was searched,and the enzymatic digestion was set to none.
2.5 Prediction of umami peptides
The iUmami-SCM is a method for predicting the umami taste of peptides based on the target peptides’ primary sequence information.The Scoring Card Method (SCM) and propensity scores of peptides were combined in this approach.The accuracy and the Matthews correlation of the iUmami-SCM were 0.865 and 0.679,respectively,which indicated that this method was an excellent and outstanding method for predicting the umami taste of peptides.In this method,if the scoring function value S(P) of the target peptide was higher than 588,the peptide was recognized as the umami peptide,and vice versa.For preliminary screening,the umami potential of all the peptides identified by nano-HPLC-MS/MS was calculated by the iUmami-SCM.
2.6 Molecular docking of umami taste peptide and T1R1/T1R3
To obtain the 3D structure of the major umami receptor T1R1/T1R3,homology modeling techniques were utilized based on the previous study[8].The amino acid sequences of T1R1/T1R3 were gained from UniProtKB (https://www.UniProt.org/).The homology model used was the metabotropic glutamate receptor (PDB ID: 1EWK)by Discovery Studio 2019 (NeoTrdent Technology Ltd.,Beijing,China).Molecular docking was carried out in CDOCKER program of Discovery Studio,referring to the reported with some changes[23].The 3D structure of the peptides was constructed using Discovery Studio.Homologous receptors and peptides were optimized using Minimization protocol.The solvent molecules were removed,and hydrogen atoms were added.
The CDOCKER program can test the ability of peptides to bind to the T1R1/T1R3 umami receptor,and the “-CDOCKER_INTERACTION_ENERGY” is more accurate for evaluating the binding affinity between the umami peptides and T1R1/T1R3[21].Therefore,the screening and prediction of umami peptides were conducted using “-CDOCKER_INTERACTION_ENERGY”as the indicator.
2.7 Solid-phase synthesis of umami peptides
The three peptides (FLNQDEEAR,FNKEE,EEFLK) with high S(P) values and perfect “-CDOCKER_INTERACTION_ENERGY”were synthesized by the solid-state synthesis method through China Peptides Co.,Ltd.(Shanghai,China) with purity not lower than 95%.
2.8 Sensory evaluation
The sensory evaluation was conducted in a sensory analysis lab at a temperature of (22.5 ± 2.5) °C under normal lighting situations.The internal panelists comprised 4 females and 6 males,aged between 25 and 35 years old,with no smoking and excessive drinking habits.Before the experiment,10 internal panelists were trained to taste reference solutions and record sensory descriptions,ensuring they could identify the five flavors[8,20,24].Referring to the previous methods with few alterations[8],MSG solution (3.5 mg/mL),sucrose solution (10 mg/mL),sodium chloride solution (3.5 mg/mL),L-isoleucine solution (2.5 mg/mL),and citric acid solution (0.8 mg/mL)were used as reference standards for umami,sweet,salty,bitter,and sour tastes,respectively.
Three-digit random codes were used to number the samples.The panelists were instructed to swirl the sample for ten seconds and expectorate.They were asked to rinse their mouths with ultrapure water at least twice and take a 2 min break to prevent fatigue and legacy effects.For each sample,the sensory evaluation experiment was repeated three times.The work described was carried out following the Code of Ethics of the World Medical Association(Declaration of Helsinki) for experiments involving humans.
2.8.1 Sensory evaluation of fractions eluted by silica C18 packing
The lyophilized samples of 0%-E,20%-E,and OY-W were dissolved in distilled water at a concentration of 5 mg/mL.The samples were graded on a scale of 10-point,with 0 and 5 representing the sensory intensity of distilled water and the reference solutions,respectively,and 10 representing an intense taste.
2.8.2 Sensory evaluation of synthesized peptides
The synthesized peptides were dissolved in distilled water at a concentration of 1 mg/mL for sensory evaluation.According to the method described in section 2.8.1,the panelists were asked to evaluate the sensory attributes of the synthetic peptide solutions.The peptides were conducted in a triangle test to determine the taste threshold[25,26].The peptide solution was stepwise diluted in a ratio of 1:1 (V/V) with distilled water and labeled,and submitted to internal panelists in the order of concentration increase.
2.9 Taste analysis by electronic tongue (E-tongue)
The taste characteristics of the samples were determined using the TS-5000Z taste sensor system (Insert,Tokyo,Japan),referring to the reported method with some alternations[27].The four fractions,0%-E,20%-E,100%-E,and OY-W,were dissolved in ultrapure water at a concentration of 5 mg/mL,while the aqueous solution of synthetic peptides in ultrapure water was prepared at a concentration of 0.6 mg/mL.MSG in ultrapure water at concentrations of 2 mg/mL was detected as the reference.Sensory probes,CA0,C00,AE1,AAE,and CT0,were used to detect sourness,bitterness,astringency,umami and saltiness,respectively.Two reference probes were self-checked with the reference solutions (2.236 5 mg/mL KCl and 0.045 mg/mL tartaric acid).At room temperature of 25 °C,sensors and reference probes were immersed in the sample solution,and the membrane potential changes generated by the solution were recorded and analyzed.
2.10 Statistical analysis
All the experiments were carried out in triplicate.SPSS 19 (SPSS Inc.,Chicago,IL,USA) was applied to analyze the differences in mean values of the data using a one-way analysis of variance(ANOVA).The figures in this article were constructed by Origin 2019 (Origin Lab Co.,Northampton,MA,USA).
3. Results and discussion
3.1 Taste characteristics of oyster alcohol extract components
The sensory evaluation and e-tongue test results of 0%-E,20%-E,100%-E and OY-W were displayed in Fig.1.As illustrated in Fig.1,the two fractions,OY-W and 0%-E,exhibited similar overall tastes to each other,regarding sourness,sweetness,saltiness,bitterness and umami.This is probably because most of the substances in OY-W are water-soluble,and they are retained in 0%-E during the elution with distilled water.The 100%-E fraction was not subjected to the sensory evaluation test attributed to its unacceptable off-flavor.According to the e-tongue test results (Fig.1B),the fraction 100%-E had a more intense flavor in umami,saltiness and bitterness compared with OY-W and 0%-E,but much weaker than that of 20%-E.The umami taste of the fraction 20%-E was the strongest (1.50) compared with 0%-E and 100%-E (Fig.1B).Similarly,the 60% ethanolic extract fraction fromTakifugu obscurusmuscle had a higher umami intensity compared with the ultrapure water extract[28].As the fraction 20%-E showed the strongest umami potential,it was consequently collected and freeze-dried to excavate the umami peptides.

Fig.1 The taste characteristic profiles of the alcohol extract of the Pacific oyster (Crassostrea gigas).(A) Sensory evaluation results.(B) E-tongue test results.OY-W: spray-dried oyster powder;0%-E,20%-E and 100%-E: eluent components of oyster power in ultrapure water by 0%,20% and 100% (V/V) ethanol solutions,respectively.Standard material: MSG solution (3.5 mg/mL),sucrose solution (10 mg/mL),sodium chloride solution (3.5 mg/mL),L-isoleucine solution(2.5 mg/mL),and citric acid solution (0.8 mg/mL),used as reference standards for umami,sweet,salty,bitter,and sour tastes,respectively.The concentration of 0%-E,20%-E,100%-E and OY-W was 5 mg/mL.Sensory evaluation of the sample 100%-E was not carried out due to its unacceptable off-flavor.
3.2 Identification and prediction of umami peptides
Using nano-HPLC-MS/MS,2 744 peptides were identified from 20%-E.Among them,1 262 peptides with S(P) values ≥ 588.00 were judged to be umami peptides by scoring with the iUmami-SCM algorithm.These peptides could be divided into different groups according to their amino acid numbers: 3-9 amino acids (256),10-19 amino acids (766),20-29 amino acids (213),30-39 amino acids(18),and 40-45 amino acids (8),and 57 amino acids (1).Some researchers have demonstrated that umami peptides are mainly short peptides (4-9 amino acids)[29].Hence,13 short peptides with the highest S(P) scores were chosen for molecular docking (Table 1).They were FLNQDEEAR,FNKEE,FSSVTLST,EEFLK,AVTTL,TLLT,LSWV,LSYF,LSFY,VPDGDLS,IWT,LWT,and LSPL,respectively.
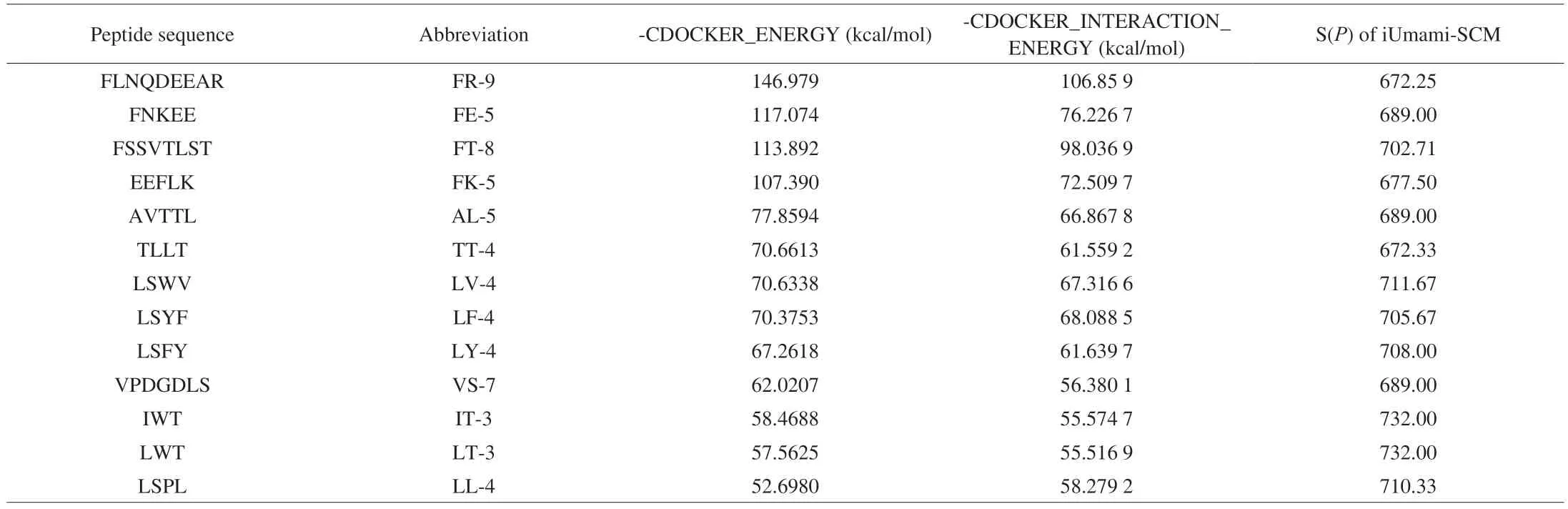
Table 1 Molecular docking energy of umami peptides with T1R1/T1R3 and score on the iUmami-SCM website.
Presently,molecular docking is an advanced technique to simulate the recognition process between receptor and ligand molecules by computer[20].Previous research has used molecular docking to simulate the binding of umami peptides and receptors[8].In our study (Fig.2),the RAMAN analysis of the final homology modeling of receptor T1R1/T1R3 showed that 98.0% of the residues were found in the allowed areas,of which 68.5% residues were in the optimal region,26.2% residues were in the acceptable region,and 3.3% residues were in the general allowable region.According to the 90% criticality evaluation principle,this model was logical in dihedral distribution and spatial collision.The program CDOCKER of Discovery Studio was used to simulate the interactions of 13 peptides with the umami receptor T1R1/T1R3.The molecular docking energy according to the optimal docking posture was shown in Table 1.

Fig.2 Homology model and the molecular docking binding sites of T1R1/T1R3 receptors.(A) Homologous structure of the umami receptor T1R1/T1R3:green,β-turn;blue, β-sheet;red,α-helix;gray,random coil.(B) Ramachandran plot for the homology model of T1R1/T1R3.
Among the 13 peptides,FLNQDEEAR (FR-9) exhibited the highest -CDOCKER_INTERACTION_ENERGY (106.859 kcal/mol),followed by FSSVTLST (FT-8,98.036 9 kcal/mol),FNKEE (FE-5,76.226 7 kcal/mol),and EEFLK (FK-5,72.509 7 kcal/mol).The LWT(LT-3) had the lowest energy (55.516 9 kcal/mol).According to Table 2,the interaction energy generated by docking did not show a clear correlation with the peptide length.However,peptides FLNQDEEAR and FSSVTLST had a higher -CDOCKER_INTERACTION_ENERGY compared to the other peptides with shorter lengths.This might be because these two peptides had appropriate amino acid numbers which can shorten the docking distance with the umami receptor.In addition,the long-chain peptides might be more flexible to bind to the umami receptor[30].Based on the results from iUmami-SCM and molecular docking,the three peptides FLNQDEEAR,FNKEE and EEFLK with two Glu residuals at different positions of the peptide chain were selected and synthesized to further verify their taste characteristics and their interaction with the umami receptor,to primarily investigate the underlying mechanism between peptide structure and umami intensity.

Table 2 The umami thresholds and sensory description of the three umami peptides.
3.3 Sensory characteristics of synthetic peptides
The sensory evaluation and the e-tongue analysis results of the synthetic peptides were shown in Fig.3.As shown in Figs.3A &B,the peptide FE-5 had a stronger umami taste than EK-5 (2.2),despite the same amino acid length of these two.This might be attributed to Glu residual at the C-terminus of the peptide FE-5.It has been reported that,when Glu is located at the C-terminus of the peptide,this peptide is more likely to have a strong umami taste[31].For the peptide FR-9,it had the lowest umami taste,but the strongest bitterness,probably because the bitterness masked its umami taste[32].Furthermore,the Glu of FR-9 was situated in the middle of the peptide chain,which means it couldn’t make a strong bond with the umami receptor compared with Glu residual at the edge,resulting in a lower umami taste than FE-5 and EK-5.The e-tongue results were consistent with the sensory evaluation,demonstrating that the electronic tongue can analyze the taste of peptides,which is beneficial for comprehending the full taste characteristics of peptides.
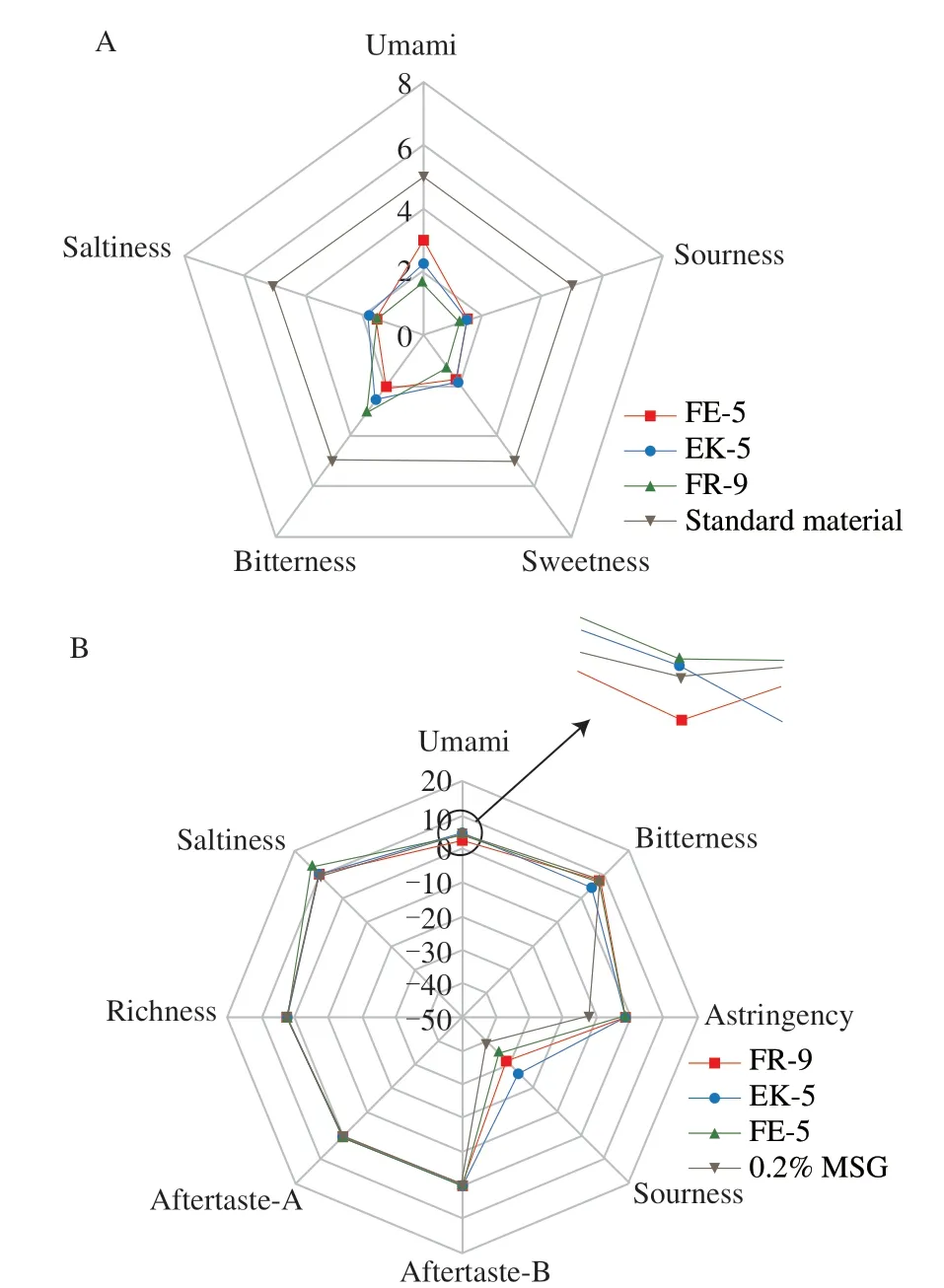
Fig.3 The taste characteristic profiles of the synthetic peptides.(A) The sensory taste profiles of the synthetic peptides.The concentration of peptides was 1 mg/mL.(B) The E-tongue taste profiles of the synthetic peptides.The concentration of peptides was 0.6 mg/mL.The 2 mg/mL MSG was detected as the reference solution.
The umami taste thresholds of the three peptides were shown in Table 2.The umami thresholds for the peptides FE-5,EK-5 and FR-9 were 0.38,0.55 and 0.58 mg/mL,respectively,much lower than that of the peptides with similar amino acid length found inRuditapes philippinarum,such as the peptides SEEK (1.00 mg/mL),TYLPVH(2.00 mg/mL),TGDPEK (2.00 mg/mL),KEMQKN (0.99 mg/mL)[33].This result indicated that the umami peptides found in our study may have a great potential to be used as potent umami additives in the food industry.
3.4 Computational molecular docking of umami peptides into T1R1/T1R3
Fig.4A-C showed the 2D-diagrams of the molecular docking pose,including the optimal docking poses and docking sites for docking with the umami receptor.The receptor binding sites of umami peptides were given in Fig.4D.As shown in Fig.4,the binding sites of the three umami peptides with the VFTD of the T1R3 subunit included Arg591,Phe592,Gly695,Gln853,His915,Pro968,Arg712 and Pro715.All three peptides could bind to that the Gln853 and Phe592 binding sites,which were the most critical binding sites.Some scholars believe that umami peptides are more likely to bind the umami receptor T1R1/T1R3 when they contain one or two acidic amino acids (Asp and Glu)[27,34].It was discovered in this study that the binding forces between umami peptides and T1R3 subunit were mainly hydrogen bonding,electrostatic interaction,van der Waals force and hydrophobic interaction.The hydrogen bonds were the most in this study,followed by electrostatic interaction,where the salt bridge was an important bond.

Fig.4 The molecular docking poses and binding sites of umami peptides interacting with T1R1/T1R3.(A) FNKEE peptide;(B) EEFLK peptide;(C) FLNQDEEAR peptide;(D) The number of molecular docking binding sites.Different colors represent different numbers of binding sites between peptides and T1R1/T1R3 during the molecular docking process: Green,0;Light blue,2;Deep blue,3.
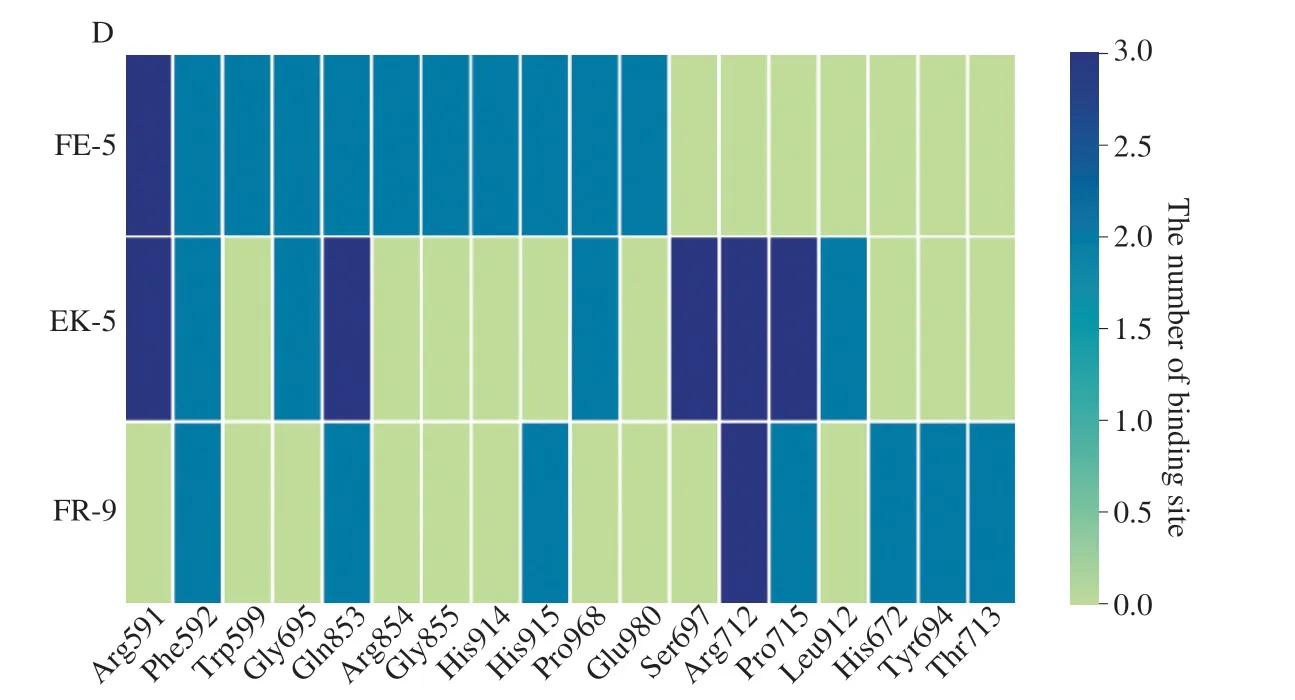
Fig.4 (Continued)
The C-terminal of the peptide FE-5 with the highest taste score was composed of two Glu acid groups,which formed strong salt bridges with Phe592,Arg712,and His915,while the N-terminal Asn could form hydrogen bonds with Gln853 and Thr713.
In contrast to FE-5,the N-terminal of EK-5,with a lower taste score than FE-5,was two Glu residuals,which formed hydrogen bonds with Ser697,Pro715,and Gly695 respectively.It was worth mentioning that the terminal Glu formed unfavorable interactionswith Glu828 and Asp717.This might be the reason why EK-5 had a less umami taste than FE-5.Interestingly,the amino acid residual Lys was found at the C-terminus of EK-5 and strongly adhered to the binding site Arg591 by a salt bridge.For the peptide FR-9 with the lowest umami score,its C-terminal amino acid residual Arg built a salt bridge with Arg591,and its middle Glu residual formed hydrogen bonds with Typ599 and Phe592.It has been reported hydrogen bonding and electrostatic interactions frequently play a role in the interaction between umami receptors and peptides[21]consistent with the findings in this study.
4. Conclusion
In summary,the computer algorithm (iUmami-SCM and molecular docking) was used to screen and predict the umami taste of oyster-derived peptides for the first time.Three novel umami peptides,FLNQDEEAR,FNKEE,and EEFLK,were identified from the Pacific oyster (Crassostreagigas).Their taste characteristics and underlying interaction with the umami taste receptor were investigated by sensory analysis,e-tongue test and molecular docking.The interaction between the umami receptor and peptides was peptide-specific and peptides with two Glu residuals at the terminals had a stronger umami taste than the peptide with two Glu residuals in the middle.More research is needed towards revealing the relationship between peptide structure and umami intensity.
Conflict of interests
Ming Du is an editorial board member forFood Science and Human Wellnessand was not involved in the editorial review or the decision to publish this article.All authors declare that there are no competing interests.
Acknowledgments
This work was supported by the National Key Research and Development Program of China: Investigate the mechanism of formation and control technologies of Chinese traditional and ethnic food quality (2021YFD2100100).
Appendix A.Supplementary data
Supplementary data associated with this article can be found,in the online version,at http://doi.org/10.26599/FSHW.2022.9250012.
杂志排行
食品科学与人类健康(英文)的其它文章
- GUIDE FOR AUTHORS
- Targeting gut microbiota in osteoporosis: impact of the microbial based functional food ingredients
- Weizmannia coagulans: an ideal probiotic for gut health
- Natural sources,refined extraction,biosynthesis,metabolism,and bioactivities of dietary polymethoxyflavones (PMFs)
- A review of salivary composition changes induced by fasting and its impact on health
- Minerals in edible insects: a review of content and potential for sustainable sourcing
