Food nutrition and toxicology targeting on specific organs in the era ofsingle-cell sequencing
2024-02-16XiaofeiWangXiaowenChengHuilingLiuXiaohuanMuHaoZheng
Xiaofei Wang,Xiaowen Cheng,Huiling Liu,Xiaohuan Mu,Hao Zheng*
National Engineering Technology Research Center for Fruit and Vegetable Processing,Key Open Laboratory of Fruit and Vegetable Processing,Ministry of Agriculture and Rural Affairs,Beijing Key Laboratory of Food Non-Thermal Processing,College of Food Science and Nutritional Engineering,China Agricultural University,Beijing 100083,China
Keywords:Dietary food Cellular heterogeneity Single-cell RNA sequencing Food nutrients Food toxicants
ABSTRACT Due to the complex natures of dietary food components,it is difficult to elucidate how the compounds affect host health.Dietary food often selectively presents its mechanism of action on different cell types,and participates in the modulation of targeted cells and their microenvironments within organs.However,the limitations of traditional in vitro assays or in vivo animal experiments cannot comprehensively examine cellular heterogeneity and the tissue-biased influences.Single-cell RNA sequencing (scRNA-seq) has emerged as an indispensable methodology to decompose tissues into different cell types for the demonstration of transcriptional profiles of individual cells.ScRNA-seq applications has been summarized on three typical organs (brain,liver,kidney),and two representative immune-and tumor related health problems.The everincreasing role of scRNA-seq in dietary food research with further improvement can provide sub-cellular information and the coupling between other cellular modalities.In this review,we propose utilizing scRNAseq to more effectively capture the subtle and complex effects of food chemicals,and how they may lead to health problems at single-cell resolution.This novel technique will be valuable to elucidate the underlying mechanism of both the health benefits of food nutrients and the detrimental consequences food toxicants at the cellular level.
1. Introduction
One of every five deaths in the world is attributed to a suboptimal diet,more than any other risk factor[1].An altering diet can shift the expression states of genes,and these alterations might lead to phenotypic transition,increasing or decreasing disease risks[2].Studies in animal models primarily drove this modern notion.The connections between diet and gene expression alteration,on the one hand,and between gene expression alteration and health problems,on the other,are supported by observational studies in human and animal models[3].Most studies attempting to link dietary components to health and disease have targeted the host phenotype,gene expressions,and metabolic pathways.Changes are evident at various molecular levels,including DNA,the transcript,mRNA,and proteins in physiology[4].However,the mechanisms behind how diet can impact host health at the molecular and cellular level are far from certain due to the complex natures of these compounds and the challenges of establishing biologically-relevant experimental models.Therefore,the precise determination of the molecular mechanisms underlying the state of health and disease after diet intervention needs further exploration.
Cells are the basic building blocks of organisms,and each cell is unique.Due to organism complexity and cell heterogeneity,conducting these dietary nutrients and toxicants experimentations on a small number of cells and even at the resolution of a single cell is necessary.Single-cell RNA sequencing (scRNA-seq) has emerged as an integral approach to decomposing tissues into cell types and/or cell states and demonstrating individual cell transcriptional profiles[11].This technology avoids the cell averaging seen in bulk RNA-seq.It offers enormous potential fordenovodiscovery,which may significantly enhance human well-being prospects.One of the major challenges in human health is the patient-specific drug response due to the patient-specific cell-to-cell heterogeneity,highlighting the growing importance of single-cell techniques in precision nutrition and precision medicine[12].The application of scRNA-seq to nutrition and health research has expanded in recent years,including comparing healthy and disease-related tissues at single-cell resolution and developing personalized nutrition strategies to understand the link between diet and health[13].At this juncture,dietary nutritional and toxicological research based on sing cell RNA technology is a very young science.Here,we concentrate on the subjects likely to yield information that will prove useful in the dietary nutrients arena as well as in food toxicants.
In this review,we briefly discuss the single-cell RNA sequencing data on diet and health and the link between diet and disease.We summarize single-cell RNA sequencing applications on three typical organs (brain,liver,kidney),and two representative immune-and tumor related health problems (Table 1).We anticipate an everincreasing role of single-cell RNA sequencing in dietary food research with further improvement in providing sub-cellular information and coupling to other cellular modalities (Fig.1).This technology may decipher the variation of cell specifications for diet interventions,and comparing healthy and disease-related tissues at single-cell resolution,with an ultimate goal to enhance our understanding of the links between diet and health.Still,we strongly encourage food scientists to think about how scRNA-Seq can be applied to food nutrition and toxicological values of interest.

Fig.1 Schematic diagram of the application of scRNA-Seq in food nutrition and toxicology.
2. Applications of scRNA-Seq in dietary food metabolism and host health

Dietary molecules must be absorbed and metabolized before their final uses as sources of energy,catalysts for reactions,ligands for signaling receptors,or structural components of cells.Modulation of their metabolism could result in beneficial or detrimental consequences on host health,probably due to the levels of gene expression and the alterations in the landscape of metabolites.Dietary nutrients and toxicants usually have “target” organs,which might act locally in the brain,liver,kidney,gut,or systemically in other organs.There seemingly homogeneous organs are complex tissues containing numerous cells with specialized characteristics in morphology,connectivity,and functions,and their cells can be classified by location,morphology,molecular markers and gene expression patterns[14].For instance,the brain cells may explain issues relating cells to learning,memory,and other cognitive functions.Therefore,the complexity of organs and the heterogeneity of cells necessitate that these dietary experiments be performed at the resolution of a small number of cells or even single cells.Although single-cell transcriptomics has a wide range of applications in biological research to compare healthy and disease-related tissues,this approach is still in its infancy in food nutrition and toxicology[13].
2.1 Brain foods——how to nourish the brainpower
There is a voluminous body of evidence from observational and intervention studies regarding the importance of diet with“memorable” food for brain health and cognitive functions.The consumption of diets rich in antioxidants (polyphenols,carotenoids,vitamin C,etc.) may prevent neurodegeneration and slow down a progression to full-blown disease[15–17].The important role of trace elements such as selenium,copper,iron,lithium,and zinc on brain health is now well-recognizedin vitroandin vivostudies[18–22].The neuroprotective role of polyunsaturated fatty acids (PUFAs)is documented in nutritional studies,prospective populationbased surveys and clinical trials.On the other hand,deleterious dietary behavior (overfeeding,a high caloric/low dietary fiber diet),environmental factors (alcohol,drugs,and exposure to pesticides)might speed up neurodegeneration[23].Researches about how these “memorable” food fight through neurodegeneration include epigenetic mediation,gut microbiota modulation,reversal of lowgrade inflammation and anomalies in lipid metabolism,however the molecular mechanisms underlying their potential therapeutic properties remain unknown[6].
The restricted access to brain tissue and neuronal cell types presents the major hurdle to understand neurodegenerative disease mechanism,and to develop therapeutic drug and neuroprotective diets[24].Dietary intake signals map exclusively to specific brain regions and are enriched for genes expressed in specialized subtypes of GABAergic,dopaminergic and glutamatergic neurons.Tea polyphenols,rich in antioxidants,are bioactive polyphenol that have been shown to alleviate motor impairment and dopaminergic neuronal injury,indicating their preservation of dopaminergic neurons[25].Unlike neuron,glial cells (oligodendrocytes,astrocytes,ependymal cells,microglial cells,satellite cells and Schwann cells)are often thought of as the supporting cast of the nervous system.Nevertheless,Yan et al.performed scRNA sequencing to evaluate the effects of oolong tea polyphenols in hypothalamus cell types in mice,with significant increase of the numbers of astrocytes.Other than polyphenol,PUFA-DHA,could enhance astrocyte-like morphologic formation in rat astrocytes[26].DHA supplementation increased the expression of glial fibrillary acidic protein,elevated the production of glial cell-derived neurotrophic factor in the astrocytes,demonstrating that the participation of astrocyte cell and the process of gliogenesis may be relevant to therapeutic effects ofn-3 PUFAs[27].It can be demonstrated that brain food usually has target cell type,traditional food nutritional experimentsin vivoandin vitroprefer taking the whole brain as the material to explore its mode of action.However,it is conceivable that mechanisms that might focus on astrocyte cells but would not be detected if the whole brain were examined.Moreover,even for one single type of cells,each individual cell may not respond in the exactly same way to exogenous diet stimulation.Brain cell heterogeneity would shed light on possible differences in the function,nutritional evaluation and underlying mechanisms of memorable food at the single cell level.
Other than memorable food,heavy metal in the environment may cause neurotoxicity and potentially interfere with neurogenesis,such as Cadmium.Single-cell RNA sequencing showed that the cadmium exposure led to an increase in the differentiation of neural stem cells into astrocytes while a decrease into neurons[28].In the context of the central nervous system,scRNA-seq can discriminate different cell types between samples at a population level,which paves the way towards a more precise understanding of neurological and neurodegenerative diseases[29].Zeisel et al.sequenced 3 005 singlecells in the mouse somatosensory cortex and hippocampal region,revealed 9 major classes of cells (interneurons,mural,ependymal,S1 pyramidal,CA1 pyramidal,endothelial,astrocytes,microglia and oligodendrocytes),and identified numerous marker genes for alignment with known cell types,morphology and location[30].As one of the most common neurodegenerative diseases,Alzheimer’s disease(AD) starts with mild memory loss,with pathological hallmarks of extracellular amyloid plaques deposition and intracellular hyperphosphorylated tau proteins accumulation[31].In order to understand the molecular complexity that drives AD pathogenesis,numerous gene expression profiles have been performed[32–34].Nevertheless,bulk samples of AD brain can not practically provide precise transcriptional changes across different cell types,particularly minor cell populations.Analyzing single cell transcriptomes showed gene expression alterations occurred in prefrontal and entorhinal cortex of AD brain,corresponding to cell types of excitatory neurons,inhibitory neurons,microglia,astrocytes and oligodendrocytes between AD and healthy individuals[35,36].Therefore,the ability to identify individual cells opens the possibility of finding pathogenic cells and pathways that can be exploited for functional food nutrient and toxicant discovery.
2.2 Liver detox food——how to repair the liver
The liver is the largest solid organ in the body,and performs vital functions of the detoxification,lipolysis,gluconeogenesis and the secretion of bile acid to enable food digestion.There are mainly two epithelial cell types (hepatocytes and cholangiocytes) within the liver,the former comprise the bulk of the liver mass with functions of bile production,detoxification,carbohydrate and lipid metabolism,while the latter act as biliary epithelial cells contributing to bile secretion[37,38].Halpern firstly utilized single-cell transcriptomic technology dissected hepatocyte zonation across liver lobule with zonated liver gene expression,providing further zonation study in other hepatic cell lineages[39].Previously unknown molecular patterns of metabolic zonation of hepatocytes,endothelial cells and hepatic stellate cells across the liver lobule were discovered through scRNA[40].This methodology further provides novel insights into critical pathophysiological changes during hepatic fibrosis,with distinct populations of macrophages,endothelial cells and mesenchymal cells to promote scar formation.
Due to its high prevalence,liver diseases,including non-alcoholic fatty liver disease (NAFLD),cirrhosis and fibrosis,are becoming increasingly important health issues.Current treatment options for patients are limited to removal of the underlying cause,if possible,or liver transplantation.These liver injury conditions are associated with alterations in the significantly increased expression of the proinflammatory genes (TNF-α,IL-6,NF-κB etc.),and liver adipogenesis related genes (ACC,PPAR-g,SREBP-1 etc.).The pathways of fatty acid synthesis,oxidation and secretion pathways are disturbed.Diet intervention is the first-line treatment for majority of live diseases.Thus,deciphering the molecular mechanism and developing effective therapies would be likely to have a considerable benefit on morbidity and mortality.
Facing the child s clear and glistening4() eyes, the hostess was stupefied, leaning on the door feebly() and holding her face with two hands, afraid to look him in the eye.
In animal models,short-term high-fat diets lead to hepatic steatosis.High fat diet altered single-cell transcriptome profile of the entire hepatocyte population in liver with zonation-independent heterogeneity,through fat accumulation and ketogenic pathway[41].Western diet (high fat,high sugar,high cholesterol) induced liver injury in mice with injury-associated macrophage clusters,identifying a distinct population of monocyte-derived macrophages that expanding during liver fibrosis with accelerated scar deposition.Moreover,distinct population of endothelial cells also expand in liver fibrosis and are located in the fibrotic niche.On the contrary,dietary polyphenols,spices,prebiotics,vitamins,n-3 PUFA have been documented to ameliorate progression of liver diseases.Integrating scRNA-seq data revealed that Curc-mPEG454,a curcumin derivative,effectively inhibited the expansion of scar-associated macrophage subpopulation and scar-producing myofibroblasts in the damaged liver,and remodeled the fibrotic niche via regulation of ligandreceptor interactions including platelet-derived growth factor-B/platelet-derived growth factor receptor-α signaling.The study of scRNA analysis provides a molecular framework for dietary intervention as the therapeutic targeting of key pathogenic cell populations.
2.3 Renal diet——how to avoid for kidney failure
The kidney is a highly complex organ with anatomically discrete segments,including the glomerulus and renal tubules,which are important components of the nephron.The functional complexity of kidney appears to be associated with different cell types in different structures[42].Glomerular endothelial cell and parietal epithelial cell are two common glomerular cell types,and the former synthesize the basement membrane and prevent the loss of proteins into urine,while the latter contributes to glomerulosclerosis,crescent and pseudocrescent formation.Renal tubule consists of the proximal and the distal tubule,regulating acid-base balance and electrolyte transport,respectively.Previous studies performed bulk RNA sequencing of different components of the kidney with transcriptome results of different segments.However,these outcomes can only reflect the overall average RNA expression,not the single-cell level.
Park et al.identified a novel cell type,transitional cell,in the kidney collecting duct in adult mice,and they found the inherited kidney diseases raised from distinct genetic mutations actually share the same cell of origin with similar phenotypic manifestations[43].Initially,the collecting duct was recognized with only one role in water reabsorption,while in recent years the identification of novel cell types and functions in the kidney collecting duct has become greatly enhanced.This compartment is comprised of more than three distinct cell types: the principal cells responsible for sodium,water reabsorption and potassium secretion;and the alpha and beta intercalated cells,which are responsible for acid and alkali secretion to balance acid-base status;and a third distinct transitional cell type with low expression levels of stress response genes and cell cycle genes,suggesting that the principal cells and the intercalated cells may undergo cellular transitions[44].
Oral administration of the probioticLactobacillus casei Zhangreduced the damage to renal tubular epithelial cells and alleviated kidney injury,potentially indicating slowing the progression of acute and chronic kidney disease[45].Other than probiotic diets,food toxicants may cause kidney injury.Ochratoxin A,a common food contaminant in improperly stored food products,is reported to cause seriously toxicological effects of nephrotoxicity,and scRNA reveals the tubule epithelial cells are the main target cell types,with potential markers of sensitive genes (Slc5a2,Cyp4a8,and Havcr1 etc.).
2.4 Immunity food——how to boost the immune system
Dietary food plays an essential role in developing,maintaining,and modulating the immunity of an individual.Essential nutrients,such as amino acids and fatty acids produce biosynthetic precursors(antibodies,cytokines,lipid immunologic mediators and so on)involved in the immune response.Other foods for a healthy immune system mainly include probiotics and prebiotics,antioxidants,vitamins,micronutrients and so on.The immunological mechanisms underpinning probiotics and prebiotics on modulating host immunity should continue to be better described for dendritic cells (DCs),epithelial cells,T regulatory cells,effector lymphocytes,natural killer T cells,and B cells[46].The immune food works either by entering into the defensive mechanism directly through interfering with the target viruses,or indirectly through activating the cells associated with the adaptive immune system.Vitamin C can support the function of neutrophils,natural killer (NK) cells,whereas zinc,copper and selenium promote monocyte and macrophage activity.In order to the“classic” nutrients,bioactive compounds,such as plant polyphenol and polysaccharide can indirectly enhance host immunity or directly inhibit and disrupt the virus particle,activating NK cells,DC and T lymphocytes[15,47–49].The above-mentioned nutrients increase the immunity of people and may work as therapeutic potential against viral infection;however,the underlying cellular mechanisms remain unclear.Therefore,in order to analyze complex cell mixtures and correct to a single cell and single molecule,it is qualified to analyze immune food reactions with scRNA method.
As a specialized defense mechanism of our body,the immune system is composed of multiple cell types in primary and secondary lymphoid organs and tissues,responding to foreign challenges in order to monitor and maintain health.Innate immunity mainly contains DCs,innate lymphoid cells,etc.,while the adaptive immunity includes T and B lymphocytes[50].Characterizing this complex heterogeneity of cells,and their interconnectedness requires studies at single-cell resolution.One of the key hallmarks of aging is the deterioration of the immune system,more prone to infections and inflammatory disorders.When exploring CD4 T cells from young and old mice,sc-RNA sequencing implied the CD4 T cell subsets—exhausted,cytotoxic,and activated regulatory T cells appear rarely in young mice but gradually accumulate with age[51].Moreover,single cell RNA sequencing enables the discovery of pathogenic immune cell subsets in health and diseases,and their developmental ‘trajectories’.Jaitin et al.classified splenic cells into transcriptional B cell,NK cell,macrophage,monocyte and plasmacytoid DC subsets,demonstrating immune cells can respond to infection by activating cell type-specific program[52].Similarly,cell subsets of innate lymphoid cells expressed NKp46 has been documented in human tonsil and small intestine[53].LPS stimulation activated only a small subset of bone marrow-derived DCs during the early stage of infection,whereas in the later stage genes expressed uniformly,indicating the dynamic regulation of the activation of signaling circuits.In the specific case of the colonic mucosa,the inflammatory fibroblasts,inflammatory monocytes,microfoldlike cells,and T cells co-expressing CD8 and IL-17 expand with ulcerative colitis[54].Abnormal immune responses have been reported in Coronavirus Disease 2019 (COVID-19),and scRNA-seq revealed that critical COVID-19 infection contained higher proportions of macrophages and neutrophils and lower proportions of myeloid DCs,plasmacytoid DCs and T cells,implicating potential mechanisms underlying pathogenesis and recovery in COVID-19[55].
2.5 Anti-cancer diet——how to lower the cancer risk
Tumor is often viewed as an abnormal development process,and the extent to which its cellular heterogeneity and transcriptional diversity on the differentiation mechanism remains unknown.Singlecell sequencing improved our understanding of premalignant invasion,intratumor heterogeneity,tumor microenvironment (TME),metastasis dissemination and therapeutic resistance[56].A major question during cancer research is the growing progress from premalignant to malignancies,while the decrease of pro-inflammatory immune cells and increase of re-programmed myofibroblasts has been demonstrated between premalignant and advanced pancreatic cancer[57].Tumor cells exhibit diverse phenotypes in proliferation,demonstrating differentiation trajectories of tumor cells in early-stage gliomas(astrocytic,oligodendrocytic) as well as dynamic plasticity between cell states in invasive glioblastoma[58].The TME can be divided into 1) stromal components,as scRNA studies revealed heterogeneous cell subtypes of carcinoma-associated fibroblasts and tumor endothelial cells[59],2) immune cells due to immunotherapy in cancer treatment,as decreased activated T cells and increased peripheral macrophage programs accompanying by progression in cancers[60].Single cell sequencing resolve the gaps during metastasis,from clone evolution,to metastatic intermediates,to cancer cell dissemination,for example,previous study identified stem-like cells initiating metastasis and implicatedMYCexpression in high-burden disease[61].Circulating tumor cell (CTC) has been explored by scRNA-seq,and CTC gene expression signatures have correlated with therapeutic response and metastasis risks in numerous cancers,paving the way for therapeutic applications[62,63].Moreover,the single cell technologies provide new opportunities for clinical translation in early tumor detection and target drug discovery.
On the other hand,tumors reply on nutrients intaken by the host for their survival,and diet modification to the host can change the metabolic activity,tumor proliferation rate and oncogenic signaling[64].Fasting and fasting-mimetic diet (low-calorie,low-protein,etc.) resulted in anti-cancer effect,preventing and delaying the development of sarcoma and lymphoma[65,66].As a central nutrient,glucose is pro-tumorigenic by providing energy for the synthesis of biomolecules and inducing pro-oncogenic signaling that sustain the high proliferation rate of cancer cells,while reductions in dietary glucose can decrease circulating insulin and lessen activity in the proproliferative PI3K and mTOR signaling pathways,leading to cancer progression inhibition[67].When glucose is limited,depriving patients of fatty acids may starve specific types of cancer (oral,cervical,gastric carcinoma,etc.) by deactivating fatty acid oxidation in the context of overexpression ofMYCor carnitine palmitoyltransferase[68,69].Pascual et al.showed that dietary palmitic acid,but not oleic acid or linoleic acid,can promote metastasis in oral carcinomas and melanoma in mice.However,it remains unknown whether palmitic acid induce prometa-static statedenovoor whether it enhance the potential of cells already present within a tumor.Single-cell RNAsequencing analyses indicate that dietary palmitic acid stimulated intratumoural Schwann cells,(not basal epidermal,vascular/lymphatic endothelial cells,erythrocytes,or smooth muscle cells)to promote prometastatic memory[70].Since the major challenges for cancer drug therapy is inter-and intra-tumoral heterogeneity,scRNA-seq identified different drug perturbations: etoposide has a conserved impact on proliferating tumor cells,while panobinostat treatment affects both,including unexpected effects on the immune microenvironment[71].Therefore,tumoral heterogeneity needs single cell sequencing to underly the bio-characteristics of cancer tissues,to explain how diet effect the oncological processes from tumorigenesis through progression to metastasis and therapy resistance.
3. Design and workflow of scRNA-seq in food nutrition and toxicology
Cells are the building blocks of tissues and organs,and their unknowns and complexity limit the complete interpretation of the cell diversity underlying the biological systems.The primary determinant of cell function is the transcription process.Traditionally,gene expression measurements are carried out on “bulk” samples containing thousands of different cells.While experimental bias in bulk RNA-sequencing is well studied,both single cell RNAseq and single nucleus or nuclei RNA-seq (snRNA-seq) are relatively new techniques.Over the last decade,the field of singlecell transcriptomics has progressed rapidly since the development of the first single-cell RNA sequencing on mammalian cells[72].Subsequently,individual cell heterogeneity that was previously masked in bulk measurements can be revealed.In certain cell types,nuclei can be easily isolated without the use of enzymes,and they are more resistant to physical stresses compared to cells.However,although most genes show similar relative abundances in cells and nuclei,a small population of genes is depleted compared to whole cells.For example,snRNA-seq is not suitable for the detection of microglial activation genes and cellular state in humans[73].Therefore,caution should be applied when choosing snRNA or scRNA-seq for the assessment of targeted cells/organs related to the functional activity of dietary food.To give a general understanding of scRNAseq concepts,we provide a brief overview of the experimental workflow to discuss potential applications of this technology in food science.Although a comprehensive review of this complicated process is not listed here,the experimental design and workflow have been provided,mainly with four steps: single-cell isolation,library construction,sequencing,and bioinformatic data analysis (Fig.2).
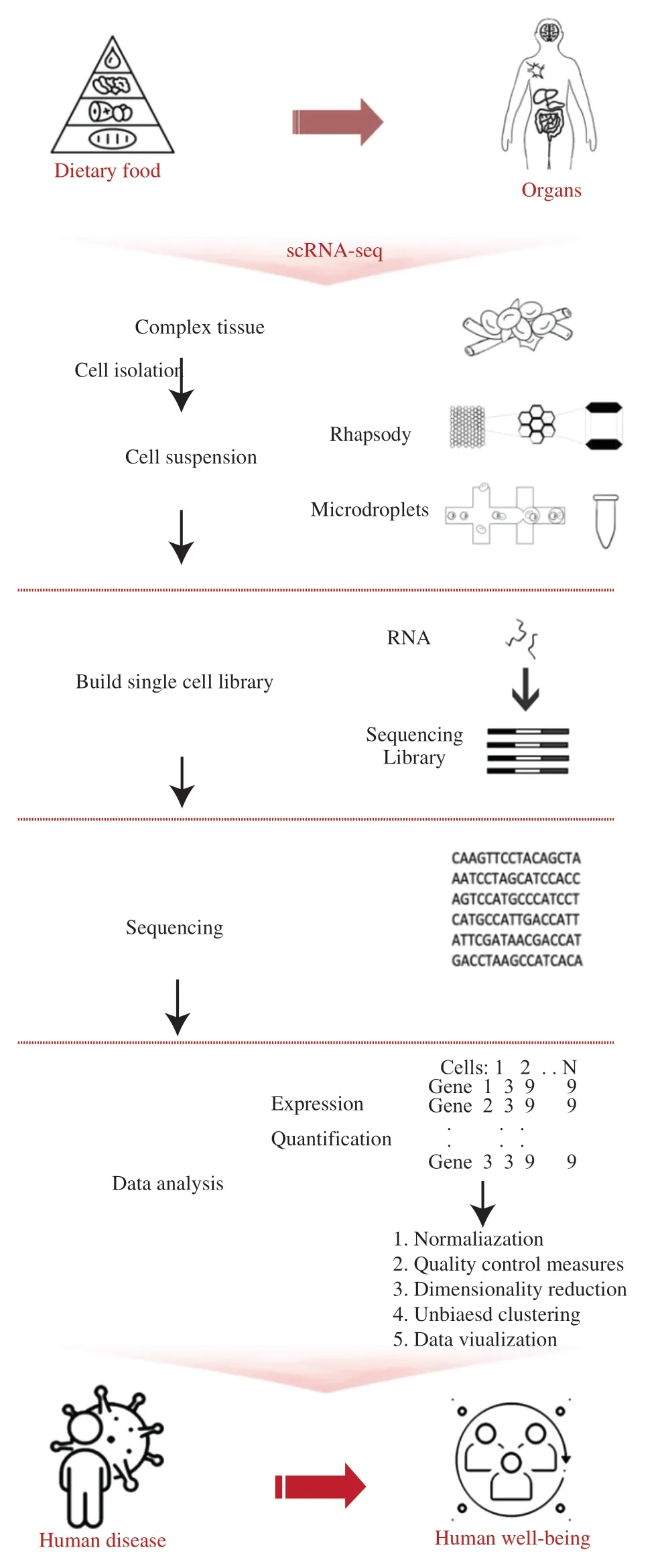
Fig.2 Design and workflow of scRNA-seq in food nutrition and toxicology.
The first step is to receive individual cell,namely single cell isolation to prepare a cell suspension.Before starting the whole process,sample preparing is critical for producing reliable individualcell transcriptome,especially in terms of dense tissue and threedimensional-like organ/animal experiments.To further understand the molecular mechanism of dietary food nutrients or toxicants to targeted cell types,specific organs can be collected as basic experimental samples.According to the characteristics of cell and tissue samples,rigorously choose proteolytic enzymes (e.g.,trypsin,collagenase,and free enzymes) and digestion time to maximize individual cell production and minimize apoptosis.Once a cell received full dissociation,it can achieve the isolation to an individual cell by various cell capturing technologies,commonly limiting dilution,micromanipulation,fluorescent activated cell sorting (FACS) and microfluidics technology[74].Limiting dilution and micromanipulation are time-consuming and low efficient.FACS uses fluorescent monoclonal antibodies to label cells,which recognize specific surface markers.Based on predetermined fluorescent parameters,a charge is applied to a cell with electrostatic deflection system,and cells are isolated magnetically,thus enabling the sort of distinct populations.This method is preferred when the target cell expresses low level of the biomarker[75].Isolation of single cell can also be accomplished by microfluidic technology,either using circuits to distribute cells into nano-wells,or isolating into oil-based droplets[53].These methods allow for unbiased cell capture,as with the FACS approach,and are massively parallelized to increase the number of cells that could be profiled in one run to tens of thousands.Droplet-based platforms enable the encapsulation of thousands of single cells into individual“chambers,” each containing all the necessary reagents for cell lysis,reverse transcription,and molecular tagging,which causes minor damage to cells during the sorting process[76].
After the isolation and lysis of single cells,scRNA-seq subsequently requires the conversion of their RNA into cDNA,and the amplification of cDNA to generate high-throughput sequencing library.Tang et al.developed the first scRNA-seq method through polymerase chain reaction (PCR)-based amplification in 2009[72].SMART (switching mechanism at 50 end of RNA template) ensured the synthesis of full-length cDNA and improved read coverage across transcripts,generating robust and quantitative transcriptome data[77].However,the above-mentioned methods were both very low throughput.The emergence of barcode labeling and sample pooling overcome the challenges of samples with limited material,with the occurrence of single-cell tagged reverse transcription sequencing(STRT-Seq),making high sensitive throughput analysis possible[78].Subsequently,with continuous improvement in nucleic acid amplification,novel scRNA-seq technologies surged,including cell expression by linear amplification sequencing (CEL-Seq),massively parallel RNA single-cell sequencing framework (MARS-Seq)[79].Although barcoding and pooling allows for multiplexing and studying many different single cells at a time,they result in the loss of accuracy due to PCR bias[80].Researchers have effectively eliminated the bias of chain amplification by adding unique molecular identifiers (UMI),as to distinguish the original template from the amplified sequence and result in evaluated accuracy[81].The newly developed Drop-Seq encapsulates individual cells into nanoliter-sized droplets together with DNA-barcoded beads,enabling highly parallel analysis of individual cells[82].The combination of the sensitivity (i.e.,the probability to covert mRNA transcript present in a single cell into a cDNA molecule present in the library),the accuracy (i.e.,the degree to which the read quantification corresponds to the actual concentration of mRNAs)and the precision (i.e.,the technical variation of the quantification)together determine the power to indicate the relative differences in expression levels and the standard to choose sequencing method[83].Subsequently,the amplified products can be sequenced downstream,along with two important questions of “how many cells to collect”and the sequencing depth (that is,the sequencing capacity spent on a single sample,reported as the number of raw reads per cell)[76].Although the ability to sequence a larger number of cells allows for a more comprehensive picture of transcriptomic profile,the answers are actually intertwined,since high-throughput methods generally involve sequencing to lower depths (<100 000 reads per cell)[84].The library is usually not sequenced to saturation,while those expressed at lower levels can benefit from further increases in the sequencing depth.
Due to scarcity of RNA in a single cell and the nature of stochastic transcription,scRNA-seq data often faces challenges of high cell-tocell variability.The results can be difficult to interpret without proper study design,since above-mentioned single-cell technologies may be affected by technical variation,and generation of biological and technical replicates whenever possible is strongly recommended[85].Unlike bulk RNA-seq,scRNA-seq analysis involves an additional cell quality control and quantitative standardization computationally to mitigate batch variation[86].The dimensionality reduction and clustering of the samples can be used to perform difference analysis,compare with the existing single-cell data identification results to obtain the types of cell subpopulations,and further determine the highly expressed genes in these cell clusters.Therefore,minimizing the RNA loss and maintaining the fidelity of information is a critical challenge in scRNA-seq technologies.scRNA-seq results should thus be interpreted with caution and functional validation is often recommended.For example,Mickelsen developed a comprehensive census of molecularly distinct cell types in the mouse lateral hypothalamic aera using scRNA-seq,and further characterized a novel population of somatostatin-expressing neurons through neuroanatomical tracing and imaging,behavioral experiments and analyses[87].
After screening the differentially expressed genes of different cell clusters,GO (Gene Ontology) and/or KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis can be used to to identify the function of candidate marker genes,construct gene set protein interaction network,analyze the relationship between related genes and diseases.At the molecular level,the interaction between different types of cells is estimated by looking for the expression of known ligands or receptors in or between cells,and transcription factors in singlecell RNA sequencing data are analyzed[88].Calculate the possible regulatory genes of each cell or cell group based on the expression matrix,infer the gene regulatory network,and finally screen out the transcription factors that significantly regulate and play a core role,so as to understand the mechanism.Furthermore,the dynamic change process of cells can be simulated based on the single-cell transcriptome expression profile,and the possible differentiation or evolution trajectory of cells can be deduced,namely pseudotime analysis[89].By constructing the change trajectory between cells,the process of cell changes over time is reshaped,so that the specific location of the translated protein in the cell and the related biological processes involved in it can be known.
4. Conclusions and future prospects
Cells are the functional unit of an organism,multi-celled or organisms.Cell heterogeneity is recently recognized but important feature of disease that underlies various bio-characteristics of different organs/tissues.The advent of single-cell sequencing technology facilitated in-depth studies into complex molecular mechanisms of cell heterogeneity at the single cell level,thus understanding how diet affect host biology and developing effective dietary intervention.In this review,we provide a general workflow of scRNA-seq,the recent applications in the scRNA-seq discipline,and how the new technology may elucidate the arguments concerning food safety and food nutrition,and relatedly human well-being.We focus on the brain,liver,renal,immune system and tumor disease of the single cell RNA sequencing data,and the link between diet and disease.By building a framework for integrating cellular molecular profiling of dietary intervention and host metabolism,the aforementioned scRNAseq can provide an opportunity to refine our fundamental concepts of food safety and nutrition,poising to have a profound impact on a direct causal-relationship between health effects and nutrition.Hence,the neoteric single cell RNA approaches,in conjunction with other ‘omics’ cascades (transcriptomics,proteomics,metabolomic or genomics),can potentially transmogrify our perspectives towards dietary foods.
Conflict of interest
Hao Zheng is an editorial board member forFood Science and Human Wellnessand was not involved in the editorial review or the decision to publish this article.All authors declare that there are no competing interests.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (32170495),and the Emergency Project for Risk Assessment of Areca Nut (Key Project of Department of Agriculture and Rural Affairs of Hainan Province &Wanning Municipal People's Government).
杂志排行
食品科学与人类健康(英文)的其它文章
- Modifications in aroma characteristics of ‘Merlot’ dry red wines aged in American,French and Slovakian oak barrels with different toasting degrees
- Effect of different drying methods on the amino acids,α-dicarbonyls and volatile compounds of rape bee pollen
- Dynamic changes in physicochemical property,biogenic amines content and microbial diversity during the fermentation of Sanchuan ham
- A comparison study on structure-function relationship of polysaccharides obtained from sea buckthorn berries using different methods:antioxidant and bile acid-binding capacity
- Yolk free egg substitute improves the serum phospholipid profile of mice with metabolic syndrome based on lipidomic analysis
- Underlying anti-hypertensive mechanism of the Mizuhopecten yessoensis derived peptide NCW in spontaneously hypertensive rats via widely targeted kidney metabolomics
