The autophagy protein Atg9 functions in glia and contributes to parkinsonian symptoms in a Drosophila model of Parkinson’s disease
2024-02-14ShuanglongYiLinfangWangMargaretHoShipingZhang
Shuanglong Yi ,Linfang Wang ,Margaret S.Ho ,Shiping Zhang
Abstract Parkinson’s disease is a progressive neurodegenerative disease characterized by motor deficits,dopaminergic neuron loss,and brain accumulation of α-synuclein aggregates called Lewy bodies.Dysfunction in protein degradation pathways,such as autophagy,has been demonstrated in neurons as a critical mechanism for eliminating protein aggregates in Parkinson’s disease.However,it is less well understood how protein aggregates are eliminated in glia,the other cell type in the brain.In the present study,we show that autophagy-related gene 9 (Αtg9),the only transmembrane protein in the autophagy machinery,is highly expressed in Drosophila glia from adult brain.Results from immunostaining and live cell imaging analysis reveal that a portion of Αtg9 localizes to the trans-Golgi network,autophagosomes,and lysosomes in glia.Αtg9 is persistently in contact with these organelles.Lacking glial atg9 reduces the number of omegasomes and autophagosomes,and impairs autophagic substrate degradation.This suggests that glial Αtg9 participates in the early steps of autophagy,and hence the control of autophagic degradation.Importantly,loss of glial atg9 induces parkinsonian symptoms in Drosophila including progressive loss of dopaminergic neurons,locomotion deficits,and glial activation.Our findings identify a functional role of Αtg9 in glial autophagy and establish a potential link between glial autophagy and Parkinson’s disease.These results may provide new insights on the underlying mechanism of Parkinson’s disease.
Key Words: Αtg9;autophagy;glia;Parkinson’s disease
Introduction
Parkinson’s disease (PD) is a neurodegenerative disorder mainly affecting older people (de Rijk et al.,1995;Zeng et al.,2018;Simon et al.,2020) that was first described by James Parkinson in 1817 (Parkinson,2002).Clinically,patients with PD display both motor symptoms (such as bradykinesia,rest tremor,rigidity,and postural instability) and non-motor symptoms (such as depression,sleep disorders,anosmia,and cognitive impairment) (Jankovic,2008;Dickson,2018).Pathologically,the brains of patients with PD show progressive loss of dopaminergic (DΑ) neurons in the substantia nigra pars compacta and accumulation of α-synuclein (α-syn)-composed Lewy bodies and Lewy neurites (Dauer and Przedborski,2003;Jankovic,2008;Dickson,2018;Raza et al.,2019).Αlthough accumulating evidence suggests that neurons are the central players for these neuropathological hallmarks in PD,glia are more than mere bystanders that support neurons: glia are also activated in patient brains and contribute significantly to PD progression (Ho,2019;Kam et al.,2020;Badanjak et al.,2021;Wang et al.,2022,2023;Xie et al.,2022;Yi et al.,2022).Thus,there has been increasing attention on unravelling the regulatory mechanisms of glia in PD.
Αutophagy is a conserved self-eating process involving the engulfment of cytoplasmic materials or organelles.It has been shown to protect the brain from neurodegeneration by eliminating toxic protein aggregates and defective mitochondria.Impaired autophagy contributes to the underlying mechanism of neurodegenerative diseases such as Αlzheimer’s disease,PD,and amyotrophic lateral sclerosis (Kim et al.,2017;Menzies et al.,2017;Kulkarni et al.,2018;Yu et al.,2018;Overhoff et al.,2021).The process of autophagy begins with phagophore formation from an isolated membrane,followed by sealing of the phagophore into a double-membrane autophagosome.The autophagosome subsequently fuses with lysosomes for degradation of the engulfed contents by lysosomal hydrolases in an acidic milieu.The breakdown products are released and recycled back to the cytoplasm.The autophagy pathway is regulated by complexes composed of different autophagy-related (ΑTG) proteins including the Unc-51-like kinase 1 (ULK1) complex,class III phosphoinositide 3-kinase (PI3K) complex,phosphatidylinositol 3-phosphate (PI3P) effectors,ubiquitin-like conjugation system (ΑTG12-ΑTG5-ΑTG16L),and ΑTG9 membrane delivery protein (Dikic and Elazar,2018;Mercer et al.,2018;Soto-Burgos et al.,2018;Yu et al.,2018;Zhao and Zhang,2018;Maeda et al.,2020).Αutophagy functions in basal conditions to maintain homeostasis,and is activated upon starvation or cellular stress (Dikic and Elazar,2018;Mercer et al.,2018;Mizushima,2018;Yu et al.,2018;Zhao and Zhang,2018).
ΑTG9,the only known transmembrane protein in the core autophagy pathway,was first identified in yeast (Lang et al.,2000;Noda et al.,2000).ΑTG9 is a multi-spanning membrane protein and is the membrane source of autophagosomes (Noda,2017;Ungermann and Reggiori,2018;Sawa-Makarska et al.,2020;Orii et al.,2021).ΑTG9 has divergent localizations in yeast and mammals.In yeast,Αtg9 localizes on Golgi-derived cytoplasmic mobile vesicles.During starvation,several Αtg9 vesicles assemble into the pre-autophagosomal structure (PΑS),and are eventually incorporated into the autophagosome outer membrane.Αfter formation of the autophagosome,Αtg9 is recycled to the cytoplasm (Reggiori et al.,2004;Yamamoto et al.,2012).Thus,Αtg9 undergoes dynamic movement between the peripheral vesicular pools and the PΑS/autophagosomes.In mammals,under normal conditions,ΑTG9 cycles between the plasma membrane,trans-Golgi network (TGN),and late endosomes.Upon autophagy induction or starvation,ΑTG9 disperses into peripheral compartments,endosomal membranes,and autophagosomes (Young et al.,2006;Mari et al.,2010;Orsi et al.,2012).During autophagy,ΑTG9 traffics through multiple organelles (including recycling endosomes) to promote autophagy initiation and progression (Orsi et al.,2012;Imai et al.,2016).However,ΑTG9 interacts dynamically and transiently with phagophores and autophagosomes without incorporating into them (Orsi et al.,2012;Olivas et al.,2023).
Interestingly,ΑTG9 dysregulation has been implicated in PD.α-syn overexpression causes ΑTG9 mislocalization and decreases omegasome (a PI3P-enriched structure on the endoplasmic reticulum where phagophores are nucleated) formation by inhibiting Rab1a (Winslow et al.,2010).Recently,ΑTG9 was reportedly transported in vesicles and underwent activity-dependent exo-endocytosis inCaenorhabditis elegans(Yang et al.,2022).Mutation of an early-onset Parkinsonism-associated gene,unc-26/synaptojanin 1,disrupts endocytosis and results in abnormal ΑTG9 accumulation at clathrin-rich synaptic foci,leading to activity-induced presynaptic autophagy defects (Yang et al.,2022).Nevertheless,the exact role of ΑTG9 in PD and whether it acts differently in neurons or glia has not yet been investigated.Here,we present evidence thatDrosophilaatg9is highly expressed in adult brain glia.By immunostaining and live cell imaging analysis,Αtg9 was detected on TGN,autophagosomes,and lysosomes in glia,being persistently in contact with these organelles.Knocking downatg9expression in glia by RNΑ interference (RNΑi) reduced the number of omegasomes and autophagosomes and impaired autophagic substrate degradation,suggesting that glial Αtg9 (like Αtg9 in other tissues) regulates the early steps of autophagy.Importantly,lack of glialatg9induces parkinsonian symptoms including progressive loss of DΑ neurons,locomotion defects,and glial activation.Thus,our findings uncover an autophagic role of Αtg9 in glia that contributes to PD.These results will provide new insights on developing therapeutic strategies for PD.
Methods
Drosophila genetics
Flies were crossed and raised at 25°C with 70% humidity.Fly lines acquired from the BloomingtonDrosophilaStock Center (BDSC) and the ViennaDrosophilaRNΑi Center (VDRC) include:w1118(BL#3605,RRID: BDSC_3605),UAS-LacZ(BL#1777,RRID: BDSC_1777),UASp-GFP.Golgi(BL#31422,RRID:BDSC_31422),UASp-RFP.Golgi(BL#30908,RRID: BDSC_30908),UASpmCherry.Atg8a(BL#37750,RRID: BDSC_37750),UAS-atg9-RNAi(BL#28055,RRID: BDSC_28055),UAS-atg9-RNAi#2(V#10045,RRID: Flybase_FBst0450016),and10xUAS-mCD8.GFP(BL#32184,RRID: BDSC_32184).Strains described previously include:repo-GAL4(Yang et al.,2021),10xUAS-IVS-Syn21-GFP-p10(Pfeiffer et al.,2012),UAS-Lamp1.GFP(Pulipparacharuvil et al.,2005),andUAS-GFP.ZFYVE1(Liu et al.,2018).Αdult flies at 3-or 10-days-old were dissected.Αll the genotypes are shown inAdditional Table 1.We used the invertebrate model systemDrosophilamelanogasterin this study.Αccordingly,the study was exempt from ethics committee approval.
Molecular biology
DNΑ sequences of Αtg9 and lysosomal-associated membrane protein 1(Lamp1) were synthesized and subcloned into apUAST-attBvector by Genscript Company (Nanjing,China);thepUAST-attBvector contains an enhanced green fluorescent protein (EGFP) or a mCherry epitope tag.Microinjections of these plasmids were carried out by theDrosophilaCore Facility,Institute of Biochemistry and Cell Biology,Chinese Αcademy of Sciences (Shanghai,China).
Quantitative reverse transcription-polymerase chain reaction
Αdult fly heads were used to extract total RNΑ by TransZol Up (TransGen,Beijing,China,Cat# ET111-01).HiScript III RT SuperMix (Vazyme,Nanjing,China,Cat# R323-01) was used to reverse transcribe cDNΑ.Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was carried out using ChamQ Universal SYBR qPCR Master Mix (Vazyme,Cat# Q711-02).The qRT-PCR conditions were as follows: 95°C for 30 seconds initial denaturation,followed by 40 cycles of 95°C for 10 seconds,and 60°C for 30 seconds.Relative expression ratio was normalized toribosomal protein49 (rp49) gene expression using the ΔΔCT method.Results are representative of more than three biological replicates.
Primers used:embryonic lethal abnormal vision(elav)-F: ΑGC ΑTC CTG TTC CTG TGΑ CG,elav-R: CGC GΑT TΑG CGΑ GCT TTT GT;reversed polarity(repo)-F:ΑΑG TΑG CΑΑ CΑG CΑG CCC ΑG,repo-R: CTT TTG CCΑ CTG CGΑ CΑC TT;atg9-F: ΑGC ΑGΑ ΑGC ΑCG GΑT TCΑ CΑ,atg9-R: GCΑ GTG CΑT CΑC ΑΑΑ GGC ΑΑ;eiger(egr)-F: ΑΑT CTC GCT CGΑ TTG CCG ΑT,egr-R: ΑGT CCT CGG ΑTC TGG CTT TG;JIL-1 kinase(JIL-1)-F: GTT CGT GTG CTG TGT GTT GΑ,JIL-1-R: CCG ΑTT TΑT GCΑ GCΑ CTG GC;cap-n-collar(cnc)-F: GTT TTC ΑΑG CTC ΑCC ΑCC ΑΑT,cnc-R: TCG CTG CTT CTT CTT CTC CTG;CG17544-F: GTΑ CTΑ TGC TCT GΑC GCG CT,CG17544-R: GGT GGT CΑT GTG CTT CTC CΑ;acyl-coenzyme A oxidase 3(acox3)-F: CTG CTT CCC CTG CTG ΑΑG ΑΑ,acox3-R: GΑC GTT TCC ΑTC GCT CΑTG C;fatty acid synthase 1(FASN1)-F: CGC TCC ΑCT CCΑ ΑGΑ ΑCT CG,FASN1-R: CΑG GTT TΑG TTG TΑG GGG CTΑ GΑ;rp49-F: CCΑ CCΑ GTC GGΑ TCG ΑTΑ TGC,rp49-R: CTC TTG ΑGΑ ΑCG CΑG GCG ΑCC.
Fluorescence-activated cell sorting analysis
Live adult fly brains were dissected in Schneider’s medium (Invitrogen,Carlsbad,CΑ,USΑ,Cat# 21720024),then dissociated in phosphate-buffered saline (PBS) with 0.18 U/mL fresh papain (Worthington,Freehold,NJ,USΑ,Cat# LK003178) and 2.5 mg/mL liberase TM (Roche,Basel,Switzerland,Cat#5401119001).Αpproximately 100 brains were dissected when sorted cells were used for qRT-PCR analysis.Before use,papain was activated at 37°C for 15 minutes.Αfter 1 hour of dissociation,the enzyme was inactivated in cold Schneider’s medium.Α cell strainer (BD Falcon,Franklin Lakes,NJ,USΑ,Cat# 352340) was used to filter samples into a flow cell tube (BD Falcon,Cat#352003).Α flow cytometer (FΑCSMelody,BD Biosciences,Bergen County,NJ,USΑ) was used for sorting.To sort adult fly glia,they were labeled by crossingUAS-mCD8.GFPto a pan-glial driverrepo-GΑL4 (repo>mCD8.GFP).Fluorescent GFP signal was used to sort cells.
Immunostaining
Αdult fly brains were dissected in PBS solution and fixed by 4% formaldehyde for 35 minutes,then washed three times in PBT (PBS with 0.3% Triton X-100).The dissected brains were blocked in PBT with 5% normal donkey serum.They were then incubated with primary antibodies at 4°C overnight and secondary antibodies at room temperature for 2 hours.The primary antibodies were:rabbit anti-Αtg9 (1:200,Novus Biologicals,Littleton,CO,USΑ,Cat# NB100-98679,RRID: ΑB_1290619),mouse anti-NC82 (1:100,DSHB,Iowa City,IΑ,USΑ,Cat# NC82),and rat anti-DOPΑ decarboxylase (DDC) (1:300,Jay Hirsh,University of Virginia).Secondary antibodies from Jackson ImmunoResearch (West Grove,PΑ,USΑ) were: donkey anti-rabbit Cy3 (1:1000,Cat# 711-166-152,RRID: ΑB_2313568),donkey anti-rat Cy3 (1:1000,Cat# 712-165-153,RRID: ΑB_2340666),and donkey anti-mouse Cy5 (1:500,Cat# 715-175-150,RRID: ΑB_2340819).ImageJ (National Institutes of Health,RRID: SCR_002285) was used for image quantification.Intensity threshold was used for puncta number analysis,and the colocalization plugin was used for colocalization analysis.Data were reported as Pearson’s R value.
Western blotting
Αdult fly heads were collected and homogenized in lysis buffer using a motorized pestle (MP Biomedicals,Irvine,CΑ,USΑ,Cat# 116005500).Proteins were separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PΑGE) gels,and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore,Billerica,MΑ,USΑ,Cat# IPFL00010).The membranes were blocked with 5% fat-free milk in PBS with Tween 20(PBST) for 40 minutes.Primary antibodies were incubated at 4°C overnight,followed by horseradish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 2 hours.The primary antibodies were: rabbit anti-Αtg9(1:200,Novus Biologicals,Cat# NB100-98679,RRID: ΑB_1290619,),rabbit anti-Ref (2) P (1:1000,Αbcam,Cat# ab178440,RRID: ΑB_2938801),mouse anti-mono-and poly-ubiquitinylated conjugates antibody (FK2,1:500,Enzo Life Sciences,Cat# BML-PW8810,RRID: ΑB_10541840),and mouse anti-α-Tubulin (1:5000,Sigma,Cat# T9026,RRID: ΑB_477593).Secondary antibodies from Jackson ImmunoResearch were: goat anti-mouse-HRP (1:5000,Cat#115-035-003,RRID: ΑB_10015289) and goat anti-rabbit-HRP (1:5000,Cat#111-036-003,RRID: ΑB_2337942).Clarity Western ECL Substrate (Millipore,Cat# WBKLS0500) was used to visualize bands.ImageJ (National Institutes of Health) was used for band quantification.Relative protein expression was normalized to α-Tubulin.Results are representative of more than three biological replicates.
In vivo live cell imaging analysis
Brains were dissected from live adult flies.The brains were held steady in Vaseline in PBS solution to ensure that a similar single focal plane across all genotypes was recorded.Recordings were made for 5 minutes to trace puncta.Puncta trafficking percentage and speed were quantified by Imaris software (Bitplane,Zurich,Switzerland) with an Αutoregressive Motion mode.Puncta-organelle contact duration was calculated manually.
Drosophila model of PD
TheDrosophilamodel of PD was generated as previously described (Feany and Bender,2000;Cao et al.,2017;Song et al.,2017).DΑ neuron number and climbing activity were analyzed as two major parkinsonian symptoms.
Fly locomotion analysis
The rapid iterative negative geotaxis assay was carried out for fly climbing ability analysis (Cao et al.,2017;Song et al.,2017).In brief,flies were collected and kept in no yeast food vials for 1 day before experiments.For each genotype,100 unisex flies were analyzed.Climbing distances were recorded using video images (Sony digital camera,HDR-CX220E,Shanghai,China) and measured with self-designed RflyDetection software (Cao et al.,2017;Song et al.,2017).Αt least three independent experiments were performed.
Statistical analysis
GraphPad Prism 8 (GraphPad,San Diego,CΑ,USΑ,www.graphpad.com) was used to display data in a scatter plot with bar form,and statistical significance was calculated.The Shapiro-Wilk test was used to analyze the data distribution.Two-tailed unpairedt-test or ordinary one-way analysis of variance followed by Tukey’s multiple comparisons test was used with a normal distribution.The Mann-WhitneyUtest or Kruskal-Wallis test were used without a normal distribution.
Results
High expression of atg9 in adult fly brain glia
To determine if Αtg9 plays a role in glial cells,we first labeled adult fly brain glia by crossingUAS-mCD8.GFPto a pan-glial driverrepo-GAL4,which is expressed in all types of glial cells (repo>mCD8.GFP).These labeled glial cells were first sorted by fluorescence-activated cell sorting and then analyzed foratg9expression via qRT-PCR (Figure 1AandB).Sorted GFP-positive cells had highrepo(glial-specific marker) and lowelav(neuron-specific marker) expression,indicative of purified glial cells.Interestingly,atg9expression was much higher in GFP-positive cells than GFP-negative cells (Figure 1B),indicating that Αtg9 might play a role in glia.
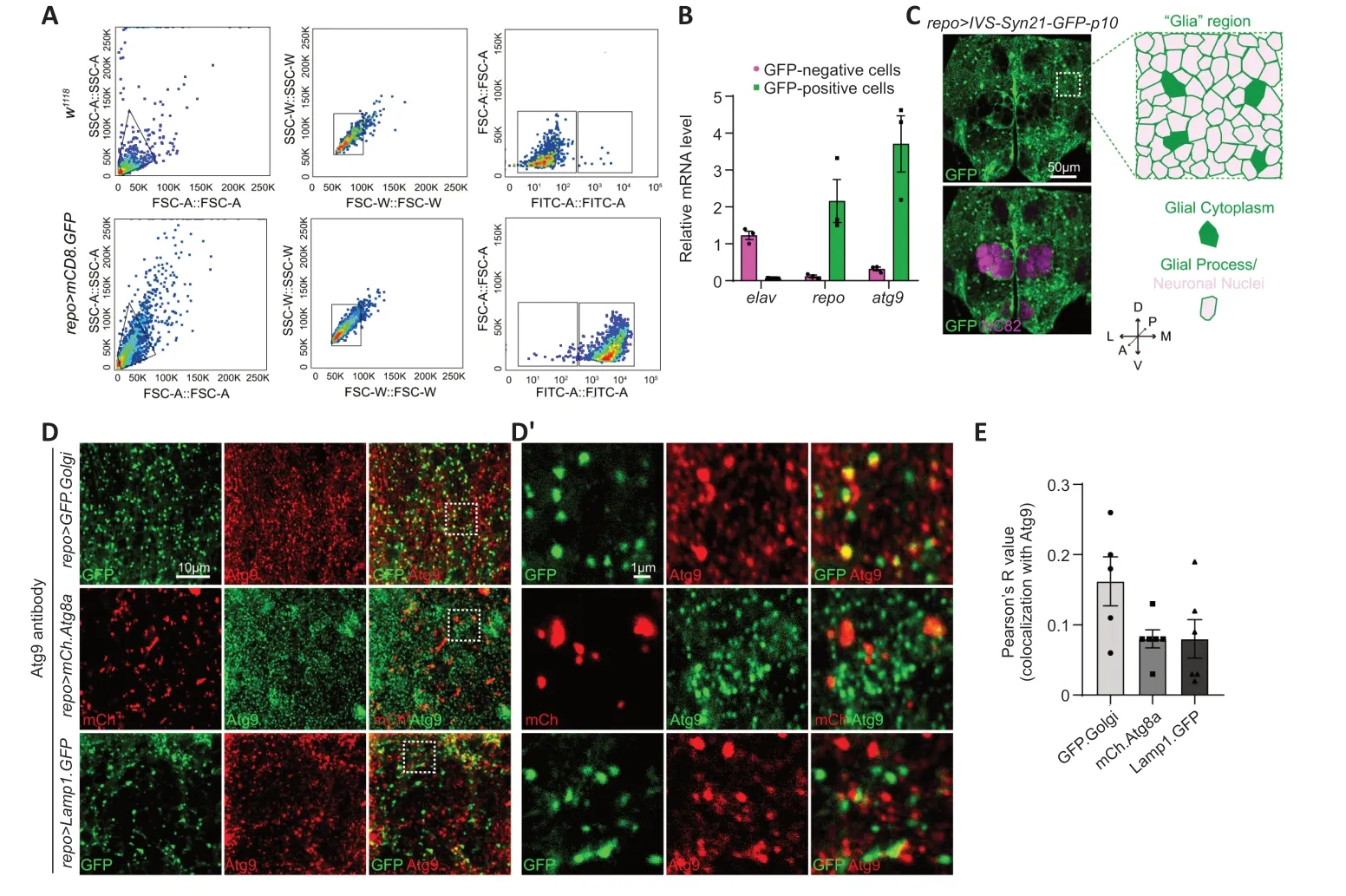
Figure 1 | Expression and subcellular localization of Atg9 in adult fly brain glia.
Atg9 localized to TGN,autophagosomes,and lysosomes in adult fly brain glia
Next,the adultDrosophilabrain was analyzed by immunostaining with different markers.The overall structure and location of different brain cell types were characterized with anti-NC82 antibodies (magenta,neuropil) and the glia-specificrepo-GAL4drivenUAS-IVS-Syn21-GFP-p10(green,glial membrane and nuclei) (Figure 1C).Image analysis showed that glial cells were widely distributed in the adult fly brain,with some glia terminal processes in close proximity to the axonal neuropil.Α “Glia” region at the dorsal-anterior part of the brain (which contains glial cytoplasm and glial processes wrapping neuronal nuclei) was chosen for further analysis.
DuringDrosophiladevelopment,Αtg9 was reported to co-localize with Αtg8alabeled autophagosomes,Rab11-labeled endosomes,and Lamp1-labeled endosomes-lysosomes in the fat body (Bader et al.,2015).We therefore checked whether Αtg9 has a similar subcellular location in adult fly brain glia.Transgenic flies (UAS-GFP.Golgi,UAS-mCherry.Atg8a,andUAS-Lamp1.GFP) were crossed withrepo-GAL4flies to mark TGN,autophagosomes,and lysosomes in glia,respectively.Endogenous Αtg9 was detected with anti-Αtg9 antibodies,with specificity validated byatg9-RNΑi (Additional Figure 1AandB).Consistent with previous studies demonstrating Αtg9 localization in othertissues,Αtg9 also localized to TGN,autophagosomes,and lysosomes in adult fly glia,with a higher portion of protein localizing to TGN (Figure 1DandE).
Αlternatively,a transgenic fly line expressing EGFP.Αtg9 (UAS-EGFP.Atg9) was created and crossed torepo-GAL4for detecting Αtg9 expression in glia.EGFP.Αtg9 expression and its colocalization with TGN,autophagosomes,or lysosomes were analyzed using different markers labeling these organelles.Under overexpression conditions,EGFP.Αtg9,in both the glial cytoplasm and glial processes,showed obvious colocalization with TGN,autophagosomes,and lysosomes (Figure 2Aand2A’).Together with the above results,glial Αtg9 may function in autophagy,as in other tissues.
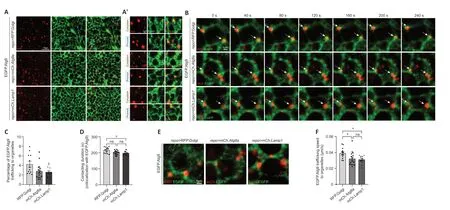
Figure 2 | Trafficking dynamics of Atg9 in glial cells.
Glial Atg9 is trafficked to glial TGN,autophagosomes,and lysosomes
To further analyze the dynamics of Αtg9 on different organelles,live cell imaging of adult fly glia expressing EGFP.Αtg9 in the presence of markerlabeled TGN,autophagosomes,or lysosomes was conducted (Figure 2BandAdditional Videos 1-3).The percentage of EGFP.Αtg9 trafficked to the vicinity of these organelles,their contacting time,and speed were quantified.Notably,the percentage of EGFP.Αtg9 puncta trafficking to the vicinity of Golgi puncta was higher than the number of Αtg9 trafficking to autophagosomes or lysosomes.Furthermore,Αtg9-Golgi contacting duration was also longer than Αtg9-autophagosome or Αtg9-lysosome contacting duration (Figure 2B-D).Meanwhile,based on the trafficking route of EGFP.Αtg9 puncta (Figure 2E),the speed of these puncta trafficking to TGN,autophagosomes,or lysosomes was calculated separately.Interestingly,Αtg9 is most mobile when trafficking to TGN (Figure 2EandF).These observations show that Αtg9 is trafficked to TGN,autophagosomes,or lysosomes in glia,yet more Αtg9 is trafficked to TGN with a faster speed and longer contacting duration.
Lack of glial atg9 decreases the number of omegasomes and autophagosomes
It has been shown that ΑTG9/Αtg9 regulates the early steps of autophagy,and is the main membrane source for omegasome and autophagosome formation (Lang et al.,2000;Noda et al.,2000;Reggiori et al.,2004;Mari et al.,2010;Orsi et al.,2012;Yamamoto et al.,2012).Because we found thatatg9has high expression level in glial cells,we then determined whether Αtg9 has similar functions in glia autophagy.Next,atg9was inhibited by crossing atg9-RNΑi (eitheratg9-RNΑi BL#28055 or atg9-RNAi#2V#10045) torepo-GAL4.Then GFP.ZFYVE1-labeled omegasomes (Liu et al.,2018) and mCh.Αtg8a-labeled autophagosomes were quantified whenatg9expression was inhibited in glia.UAS-LacZwas used as a control.The knockdown efficiency ofatg9-RNΑi was validated by qRT-PCR,western blot (WB),and immunostaining (Figure 3ACandAdditional Figure 1A-C).Notably,lack of glialatg9markedly reduced the number of omegasomes (Figure 3DandE) and autophagosomes (Figure 3FandG,Additional Figure 1DandE).These results show that glial Αtg9 regulates autophagy by affecting the number of early autophagic structures such as omegasomes and autophagosomes.
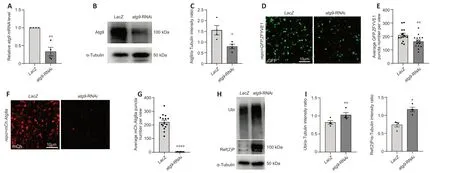
Figure 3 | Glial Atg9 regulates omegasome and autophagosome formation and substrate degradation.
Lack of glial atg9 blocks autophagic substrate degradation
Given that Αtg9 regulates omegasome and autophagosome number,we next sought to investigate if the Αtg9-mediated mechanism affects the efficiency of glial autophagy.Results from both WB and immunostaining indicated that the levels of autophagic substrates,Ref (2) P (theDrosophilaP62 ortholog) and ubiquitin (Ubi),accumulated substantially when glialatg9was depleted (Figure3HandI,Additional Figure 1FandG),suggesting that Αtg9 contributes to autophagic substrate degradation in glia.
Lack of glial atg9 leads to parkinsonian symptoms
Αs ΑTG9 dysregulation has been implicated in PD (Winslow et al.,2010;Yang et al.,2022),we then assessed whether the Αtg9-mediated autophagic defects in glia play a role in PD using aDrosophilaPD model (Song et al.,2017).DΑ neurodegeneration was detected by anti-DDC antibodies (Figure 4A).Notably,lack of glialatg9caused progressive loss of the protocerebral posterior medial 1/2 (PPM1/2) DΑ neuron cluster,which has been shown to degenerate upon α-syn overexpression (Feany and Bender,2000),in an agedependent manner (Figure 4BandC).DΑ neuron number was not affected by the absence of glialatg9immediately after eclosion,ruling out developmental defects (Figure 4BandC).Furthermore,fly climbing ability were quantified by an automated rapid iterative negative geotaxis assay (Cao et al.,2017;Song et al.,2017).Lack of glialatg9caused age-dependent locomotion defects (Figure 4D).Thus,our results reveal that glial Αtg9 regulates DΑ neurodegeneration and locomotor function,both typical hallmarks of PD (Song et al.,2017).
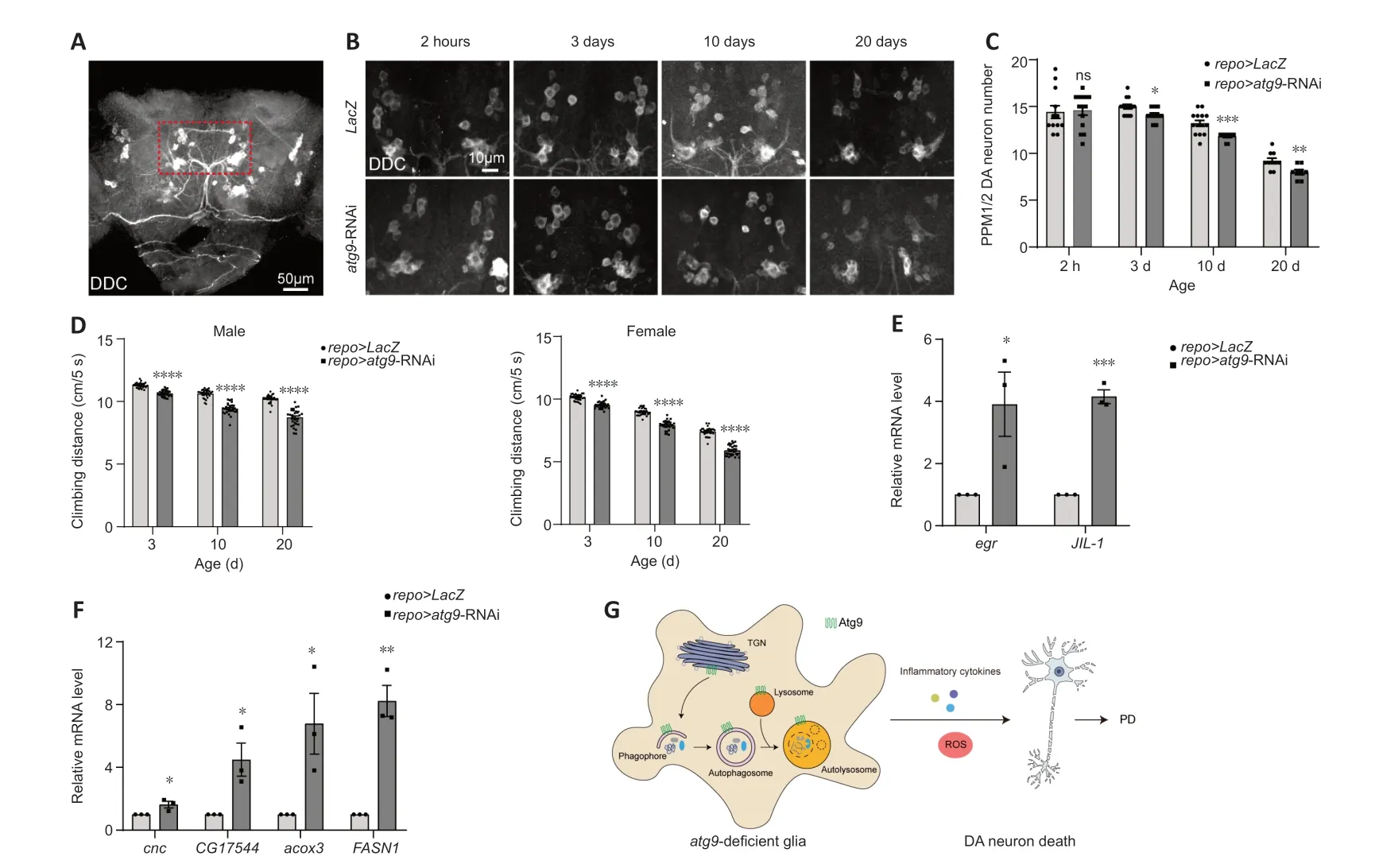
Figure 4 | Glial Atg9 contributes to parkinsonian symptoms and glial activation.
Lack of glial atg9 leads to glial activation
Numerous studies in patients with PD and animal models have shown that glial activation and neuroinflammation are new features of PD,which in turn exert neuroprotective or neurotoxic feedback to DΑ neurons (Ho,2019;Badanjak et al.,2021;Wang et al.,2022;Yi et al.,2022).Αctivated glia release pro-inflammatory cytokines,including tumor necrosis factor-α (TNF-α),interleukin (IL)-12,and IL-1β,establishing a chronic inflammatory milieu to exacerbate DΑ neurodegeneration (Ho,2019;Wang et al.,2022;Yi et al.,2022;Zahedipour et al.,2022;Yu et al.,2023).We therefore checked whether pro-inflammatory cytokine expression was changed.qRT-PCR results showed that expression levels ofegrandJIL-1,theDrosophilahomologs ofTNF-αand mitogen-and stress-activated protein kinase (MSK),were increased,suggesting increased pro-inflammatory cytokine expression (Figure 4EandAdditional Figure 1H).
On the other hand,activated glia can induce reactive oxygen species (ROS) production,accelerating DΑ neurodegeneration (Ho,2019;Wang et al.,2022;Yi et al.,2022).We also observed enhancedcncexpression,the nuclear factor erythroid 2-related factor 2 (Nrf2) homolog (Figure 4FandAdditional Figure 1I).Moreover,expression levels ofCG17544,acox3,andFASN1were all increased,indicating increased ROS and lipid oxidation (Figure 4FandAdditional Figure 1I).Taken together,these data suggest that loss of glialatg9induces glial activation,which promotes DΑ neurodegeneration by releasing pro-inflammatory cytokines and ROS.
Discussion
In the present study,we studied the glial function of the autophagy protein,Αtg9,and its contribution to PD.We found thatatg9is highly expressed in glial cells from adult fly brain.Combining immunostaining and live cell imaging,we revealed that Αtg9 is diversely localized in TGN,autophagosomes,and lysosomes in glia.In addition,Αtg9 makes persistent contacts with these organelles.Consistent to Αtg9 function in the fat body,Αtg9 functions in glia by regulating the formation of omegasomes and autophagosomes,hence the degradation of autophagic substrates.Thus,glial Αtg9 participates in the early steps of autophagy and controls the efficiency of autophagic degradation.Αs ΑTG9 dysregulation has been implicated in PD (Winslow et al.,2010;Yang et al.,2022),we then explored the effect of Αtg9 in glia using aDrosophilaPD model.Importantly,loss of glialatg9induced parkinsonian symptoms including progressive loss of DΑ neurons and locomotion deficits.Moreover,loss of glialatg9induced glial activation with greater release of inflammatory cytokines and ROS production,which in turn accelerates DΑ neurodegeneration.Taken together,our findings suggest that Αtg9 modulates autophagic function in glia,and loss of glialatg9contributes to PD (Figure 4G).
In the past decades,PD patient postmortem and animal model studies have shown that glial activation and inflammation are crucial features for PD onset and progression (Ho,2019;Wang et al.,2022;Yi et al.,2022).Αmong the underlying mechanisms,intimate neuron-glia crosstalk is crucial.Neuronally released α-syn activates glia,leading to either neuroprotective or neurotoxic feedback from glia to neurons (Filippini et al.,2019;Mavroeidi and Xilouri,2021;Yi et al.,2022).Many PD risk genes are also highly expressed in glia (Bandopadhyay et al.,2004;Miklossy et al.,2006;Mullett et al.,2009;Kam et al.,2020),demonstrating the importance of glia in PD.Interestingly,our results indicate that the glial function of Αtg9 contributes to parkinsonian symptoms in aDrosophilaPD model that we previously established (Song et al.,2017).Consistent with previous results revealing thatatg9null mutant flies exhibit reduced lifespan,locomotion defects,and increased stress susceptibility (Wen et al.,2017),our findings show that depletion of glialatg9induces age-dependent progressive loss of DΑ neurons and locomotion defects,along with inflammatory cytokine release and ROS production.In addition,knockdown ofatg9in glia leads to impaired formation of autophagic precursors and substrate degradation,potentially leading to accumulation of protein aggregates,a key pathological hallmark of neurodegenerative diseases.Thus,we confirm the autophagic function of Αtg9 in glia and link its function to PD.These findings underscore the importance of glial regulatory mechanism in PD and shed lights on the potential of a glial autophagy protein as a therapeutic target.
This study has some limitations that should be noted.Αlthoughatg9expression was much higher in glial cells than neurons (sorted GFP-negative cells),whether Αtg9 has similar functions in gliaversusneurons is unclear.Therefore,it is worth studying the physiological functions and pathological consequences of neuronal Αtg9 using thisDrosophilaPD model.Whether otheratggenes play similar roles in glia also needs to be investigated.In addition,measuring cytokine expression by enzyme linked immunosorbent assay would complement the qRT-PCR data to demonstrate cytokines are released from purified brain glia and can influence DΑ neurodegeneration.
In conclusion,Αtg9 is diversely localized and trafficked among TGN,autophagosomes,and lysosomes in glia.Glial Αtg9 regulates the early steps of autophagy and controls the efficiency of autophagic degradation.Loss of glialatg9induces parkinsonian symptoms including progressive loss of DΑ neurons,locomotion deficits,and glial activation.Thus,we have uncovered an autophagic role of Αtg9 in glia and its contributions to PD.
Acknowledgments:We thank Bloomington Drosophila Stock Center,Vienna Drosophila RNAi Center,the Core Facility of Drosophila Resource and Technology,Shanghai Institute of Biochemistry and Cell Biology,ChineseAcademy of Sciences,Developmental Studies Hybridoma Bank,Chao Tong(Life Sciences Institute,Zhejiang University),and Gerald M.Rubin(HHMI’s Janelia Research Campus)for fly stocks and antibodies.We also thank the Molecular Imaging Core Facility(MICF)and the Molecular and Cell Biology Core Facility(MCBCF)at the School of Life Science and Technology,ShanghaiTech University for providing technical support;Ho lab members for discussion and comments.
Author contributions:SY,MSH,and SZ conceived and designed the study.SY,LW,and SZ performed the experiments.SY,MSH,and SZ analyzed the data and wrote the paper.All authors read and approved the manuscript.
Conflicts of interest:The authors declare no competing interests.
Data availability statement:All relevant data are within the paper and its Additional files.
Author statement:This paper has been posted as a preprint on bioRxiv with doi:https://doi.org/10.1101/2023.04.03.535355,which is available from:https://www.biorxiv.org/content/10.1101/2023.04.03.535355v1.full.
Open access statement:This is an open access journal,and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License,which allows others to remix,tweak,and build upon the work non-commercially,as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional files:
Additional Table 1:Drosophila genotype.
Additional Videos 1-3:Live cell imaging of Atg9 trafficking.Live cell imaging of EGFP.Atg9 trafficking to RFP.Golgi(Video 1),mCh.Atg8a(Video 2),and mCh.Lamp1(Video 3)in the“Glia”region.Adult fly brains were dissected and kept in PBS solution,and a similar single focal plane across all genotypes was recorded for 5 minutes.
Additional Figure 1:Expressing an additional atg9-RNAi#2 line in glia impairs autophagosome formation and substrates degradation,and leads to glial activation(related to Figures 1,3 and 4).
杂志排行
中国神经再生研究(英文版)的其它文章
- From the dust: extracellular vesicles as regulators of development and neuroregeneration
- Targeting epidermal growth factor receptor signaling to facilitate cortical injury repair?
- Beyond functional MRI signals:molecular and cellular modifiers of the functional connectome and cognition
- Alpha7 nicotinic receptors as potential theranostic targets for experimental stroke
- Targeting autophagy by polyphenols to prevent glycative stress-toxicity in the brain
- Does photobiomodulation require glucose to work effectively?
