基于转录组测序的剑白香猪表皮颜色相关miRNA筛选与鉴定
2024-01-01赵德鹏覃海毕欢袁巍张依裕陈伟



摘要:【目的】对剑白香猪黑色和白色表皮组织进行转录组测序,筛选出对黑色素生成起调节作用的miRNA,为进一步研究miRNA对剑白香猪表皮颜色形成的调控机制提供理论依据。【方法】以剑白香猪黑色和白色表皮组织为 试验材料进行转录组测序,以|logFold Change|≥1且错误发现率(FDR)lt;0.05为标准,采用DESeq2筛选差异表达 miRNA,使用miRanda和RNAhybrid预测差异表达miRNA靶基因并进行GO功能注释和KEGG信号通路富集分析,构 建差异表达miRNA、靶基因和KEGG信号通路互作网络,利用实时荧光定量PCR对转录组测序结果进行验证。【结果】 从剑白香猪黑色和白色表皮中鉴定出280个已知miRNA和618个新miRNA,筛选出17个差异表达miRNA,白色表皮较黑色表皮有10个miRNA上调表达,7个miRNA下调表达。共预测获得1327个靶基因,GO功能注释分析结果显示,在生物学过程中,差异表达miRNA靶基因主要涉及上皮发育、细胞增殖和分解代谢过程等条目;在细胞组分和分子功能中,主要涉及细胞质、细胞质囊泡、ATP结合和催化活性等条目。KEGG信号通路富集结果显示,靶基因在磷脂酰 肌醇信号系统、黑色素生成和细胞色素P450等信号通路富集,其中ssc-miR-615靶基因包括CALM3、CREBBP、FZD5和 DVL2,其富集通路均与表皮色素沉着有关,ssc-miR-221-3p靶基因TYRP1和ssc-miR-224靶基因RAB23富集的通路与 黑色素生成有关。选取7个差异表达miRNA与靶基因TYRPI和RAB23进行实时荧光定量PCR验证,结果表明差异表达miRNA的表达变化与转录组测序分析的变化趋势一致,TYRPI和RAB23在剑白香猪黑色表皮中的表达量极显 著高于白色表皮(Plt;0.01)。【结论】ssc-miR-615、ssc-miR-221-3p和ssc-miR-224是影响剑白香猪表皮颜色的候选基因, 可能对剑白香猪表皮的黑色素形成有重要影响。
关键词:miRNA;剑白香猪;表皮;黑色素;转录组
中图分类号:S828.89
文章编号:2095-1191(2024)03-0670-10
文献标志码:A
Screening and identification of miRNAs related to epidermal color in Jianbai Xiang pigs based on transcriptome sequencing
ZHAO De-peng, QIN Hai, BI Huan, YUAN Wei, ZHANG Yi-yu, CHEN Wei*(College of Animal Science, Guizhou University/Key Laboratory of Animal Genetics,Breeding and Reproduction of Plateau Mountain Areas, Ministry of Education/Key Laboratory of Animal Genetics and Breeding of GuizhouProvince, Guiyang, Guizhou 550025, China)
Abstract:[Objective]The purpose of the study was to sequence the transcriptome of black and white epidermal tissues of Jianbai Xiang pig, to screen out miRNA that regulated melanin production, and to provide a theoretical basis for further study on the regulatory mechanism of miRNA on epidermal color formation of Jianbai Xiang pig. 【Method】The black and white epidermal tissues of Jianbai Xiang pigs were used as experimental materials for transcriptome sequencing.DESeq2 was used to screen differentially expressed miRNAs with |log2 Fold Change| ≥1 and 1 discovery rate (FDR) ≤0.05 as the standard. The target genes of differentially expressed miRNAs were predicted by miRanda and RNAhybrid,and the enrichment analysis of KEGG signaling pathway was performed by GO functional annotation. The interaction network of differentially expressed miRNAs, target genes and KEGG signaling pathway was constructed, and the results of transcriptome sequencing were verified by real-time fluorescence quantitative PCR. [Result] A total of 280 known miRNAs and 618 new miRNAs were identified from the black and white epidermis of Jianbai Xiang pig, and 17 differentially expressed miRNAs were screened. Ten miRNAs were up-regulated and seven miRNAs were down-regulated in the white epidermis compared with the black epidermis. A total of 1327 target genes were predicted and obtained. GO functional annotation showed that in biological processes, the differentially expressed miRNA target genes were mainly involved in epithelial development, cell proliferation and catabolic processes. In cell components and molecular functions, they were mainly involved in cytoplasm, cytoplasmic vesicles, ATP binding and catalytic activity. The enrichment results of KEGG signaling pathway showed that the target genes were enriched in phosphatidylinositol signaling system, melanin production and cytochrome P450 signaling pathways. Among them, ssc-miR-615 target genes included CALM3, CREBBP, FZD5 and DVL2, and their enrichment pathways were all related to epidermal pigmentation. The enrichment pathways of ssc-miR-221-3p target gene TYRPI and ssc-miR-224 target gene RAB23 were related to melanin production.Seven differentially expressed miRNAs and target genes TYRPI and RAB23 were selected for real-time fluorescence quantitative PCR verification. The results showed that the expression changes of differentially expressed miRNAs were consistent with the changing trend of transcriptome sequencing analysis. The expression of TYRPl and RAB23 in the black epidermis of Jianbai Xiang pigs was extremely significantly higher than that in the white epidermis (Plt;0.01).
【Conclusion】 The ssc-miR-615, ssc-miR-221-3p and ssc-miR-224 are candidate genes affecting the epidermal color of Jianbai Xiang pig, which may have an important effect on the formation of melanin in the epidermis of Jianbai Xiang pig.
Key words: miRNA; Jianbai Xiang pig; epidermis; melanin; transcriptome
Foundation items: Guizhou Science and Technology Plan Project(QKHZC[2022〕 Yiban 087) ;Guizhou Science and Technology Support Plan Project (QKHZC [2021〕Yiban 147) ; Guizhou Seed Industry Development Project (2024) ; Gui- zhou Pig Seed Industry Development Project(Qiannongjicai(2022)10)
0 引言
【研究意义】剑白香猪是我国地方猪种之一,主 产于贵州省剑河县,具有“两头乌”的特点,头部和尾 部为黑色,中部为白色。由于猪与人表皮的相似性, 剑白香猪可作为研究表皮黑色素细胞及黑色素沉积 机制的理想模型(Vodicka et al.,2005)。肤色是动物 的基本特征,明确猪肤色性状的分子遗传基础对猪 的育种和品种纯度鉴定具有重要意义。【前人研究进 展】皮肤作为动物机体的保护屏障,对外界环境造成 的化学刺激、微生物损伤、过敏原刺激和紫外线辐射 等具有保护作用(Lai and McGrath,2021)。动物肤色不同的原因是皮肤中黑色素细胞数量和黑色素 种类(真黑色素和褐黑色素)的不同(Solano,2020;Liang et al.,2021)。黑色素细胞是位于表皮基底层 的异质性细胞,其产生的黑色素能吸收紫外线和可 见光,从而减少皮肤的紫外线损伤,黑色素还具有较 强的抗氧化活性和自由基清除能力(Cichorek et al., 2013;D'Orazio et al.,2013;Cao et al.,2021)。人体内 黑色素合成相关基因缺失会引起白化病和白癜风等 疾病(Tian et al.,2021)。miRNA是由19~25个核苷酸组成的非编码RNA,miRNA通过抑制其靶基因的翻译来调节基因表达(Syeda et al.,2020)。大量研究表明,miRNA通过多种途径介导黑色素细胞的分 化、生长和凋亡,且在黑色素生成过程中发挥重要作用(Itoh et al.,2020;Hushcha et al.,2021;Zhang et al.,2022)。Wang等(2016)研究发现,miR-21a-5p可通过抑制SRY-box转录因子5的表达,上调小眼畸形相关转录因子(MITF)和酪氨酸酶(TYR)的mRNA和蛋白表达水平,增加黑色素生成。Liu等(2019)研究发现,在羊驼黑色素细胞中miR-380-3p通过靶向SRY-box转录因子6调控黑色素形成。Zhu等(2020)研究发现,miR-233、miR-217和miR-211在不同表皮 颜色小鼠中差异表达,其与色素沉着过程中的发育 机制有关。肖敏等(2024)研究发现,miR-129-5p在黑色山羊皮肤组织中高表达,可通过调控TYR、酪氨酸酶相关蛋白1(TYRP1)mRNA和蛋白的表达影响黑色素生成。【本研究切入点】现阶段有关miRNA调 控剑白香猪表皮颜色沉着的研究较少,剑白香猪表 皮黑色素沉着的分子机制的尚未完全明确。【拟解 决的关键问题】对剑白香猪黑色和白色表皮组织 进行转录组测序,鉴定差异表达miRNA,对其靶基 因进行预测和分析,筛选出与黑色素生成相关的 miRNA,为进一步研究miRNA对剑白香猪表皮颜色 形成的调控机制提供理论依据。
1材料与方法
1.1样品采集
以3头体重、体型和外貌相似且饲养条件相同 的24月龄雌性剑白香猪为试验材料,使用皮肤采样 器采集剑白香猪背部白色表皮和耳部黑色表皮放入 冻存管,立即置于液氮罐中带回实验室,-80℃保存。 动物试验经贵州大学动物伦理委员会批准,批准号EAE-GZU-2020-7007。
1.2总RNA提取、文库构建和转录组测序
样品经研磨后采用TRIzol试剂(美国Invitrogen公司)提取总RNA,使用1.0%琼脂糖凝胶电泳检测RNA降解和污染情况。利用NanoDrop2000超微 量分光光度计(美国ThermoFisher Scientific公司)和 Qubit 2.0荧光计及Qubit RNA检测试剂盒(美国 ThermoFisher Scientific公司)检测RNA纯度和浓 度。采用Agilent 2100生物分析仪和RNA 6000 Nano检测试剂盒(美国Agilent公司)评估RNA完整性。 RNA检测合格后,使用Multiplex Small RNA Library Prep Kit for Illumina(美国NEB公司)构建small RNA文库,每个样品取1.5μgRNA,经打断、末端修复、加poly(A)尾、连接测序接头和PCR富集后,利 用Illumina HiSeq X Ten平台进行测序,测序读长为 单端50nt。
1.3测序数据处理
对测序得到的原始序列(Raw reads)进行质量控制,去除低质量Reads,去除N含量≥10%的Reads,去除不含3'接头的Reads,剪切掉3'接头序列,去除lt; 18 nt或gt;30 nt的Reads,获得有效序列(Clean reads),计算Q20、Q30、碱基组成和序列重复水平。
1.4miRNA鉴定
使用Bowtie将有效序列与Silva数据库、GtRNAdb数据库、Rfam数据库和Repbase数据库进行序列比对,过滤非编码RNA和重复序列(Langmead et al.,2009)。 以 Sscrofa11.1(ftp://ftp.ensembl.org/pub/ release-90/fasta/sus_scrofa/dna/)为参考基因组,对过 滤后的Reads进行序列比对和分析。将成功比对到参考基因组的Reads与mirbase(v21)数据库中的已知miRNA成熟体序列进行比对,序列完全相同的鉴 定为已知miRNA,利用miRDeep2(Friedländer et al., 2012)对其余有效序列进行新miRNA预测,采用Randfold对新miRNA进行二级结构预测。
1.5差异表达miRNA鉴定
采用TPM(Fahlgren et al.,2007)对miRNA表达 量进行标准化处理。采用DESeq2(Anders and Huber, 2010)筛选剑白香猪黑色和白色表皮间差异表达 miRNA,以|log2Fold Change≥1且错误发现率(FDR)≤0.05作为筛选标准。
1.6差异表达miRNA靶基因预测与分析
采用miRanda(Betel et al.,2008)和RNAhybrid (Rehmsmeier et al.,2004)预测剑白香猪黑色和白色表皮间差异表达miRNA的靶基因,对预测结果取交 集作为靶基因。利用topGO(Ashburner et al.,2000)对差异表达miRNA靶基因进行GO功能注释分析,基于KEGG数据库进行信号通路富集分析。采用Cytoscape对差异表达miRNA、靶基因和KEGG信号 通路互作网络进行可视化。
1.7实时荧光定量PCR验证
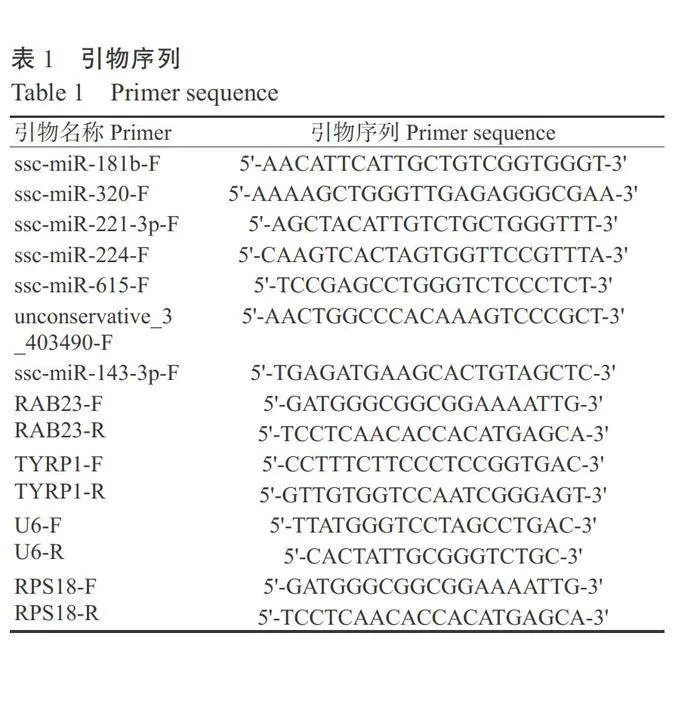
选取7个差异表达miRNA(ssc-miR-181b、ssc- miR-320、ssc-miR-221-3p、ssc-miR-224、ssc-miR-615、 unconservative_3_403490 和 ssc-miR-143-3p) 与 ssc- miR-221-3p靶基因TYRP1和ssc-miR-224靶基因 RAB23进行验证。利用NCBI和Primer Premier5.0 设计引物,以U6和RPS18为内参基因,引物序列见 表1,委托北京擎科生物科技股份有限公司合成。使 用TRIzol法提取样品总RNA,采用逆转录试剂盒(北京康为世纪生物科技有限公司)合成cDNA。使用SYBR Green试剂盒[天根生化科技(北京)有限公司]通过实时荧光定量PCR检测miRNA表达量,扩增程序:95℃预变性15min;94℃20s,60℃30s, 进行42个循环。使用2×Tag MasterMix试剂盒(北 京康为世纪生物科技有限公司)通过实时荧光定 量PCR检测基因表达量,扩增程序:95℃预变性 10min;95°℃10s,58°℃30s,进行40个循环。采用2:AAc法计算miRNA和基因相对表达量(Livak and Schmittgen,2001)。设3次重复,采用SPSS 25.0进 行t检验,分析不同样品间差异显著性,数据以平均 值±标准误表示,采用GraphPad Prism 8.0作图。
2结果与分析
2.1转录组测序结果
转录组测序结果如表2所示。黑色表皮样品文 库(BE1、BE2和BE3)和白色表皮样品文库(WE1、WE2和WE3)分别获得5803万和6108万条原始序 列。经质控后,平均每个文库获得1764万条有效序 列,Q20和Q30的平均值分别为99.51%和98.67%。平均69.07%的Reads长度为21~23 nt,其中22 nt的Reads最多。平均67.53%的Reads总比对到参考基 因组。转录组测序数据具有较高的可靠性和准确 性,可用于后续分析。
2.2miRNA的鉴定和预测结果
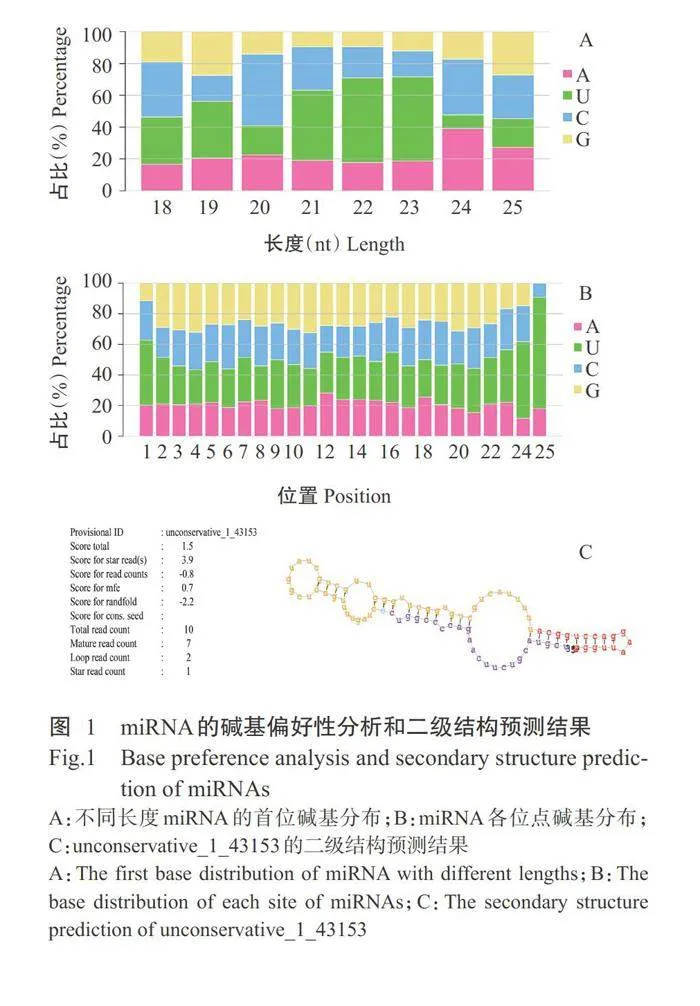
共鉴定出898个miRNA,其中280个为已知miRNA,618个为预测获得的新miRNA。对miRNA进行碱基偏好性分析,结果显示21~23nt miRNA的5'端首位碱基大部分为U(图1-A),U、G、G、G、C、U和G在2~8位碱基出现频率最高(图1-B)。预测的新miRNA前体具有典型发夹结构,unconservative_143153的二级结构预测结果如图1-C所示。
2.3差异表达miRNA筛选结果
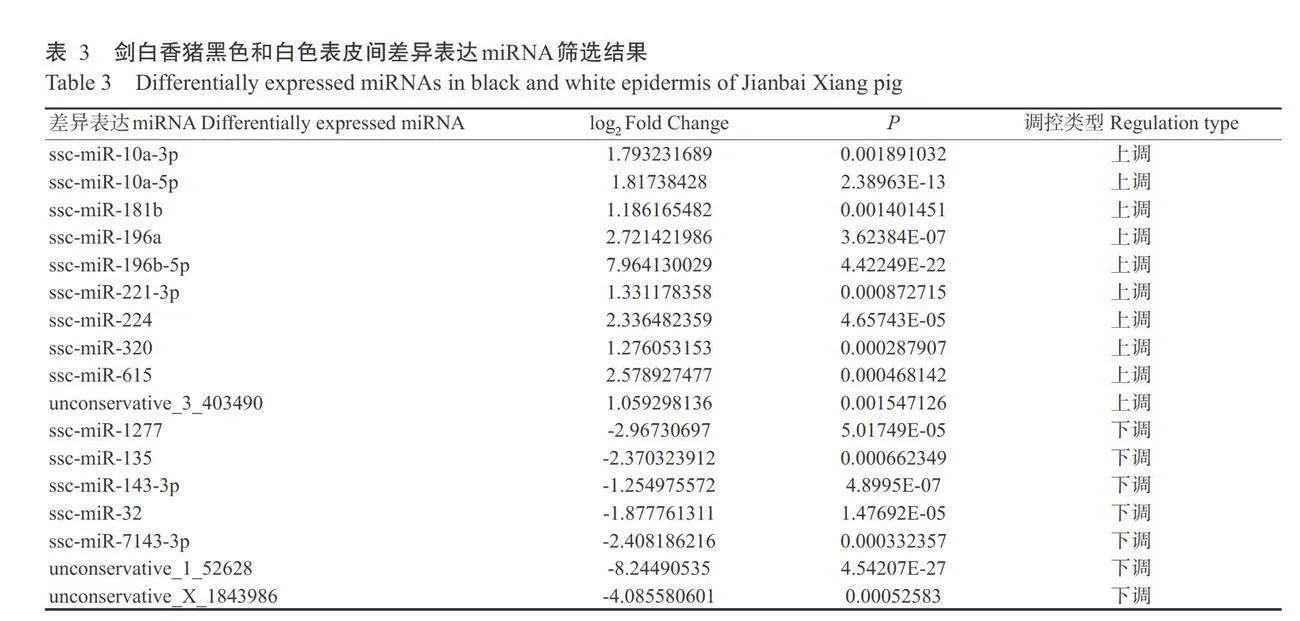
从剑白香猪黑色和白色表皮组织中筛选出17 个差异表达miRNA,白色表皮较黑色表皮有10个miRNA上调表达,7个miRNA下调表达(表3)。
2.4差异表达miRNA靶基因预测与分析结果
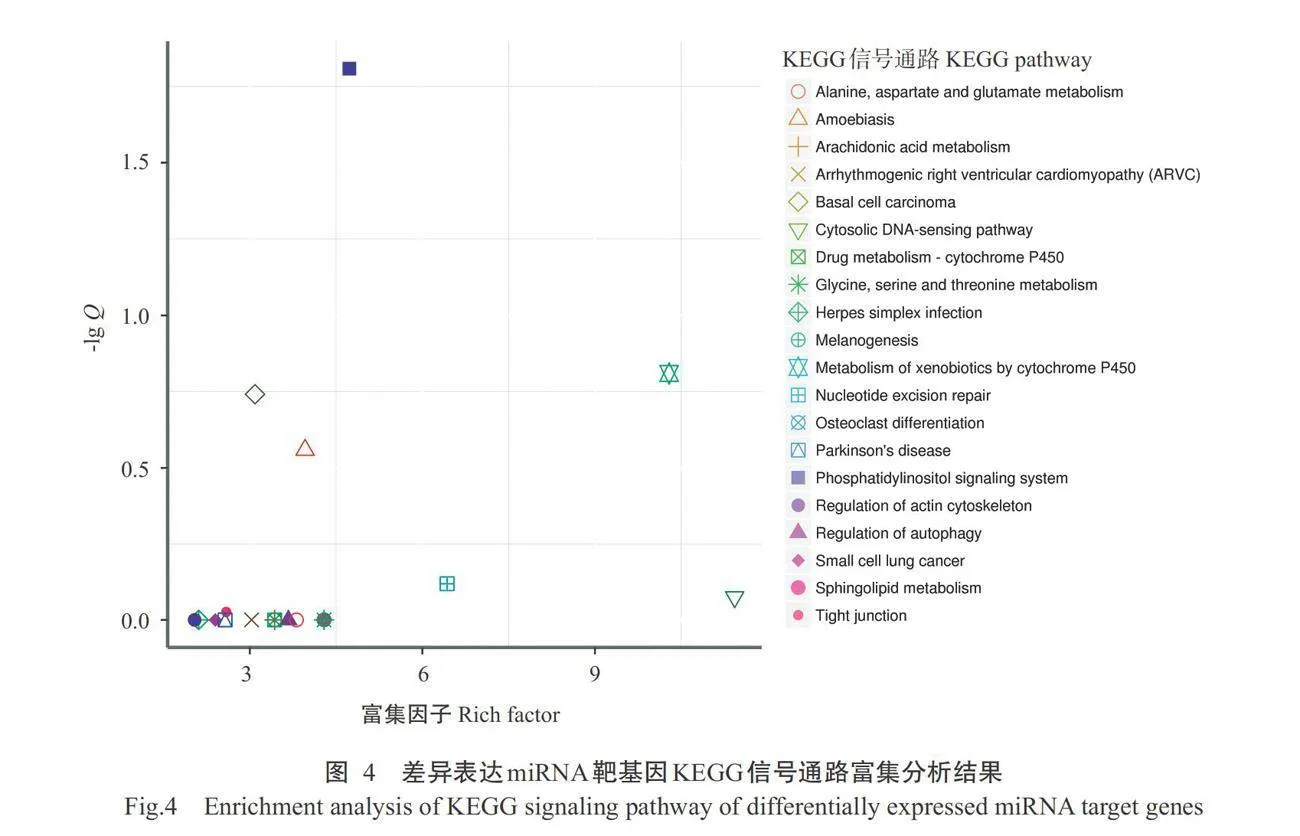
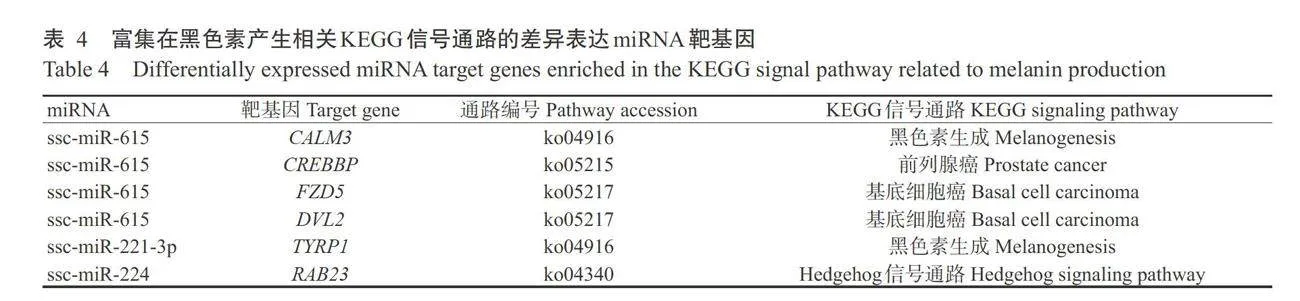
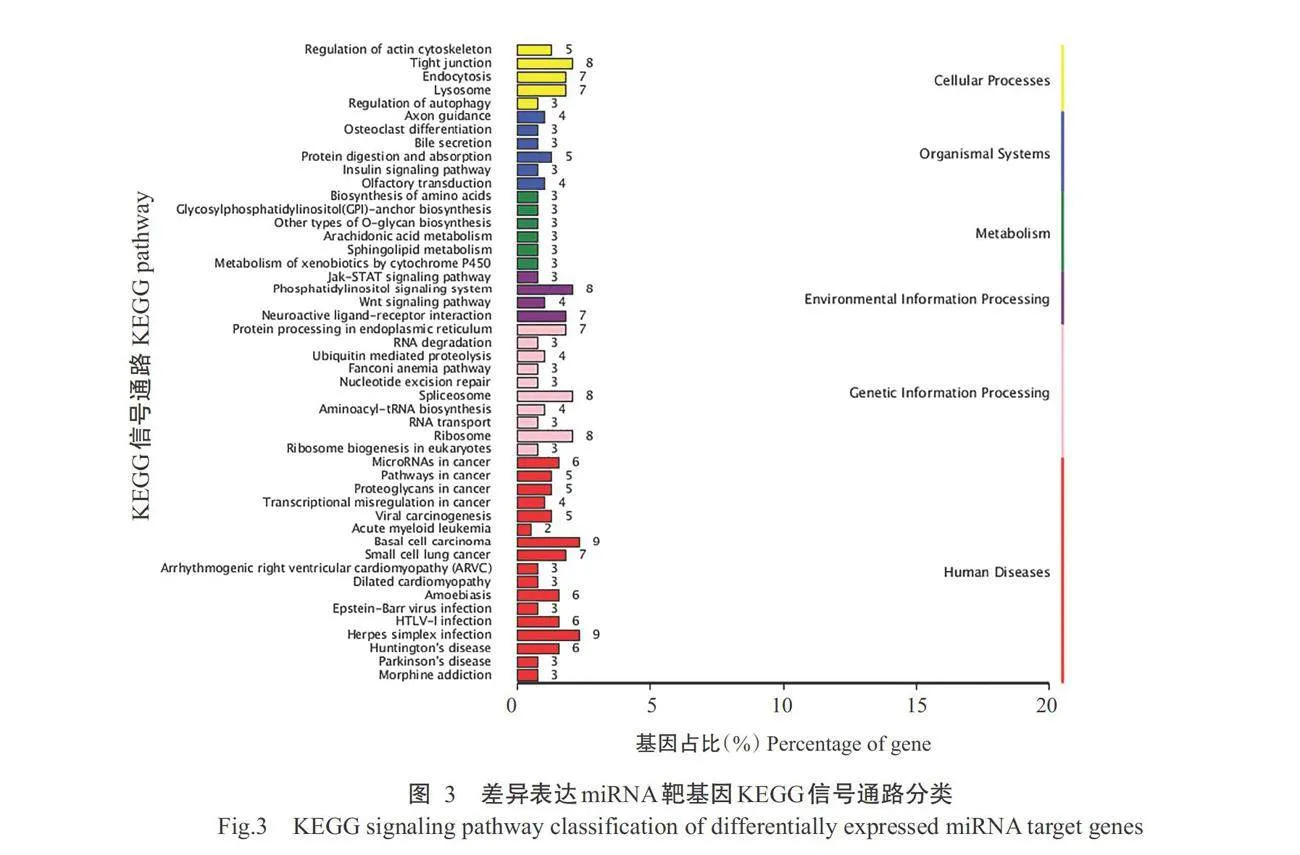
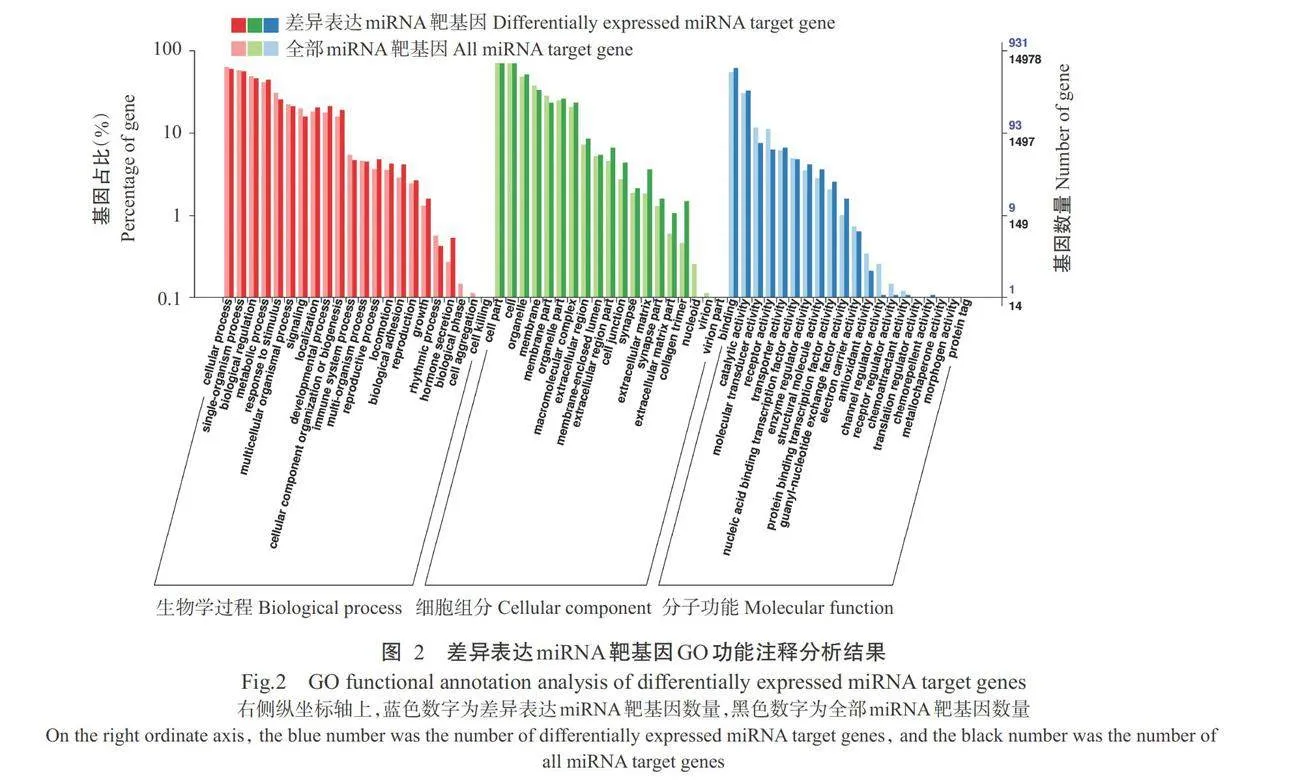
对17个差异表达miRNA靶基因进行预测,共获得1327个靶基因。对差异表达miRNA靶基因进行GO功能注释分析,结果如图2所示,在生物学过程中,差异表达miRNA靶基因主要涉及上皮发育、 细胞增殖和分解代谢过程等条目。在细胞组分和分 子功能中,主要涉及细胞质、细胞质囊泡、ATP结合和催化活性等条目。如图3所示,差异表达miRNA靶基因注释到氨基酸的生物合成、RNA转运和Wnt 信号通路等48条信号通路,如图4所示,差异表达 miRNA靶基因在磷脂酰肌醇信号系统、黑色素生成 和细胞色素P450对外源物质的代谢等信号通路显 著富集(Plt;0.05,下同)。如图5和表4所示,ssc-miR-615靶基因包括钙调蛋白3基因(CALM3)、cAMP反应元件结合蛋白(CREB)结合蛋白基因(CREBBP)、卷曲类受体5基因(FZD5)和散乱蛋白2基因(DL2),其富集通路均与表皮色素沉着有关,ssc-miR-221-3p靶基因TYRP1和ssc-miR-224靶基因RAB23富集的通路与黑色素生成有关。
2.5差异表达miRNA和靶基因的实时荧光定量 PCR验证结果
选取7个差异表达miRNA,利用实时荧光定量PCR检测其相对表达量,结果如图6-A所示,剑白香猪白色表皮中ssc-miR-143-3p下调,ssc-miR-181b、 ssc-miR-224、ssc-miR-615、ssc-miR-221-3p、 ssc-miR- 320和unconservative 3403490上调,与转录组测序 结果趋势一致。对ssc-miR-221-3p靶基因TYRPI和ssc-miR-224靶基因RAB23的相对表达量进行检测,结果如图6-B所示,TYRPI和RAB23在剑白香猪 黑色表皮中的相对表达量极显著高于白色表皮(Plt;0.01,下同)。
3讨论
黑色素形成过程复杂,除了黑色素细胞的发育、 存活和分化外,整个过程均与角质形成细胞密切相 关(Lai and McGrath,2021)。超过100个基因和多个 信号通路在黑色素形成中发挥调控作用,一些表观 遗传学事件也会影响黑色素的形成(Zhou et al.,2021)。本研究从剑白香猪黑色和白色表皮中共鉴 定出898个miRNA,其中280个为已知miRNA,618个为预测获得的新miRNA。miRNA长度多为21~23nt,且22nt占比最多,符合miRNA的长度分布特征。miRNA首位碱基偏向于U碱基,与前人研究结果一致(付雪峰等,2021),其原因与miRNA形成过 程和其对靶基因的沉默机制有关(袁钰洁等,2022)。 实时荧光定量PCR验证结果表明,选取的7个差异 表达miRNA表达变化与转录组测序分析的变化趋 势一致,表明测序结果可靠。此外,本研究筛选出17个差异表达miRNA,预测获得1327个靶基因,差 异表达miRNA靶基因显著富集在磷脂酰肌醇信号 系统、黑色素生成和细胞色素P450对外源物质的代谢等信号通路上,其中ssc-miR-615、ssc-miR-221-3p和ssc-miR-224的靶基因与黑色素生成密切相关。
TYRP1、TYR和多巴色素互变异构酶(DCT)均属酪氨酸酶家族,TYR、TYRPI和DCT基因对黑色素生成有调控作用,且与MITF有密切关系(Lai et al.,2018;Gautron et al.,2021)。TYRP1可激活和稳定酪氨酸酶(Videira et al.,2013),其具有5,6-二羟 基吲哚羧酸(5,6-dihydroxyindole-2-carboxylic acid, DHICA)氧化活性,可催化DHICA转换为真黑素(Zhou et al.,2021)。TYRP1基因的突变会导致猪皮毛颜色改变,且TYRPI基因在猪白色表皮中表达量 极低(Ren et al.,2011;Wu et al.,2016;Jin et al., 2020)。TYRP1基因还与水貂(Song et al.,2017)、绵羊(Paris et al.,2019)、兔子(Jia et al.,2021)和鸡 (Khumpeerawat et al.,2021)等多种动物皮毛颜色沉积有关。本研究中,ssc-miR-221-3p在剑白香猪白色表皮中呈低表达,其靶基因包括TYRP1,在剑白香猪 黑色表皮中TYRP1的相对表达量极显著高于白色表 皮。Yuan等(2023)研究发现,ssc-miR-221-3p模拟物转染香猪黑色素细胞后,ssc-miR-221-3p的表达显著上调,且抑制TYR、TYRPI和DCT基因和蛋白表达, 导致细胞中黑色素含量显著降低。上述结果表明, ssc-miR-221-3p为TYRP1新的调控因子,靶向并抑制TYRP1基因的表达,导致猪黑色素细胞中黑色素 含量降低。
黑素小体是一种独特的细胞器,其生成于黑色 素细胞,通过膜运输的方式到达角质形成细胞并最 终降解(Tian et al.,2021)。小分子GTP酶RAB家族成员,如RAB1A、RAB3A、RAB32、RAB38和RAB36等多个RAB异构体均参与黑素小体的生存过程,其 主要功能一是将货物蛋白(黑色素生成酶)分类并运 输至未成熟的黑素小体,二是运输黑素小体本身 (Fukuda,2021)。RAB4A参与黑素小体货物蛋白(TYR和TYRP1)的分离(Nag et al.,2018),RAB32与RAB38在TYR、TYRP1和DCT的运输中扮演重 要角色(Bultema et al.,2014)。本研究中,ssc-miR- 224在剑白香猪白色表皮中呈低表达,其靶基因 RAB23的表达量在黑色表皮中极显著升高。RAB23可通过影响上游cAMP依赖蛋白激酶(PKA)/CREB/MITF通路,改变细胞中酪氨酸酶活性和黑素小体运输效率,从而调控黑色素生成(Wei et al.,2022)。本研究中,ssc-miR-615在剑白香猪白色表皮中呈低表 达,其靶基因CALM3、CREBBP、FZD5和DVL2均与 表皮色素沉着有关。黑色素的形成与黑色素细胞内 游离Ca2+的含量有关(Jia et al.,2020),钙调蛋白是一种必不可少的Ca²*传感器蛋白,是许多下游蛋白的起点,而CALM3是编码钙调蛋白的非等位基因之一(Horigane et al.,2019)。cAMP可使CREB转录因子 磷酸化,促进MITF激活,进而促进下游TYR、TYRP1 和DCT蛋白的表达(Kim et al.,2016)。CREBBP可 特异性结合磷酸化的CREB并增强其对cAMP的转 录活性(Du et al.,2000)。黑色素的生成受cAMP/PKA和Wnt/p-catenin等多条信号通路调控,FZD5和DVL2均在Wnt/β-catenin信号通路上扮演重要角色 (Fu et al.,2020;Zhou et al.,2021)。 ssc-miR-224和 ssc-miR-615表达调控因子具有多功能性,在癌症、 神经退行性疾病、内皮细胞血管生成和组织修复 等方面具有较大潜能(Zhang et al.,2013;Icli et al., 2019;Godínez-Rubí and Ortuño-Sahagún,2020)。 但 ssc-miR-224和ssc-miR-615在黑色素生成过程中的 调控机理尚不清楚,需进一步开展功能验证研究。
4结论
通过对剑白香猪黑色和白色表皮进行转录组分 析,鉴定出17个差异表达miRNA,其中,ssc-miR-615、ssc-miR-221-3p和ssc-miR-224是影响剑白香猪表皮 颜色的候选基因,可能对剑白香猪表皮的黑色素形 成有重要影响。
参考文献(References):
付雪峰,赵冰茹,索朗达,巴贵,德吉,阿旺措吉,吴玉江,田可 川.2021.不同绒细度的西藏绒山羊皮肤组织miRNA分
析与鉴定[J].农业生物技术学报,29(11):2118-2128.
[Fu X F,Zhao B R, Suo L D,Ba G,De J, Awang C J, Wu Y J, Tian K C. 2021. Analysis and identification of miRNA in skin tissues of Tibetan cashmere goats (Capra hircus) with different cashmere fineness[J]. Journal of Agricultural Biotechnology, 29 (11) : 2118-2128.] doi: 10.3969/j. issn.
1674-7968.2021.11.006.
肖敏,赵威,孙武,娜日苏,赵乐,刘陶禄,张继攀,赵永聚.
2024.山羊皮肤组织miRNA测序与miR-129-5p调控黑
色素生成的功能研究[J].畜牧兽医学报,55(3):1019-
1029.[Xiao M,Zhao W,Sun W,Na R S,Zhao L,Liu T L, Zhang J P, Zhao Y J. 2024. The skin tissue miRNA-seq and regulation mechanism of miR-129-5p on melanogenesis in goat (Capra hircus) [J]. Acta Veterinaria et Zootechnica Sinica, 55 (3) : 1019-1029.] doi: 10.11843/j. issn.
0366-6964.2024.03.015.
袁钰洁,周婧雯,殷实,杨柳青,秦文昌,李键.2022.不同发育 阶段牦牛睾丸组织miRNA的分析及鉴定[J].中国兽医 学报,42(1):165-174.[Yuan YJ,ZhouJW,YinS,Yang L Q, Qin W C, Li J. 2022. Analysis and identification of miRNA in yak testis at different developmental stages[J].
Chinese Journal of Veterinary Science, 42 (1) : 165-174.] doi:10.16303/j.cnki.1005-4545.2022.01.27.
Anders S, Huber W. 2010. Differential expression analysis for sequence count data [J]. Genome Biology, 11 (10) : R106.
doi:10.1186/gb-2010-11-10-r106.
Ashburner M, Ball C A, Blake J A, Botstein D, Butler H, Cherry J M, Davis A P,Dolinski K, Dwight S S, Eppig J T, Harris M A, Hill D P, Issel-Tarver L, Kasarskis A, Lewis S, Matese J C,Richardson J E, Ringwald M,Rubin G M, Sherlock G. 2000. Gene ontology: Tool for the unification of biology [J]. Nature Genetics,25(1) :25-29. doi:
10.1038/75556.
Betel D, Wilson M, Gabow A, Marks D S,Sander C. 2008. The
microRNA. org resource: Targets and expression [J]. Nucleic Acids Research,36(S1):D149-D153. doi: 10.1093/nar/ gkm995.
Bultema J J,Boyle J A, Malenke P B,Martin F E,Dell'Angelica E C, Cheney R E, Di Pietro S M. 2014. Myosin vc interacts with Rab32 and Rab38 proteins and works in the biogenesis and secretion of melanosomes[J]. Journal of Biological Chemistry, 289 (48) : 33513-33528. doi: 10.1074/
jbc.M114.578948.
Cao W, Zhou X, McCallum N C, Hu Z, Ni Q Z, Kapoor U, Heil C M,Cay K S,Zand T,Mantanona A J,Jayaraman A, Dhinojwala A, Deheyn D D, Shawkey M D, Burkart M D, Rinehart J D, Gianneschi N C. 2021. Unraveling the structure and function of melanin through synthesis[J]. Journal of the American Chemical Society, 143(7):2622-2637. doi: 10.1021/jacs.0c12322.
Cichorek M, Wachulska M, Stasiewicz A, Tyminska A. 2013. Skin melanocytes: Biology and development [J]. Postepy Dermatol Alergol,30(1):30-41. doi: 10.5114/pdia.2013.33
376. D'Orazio J, Jarrett S, Amaro-Ortiz A, Scott T. 2013. UV radiation and the skin [J]. International Journal of Molecular Sciences,14(6):12222-12248. doi:10.3390/jms140612222.
Du K, Asahara H,Jhala U S, Wagner B L, Montminy M. 2000. Characterization of a CREB gain-of-function mutant with constitutive transcriptional activity in vivo [J]. Molecular
and Cellular Biology , 20 (12 ) : 4320-4327. doi : 10.1128/
MCB.20.12.4320-4327.2000.
Fahlgren N, Howell M D, Kasschau K D, Chapman E J, Sullivan C M, Cumbie J S, Givan S A, Law T F, Grant S R, Dangl J L, Carrington J C. 2007. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes[J]. PLoS One, 2 (2) : e219. doi: 10.1371/journal.pone.0000219.
Friedländer M R,Mackowiak S D, Li N,Chen W,Rajewsky N.
2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades [J]. Nucleic Acids Research, 40 (1) : 37-52. doi: 10.1093/ nar/gkr688.
Fu C H,Chen J,Lu J Y,Yi L, Tong X L,Kang L Y,Pei S Y, Ouyang Y J,Jiang L,Ding Y F,Zhao X J, Li S,Yang Y, Huang J H, Zeng Q H. 2020. Roles of inflammation factors in melanogenesis (review) [J]. Molecular Medicine Reports,21(3):1421-1430. doi: 10.3892/mmr.2020.10950.
Fukuda M. 2021. Rab GTPases: Key players in melanosome biogenesis, transport, and transfer[J]. Pigment Cell amp; Melanoma Research , 34(2) : 222-235. doi : 10.1111/pcmr.
12931. Gautron A, Migault M, Bachelot L, Corre S, Galibert M D, Gilot D. 2021. Human TYRP1: Two functions for a single gene? [J]. Pigment Cell Melanoma Research, 34 (5) : 836852. doi: 10.1111/pcmr.12951.
Godínez-Rubí M, Ortuño-Sahagún D. 2020. miR-615 fine- tunes growth and development and has a role in cancer and in neural repair [J]. Cells, 9 (7) : 1566. doi: 10.3390/cells
9071566.
Horigane S I, Ozawa Y, Yamada H, Takemoto-Kimura S. 2019. Calcium signalling: A key regulator of neuronal migration [J]. The Journal of Biochemistry, 165 (5) :401-409. doi: 10.1093/jb/mvz012.
Hushcha Y, Blo I, Oton-Gonzalez L, Mauro G D, Martini F, Tognon M, Mattei M D. 2021. microRNAs in the regulation of melanogenesis[J]. International Journal of Molecular Sciences, 22(11):6104. doi: 10.3390/ijms22116104.
Icli B, Wu W, Ozdemir D, Li H, Cheng H S, Haemmig S, Liu
X, Giatsidis G, Avci S N, Lee N, Guimaraes R B, Manica
A, Marchini J F,Rynning S E, Risnes I,Hollan I, Croce K, Yang X, Orgill D P, Feinberg M W. 2019. MicroRNA-6155p regulates angiogenesis and tissue repair by targeting AKT/eNOS (protein kinase b/endothelial nitric oxide synthase) signaling in endothelial cells [J]. Arteriosclerosis, Thrombosis, and Vascular Biology,39(7): 1458-1474. doi: 10.1161/ATVBAHA.119.312726.
Itoh T, Fukatani K, Nakashima A, Suzuki K. 2020. MicroRNA141-3p and microRNA-200a-3p regulate a-melanocyte stimulating hormone-stimulated melanogenesis by directly targeting microphthalmia-associated transcription factor [J]. Scientific Reports, 10(1) : 2149. doi: 10.1038/s41598-
020-58911-w.
Jia Q, Hu S X, Jiao D X, Li X Q, Qi S H,Fan R W. 2020. Synaptotagmin-4 promotes dendrite extension and melanogenesis in alpaca melanocytes by regulating Ca2+ influx via TRPM1 channels [J]. Cell Biochemistry and Function, 38
(3):275-282. doi:10.1002/cbf.3465.
Jia X B, Ding P, Chen S Y, Zhao S K, Wang J, Lai S J. 2021. Analysis of MC1R, MITF, TYR,TYRP1, and MLPH genes polymorphism in four rabbit breeds with different coat colors[J]. Animals,11(1):81. doi: 10.3390/ani11010081.
Jin L,Zhao L R, Hu S L, Long K R, Liu P L, Liu R,Zhou X, Wang Y X,Huang Z Q, Lin X X, Tang Q Z, Li M Z. 2020. Transcriptional differences of coding and non-coding genes related to the absence of melanocyte in skins of Bama Pig
[J]. Genes, 11(1):47. doi: 10.3390/genes11010047.
Khumpeerawat P, Duangjinda M, Phasuk Y. 2021. Factors affecting gene expression associated with the skin color of black-bone chicken in Thailand[J]. Poultry Science, 100
(11):101440. doi:10.1016/j.psj.2021.101440.
Kim Y M, Cho S E, Seo Y K. 2016. The activation of melanogenesis by p-CREB and MITF signaling with extremely low-frequency electromagnetic fields on B16F10 melanoma [J]. Life Sciences, 162: 25-32. doi: 10.1016/j. lfs.2016.08.015.
Lai X, Wichers H J, Soler-Lopez M, Dijkstra B W. 2018. Structure and function of human tyrosinase and tyrosinaserelated rroteins [J]. Chemistry - A European Journal, 24
(1):47-55. doi:10.1002/chem.201704410.
Lai C J E, McGrath J A. 2021. Structure and function of skin, hair and nails[J]. Medicine,49(6) : 337-342. doi: 10.1016/ j.mpmed.2021.03.001.
Langmead B, Trapnell C, Pop M, Salzberg S L. 2009. Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome [J]. Genome Biology, 10 (3) : R25.
doi: 10.1186/gb-2009-10-3-r25.
Liang D,Zhao P J, Si J F,Fang L Z,Pairo-Castineira E,Hu X
X, Xu Q, Hou Y L, Gong Y, Liang Z W, Tian B, Mao H
M, Yindee M, Faruque M O,Kongvongxay S, Khamphoumee S, Liu G E,Wu D D,Barker J S F,Han J L,Zhang Y.
2021. Genomic analysis revealed a convergent evolution of LINE-1 in coat color: A case study in water buffaloes (Bubalus bubalis) [J]. Molecular Biology and Evolution, 38(3):1122-1136. doi:10.1093/molbev/msaa279.
Liu X X,Du B,Zhang P Q,Zhang J Z,Zhu Z W,Liu B,Fan R
W. 2019. miR-380-3p regulates melanogenesis by targeting SOX6 in melanocytes from alpacas (Vicugna pacos)
[J]. BMC Genomics, 20 (1) : 962. doi: 10.1186/s12864-019-6343-4.
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-Aact method [J]. Methods, 25 (4) : 402-408. doi: 10.1006/ meth.2001.1262.
Nag S, Rani S, Mahanty S, Bissig C, Arora P, Azevedo C, Saiardi A, van der Sluijs P, Delevoye C, van Niel G, Raposo G, Setty S R G. 2018. Rab4A organizes endosomal domains for sorting cargo to lysosome-related organelles [J]. Journal of Cell Science, 131 (18) : jcs216226. doi: 10.
1242/jcs.216226.
Paris J M, Letko A, Häfliger I M, Ammann P, Flury C, Drögemüller C. 2019. Identification of two TYRPl loss-of- function alleles in Valais Red sheep [J]. Animal Genetics,
50(6):778-782. doi:10.1111/age.12863.
Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes
[J]. RNA,10(10): 1507-1517. doi: 10.1261/rna.5248604.
Ren J, Mao H, Zhang Z, Xiao S, Ding N, Huang L. 2011. A 6-bp deletion in the TYRPl gene causes the brown colouration phenotype in Chinese indigenous pigs[J]. Heredity,
106(5):862-868. doi:10.1038/hdy.2010.129.
Solano F. 2020. Photoprotection and skin pigmentation: Melanin-related molecules and some other new agents obtained from natural sources[J]. Molecules,25(7):1537.
doi: 10.3390/molecules25071537.
Song X C,Xu C,Liu Z Y,Yue Z G,Liu L L,Yang T G,Cong B, Yang F H. 2017. Comparative transcriptome analysis of mink (Neovison vison) skin reveals the key genes involved in the melanogenesis of black and white coat colour[J].
Scientific Reports, 7(1) : 12461. doi: 10.1038/s41598-017-12754-0.
Syeda Z A, Langden S S S, Munkhzul C, Lee M, Song S J.
2020. Regulatory mechanism of microRNA expression in cancer[J]. International Journal of Molecular Sciences, 21
(5):1723. doi:10.3390/ijms21051723.
Tian X Y, Cui Z Y, Liu S, Zhou J, Cui R T. 2021. Melanosome transport and regulation in development and disease [J].
Pharmacology amp; Therapeutics, 219: 107707. doi: 10.1016/ j.pharmthera.2020.107707.
Videira I F, Moura D F, Magina S. 2013. Mechanisms regulating melanogenesis[J]. Anais Brasileiros de Dermatologia,
88(1):76-83. doi:10.1590/s0365-05962013000100009.
Vodicka P, Smetana K Jr, Dvoránková B, Emerick T, Xu Y Z, Ourednik J, Ourednik V, Motlík J. 2005. The miniature pig as an animal model in biomedical research [J]. Annals of the New York Academy of Sciences, 1049 (1) : 161-171.
doi:10.1196/annals.1334.015.
Wang P C,Zhao Y Y,Fan R W,Chen T Z,Dong C S. 2016. MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting Sox5 in mouse skin melanocytes [J].
International Journal of Molecular Sciences, 17 (7) : 959.
doi:10.3390/ijms17070959.
Wei X J,Huang M, Yang Y, Liu Y L,Chi S M, Li C X. 2022. Silencing of Rab23 by siRNA inhibits ultraviolet Binduced melanogenesis via downregulation of PKA/CREB/ MITF [J]. Experimental Dermatology, 31 (8) : 1253-1263.
doi:10.1111/exd.14586.
Wu X Q, Zhang Y, Shen L Y, Du J J, Luo J,Liu C D,Pu Q, Yang R L,Li X W,Bai L,Tang G Q,Zhang S H,Zhu L.
2016. A 6-bp deletion in exon 8 and two mutations in introns of TYRPI are associated with blond coat color in Liangshan pigs[J]. Gene,578(1) : 132-136. doi: 10.1016/j.gene.2015.12.011.
Yuan W, Qin H, Bi H, Zhao D P, Zhang Y Y, Chen W. 2023. Ssc-mir-221-3p regulates melanin production in Xiang pigs melanocytes by targeting the TYRP1 gene [J]. BMC Genomics,24(1):369. doi:10.1186/s12864-023-09451-w.
Zhang G J, Zhou H, Xiao H X, Li Y, Zhou T. 2013. Upregulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancer [J].
Cancer Cell International, 13 (1) : 104. doi: 10.1186/1475-2867-13-104.
Zhang Z G, Shen W L, Liu W M, Lyu L C. 2022. Role of miR-
NAs in melanin metabolism: Implications in melaninrelated diseases [J]. Journal of Cosmetic Dermatology, 21
(10):4146-4159. doi: 10.1111/jocd.14762.
Zhou S H,Zeng H L,Huang J H, Lei L, Tong X L, Li S,Zhou
Y, Guo H R, Khan M, Luo L P, Xiao R, Chen J, Zeng Q H. 2021. Epigenetic regulation of melanogenesis[J].
Ageing Research Reviews, 69: 101349. doi: 10.1016/j.arr.2021.101349.
Zhu Z W, Ma Y Y, Li Y,Li P F,Cheng Z X, Li H F,Zhang L H, Tang Z W. 2020. The comprehensive detection of miRNA, IncRNA, and circRNA in regulation of mouse melanocyte and skin development[J]. Biological Research,
53(1):4. doi:10.1186/s40659-020-0272-1.
(责任编辑刘可丹)
