Expression and significance of ADAM12 in bladder cancer
2023-12-29ZHANGXiaolinWANGSheng
ZHANG Xiao-lin, WANG Sheng
Department of Urology, First Affiliated Hospital of Bengbu Medical College, Bengbu 233000, China
Keywords:Adam12 Bladder cancer Cancer metastasis EMT Immunological infiltration
ABSTRACT
1.Introduction
Bladder cancer is one of the most common cancers, and the incidence of bladder cancer will increase as the population grows and ages[1].In 2018, bladder cancer became the 12th most malignant tumor in the world[2].Approximately 70% of bladder cancers are classified as non-muscle-invasive bladder cancer (NMIBC) at the time of diagnosis, while the remaining approximately 30% are classified as muscle-invasive bladder cancer (MIBC), and highgrade carcinoma in situ in NMIBC has a tendency to invade the musculature of the bladder wall, thus becoming MIBC[3-4].At the biological and clinical level, NMIBCs are a heterogeneous cancer with a tendency to recur[5].Although the treatment of early bladder cancer can achieve good results, there is no effective treatment after the progression of intermediate and advanced bladder cancer.The prognosis of MIBC is much worse than that of NMIBC, so there is an urgent need for measures to reduce the chance of bladder cancer progression and improve the prognosis of intermediate and advanced bladder cancer.Nowadays, exploring the pathogenesis of bladder cancer is one of the hot spots in the research on tumor development and treatment[6].If effective immune agents or molecularly targeted drugs for bladder cancer can be developed,it will bring good news to bladder cancer patients.In addition, to discover markers that can be used for diagnosis or prognosis, an important direction for researchers is to clarify the pathogenesis and development mechanisms of bladder cancer.
ADAM is a family of proteins containing disintegrin and metalloproteinase domains that are involved in the regulation of a wide range of biological processes, including cell migration,adhesion, differentiation, and proliferation[7-8].ADAM12 is a family member with a gene located at 10q26.3 and a typical ADAM family structural sequence, including metalloproteinase, disintegrin,cysteine-rich, and epidermal growth factor-like domains, which may be involved in functions such as protein hydrolysis, cell adhesion,and intracellular signaling[9-11].Already has Several studies have shown that ADAM12 is highly expressed in a variety of cancers and promotes malignant tumor progression, such as breast cancer[12],colorectal cancer[10], liver cancer[13], pancreatic cancer[14], etc.Among them, the relevant literature reports that ADAM12 promotes gastric cancer progression by regulating immune tumor infiltration and epithelial-interstitial transformation (EMT)[15].Although ADAM12 has been reported in bladder cancer, its mechanism of action has not been clearly elucidated, and its relationship with tumor immune cells has not been reported[18].
Therefore, bioinformatics combined with in vitro experimental studies was used to analyze the expression of ADAM12 in bladder cancer, and the relationship between ADAM12 and clinicopathological features and immune cell invasion was also explored, so as to further study the mechanism of its possible effect on the progression of bladder cancer.
2.Materials and methods
2.1 Experimental Materials & Instruments
Normal human bladder cells SV-HUC-1 and bladder cancer cell lines UMUC3, 5637, J82 purchased from Shanghai Cell Bank were used as experimental materials.Bladder cancer cell line T24 was donated by Guo Yuanyuan’s research group from the First Affiliated Hospital of Bengbu Medical College.Main Reagents and Instruments:Media including (MEM, DMEM and 1640) were purchased from Wuhan Procell and FBS was purchased from Zhejiang Tianhang Biology.Pancreatic enzymes were purchased from Gbico.Small interference siRNA and plasmids were purchased from Shanghai Jiman Biological Company.Nanotrans was purchased from Hefei Bai Oujing Company.Primers were purchased from Anhui General Biological Company.Marker, Trizol, MonScriptTM RTlll All-in-One Mix with dsDNase, and MonAmpTM ChemoHS qPCR Mix were purchased from Wuhan Mona Biotech.Protease phosphatase inhibitors and BCA protein quantification kits were purchased from Shanghai Biyuntian Biological Company.The primary and secondary antibodies were purchased from Wuhan Proteintech and Beijing Bioss respectively.The multimode microplate reader (model: 1510)was purchased from Thermo in the United States.The real-time PCR instrument (model number: 7500) was purchased in the United States Bio-Rad.The optical microscope (model: 4J09628) was purchased from Olympus Corporation in Japan.
2.2 Source of the specimen
The Department of Pathology of the First Affiliated Hospital of Bengbu Medical College collected 30 surgical resection specimens from bladder cancer patients in 2021.Inclusion Criteria:(1) All patients with total cystectomy; (2) None of the patients underwent radiotherapy or chemotherapy before surgery; (3) It has been confirmed by pathological examination in our hospital; Exclusion Criteria:(1) Patients with transurethral local electrosection; (2)Patients who have been treated before surgery; (3) Patients with other major diseases.This experimental plan was approved by the Ethics Committee of Bengbu Medical College.
2.3 Data download and processing
Download the bladder cancer transcriptome from the official TCGA database website FPKM data, with a total of 412 bladder cancer tissue samples and 19
Normal bladder tissue sample.The downloaded gene expression data was normalized using software such as R.4.0.3 and then transformed using log2 for subsequent analysis.At the same time, the clinicopathological characteristics of bladder cancer patients were downloaded and extracted, and only the complete data of clinical information were retained for subsequent analysis.
2.4 Database and bioinformatics analysis
The TCGA database analyzes the expression of ADAM12 in tumors and normal tissues, and performs prognostic analysis based on its expression level.Gene correlation were analysised by using the public online GEPIA database (http://gepia.cancer pku.cn).gene enrichment were analysised by GSEA4.0.3 software Perform.The expression of ADAM12 in Pan-cancer were analysised and explore its relationship with EMT-related markers by Xiantao Academic Database (https://www.xiantao.love) Performed.The method CIBERSORT was used to calculate the infiltration ratio of 22 immune cells in bladder cancer samples, and P<0.05 was used as the filter standard and the filtration results are displayed using a histogram.At the same time, it is extracted and analyzed Correlation between the ADAM12 gene and immune cell infiltration, and through Scatter plot display.
2.5 ADAM12 expression and clinicopathological characteristics
According to the median expression level of ADAM12 in bladder cancer patients, bladder cancer patients were divided into high expression group and low expression group.SPSS Statistics 25.0 statistical software was used to analyze the relationship between ADAM12 expression and age, sex, T stage, N stage, M stage,Grade grade, Stage stage, etc.I investigated the prognostic value of ADAM12 in bladder cancer patients by performing univariate and multivariate cox regression analysis on survival data and ADAM12 expression in bladder cancer patients.These data are from the TCGABLCA database.
2.6 Immunohistochemistry assays
Immunohistochemical staining of paraffin sections of tumor tissues was performed using the DAB method, and after baking,deparaffinization, hydration, antigen regeneration and other processes, after the addition of 3% hydrogen peroxide to inactivate endogenous peroxidase, the samples of a primary antibody (rabbit antibody, diluted 1:200) was added, and then Incubate overnight at 4 ℃ (12 h total), followed by a universal secondary antibody (antirabbit, 1:2 000) at 37 ℃ for 30 min.Subsequently, the samples were dehydrated, mounted, and finally observed under a microscope and the results were statistically counted.Protein expression intensity was calculated as follows: 3 non-adjacent fields of view were randomly selected under 100× and 400× high-power magnification,and staining intensity was assessed according to the number of brown or brownish-yellow particles present in the cytoplasm (0:negative; 1: Weakly positive; 2: Moderately positive; 3: strong positive); Calculate the percentage of positive stained cells out of the total number of cells, and calculate the fractional score (0 points:0; 1 point: 1%~10%; 2 points: 11%~50%; 3 points: 51%~80%; 4 points: 81%~100%).The staining intensity score for each slice is calculated by multiplying the scores of the two.Scores range from 0 to 1 and indicate negative, denoted by the “-” symbol; A score from 2 to 4 indicates a weak positive, denoted by a “+” symbol; A score of 5 to 8 indicates moderate positive, denoted by the “++” symbol;A score of 9 to 12 indicates a strong positive, indicated by the “+++”symbol.
2.7 Cell culture and transfection
Cells were cultured at 37 ℃, 5% CO2, in MEM medium containing 1% bispecific antibody and 10% fetal bovine serum.UMUC3 and J82 cells were seeded separately into a 6-well culture plate.When the cell fusion rate reached 50%~60%, small interferences or plasmids were transferred into the cells according to the nanotrans instructions, and the normal complete medium was replaced after 6 h.The culture was continued for 24 h and then used for follow-up experiments.Transfection group:siNC vs.siADAM12 or NC vs.Oe-ADAM12.
2.8 qRT-PCR assays
UMUC3 cells were collected, and total RNA was extracted from each group using Trizol reagent.Based on the results of RNA quantification, total RNA was reverse transcribed into cDNA using a reverse transcription kit (reaction conditions: 37 ℃ 2 min, 55℃ 15 min, 85 ℃ 5 min).Using cDNA as a template,Configure the reaction setup using the real-time PCR kit instructions to complete the qRT-PCR reaction (reaction conditions: 95 ℃for 30 seconds, 95 ℃ for 5 s, 60 ℃ for 30 s, 40 cycles).The 2-ΔΔCTmethod was used to analyze the ADAM12 expression level.ADAM12 F:5’ - CGAGGGGTGAGCTTATGGAAC-3’;R:5’-GCTTTCCCGTTGTAGTCGAATA - 3’.GAPDH F:5’ - GAGAAGTATGACAACAGCCTCAA - 3’;R:5’ -GCCATCACGCCACAGTTT-3’.
2.9 Western blot detection
After transfection of UMUC3 and J82 cells for 24 h, the total protein was extracted using RIPA and quantified using the BCA kit.The total amount of 20~40 μg per well was loaded, 3 double wells were set up in each group, and electrophoresis was performed after the loading was completed (condition: 120 mV for 2 h).The electrophoresis PAGE gel was transfected (condition: 400 mA for 1 h), and after finishing, the PVDF membrane was placed in 5% skim milk powder and sealed at room temperature for 2 h.After successful blocking, wash the membrane 3 times using TBST and then incubate the primary antibody overnight at 4 ℃.ADAM12, E-cadherin,N-cadherin, vimentin, GAPDH and secondary antibodies were diluted at 1:500, 1:1 000, 1:1 000, 1:1 000, 1:5 000 and 1:5 000,respectively.
2.10 CCK-8 detection
Each group of UMUC3 cells was plated in 96-well plates of 5 replicate wells, 2 000 cells per well, 100 μL of complete medium,and after the cells were adherent, 10 μL of CCK-8 reagent was added to each well, and then placed in an incubator for 2 h, and then the absorbance value (OD value) of each well at 450 nm wavelength was measured using a microplate reader.And so on, record the values of each well continuously at 0, 24, 48, and 72 h.Finally, the OD value was detected in the OD450 band by microplate reader, and the multiplication rate of the cells was calculated.
2.11 Transwell detection
Migration experiment: A 24-well plate was taken first, each group of cells from successfully transfected UMUC3 was digested and centrifuged, the cells were resuspended with serum-free MEM and counted, and 5 104 cells per well were spread into each well of the chamber 100 μL of serum-free medium was added, and 500 μL of complete medium containing 20% serum was added to each well outside the chamber.Each group had 3 replicate wells and was incubated in the incubator for 24 h.Take out the chamber, fix it with 4% paraformaldehyde for 2 h, stain 1% crystal violet at room temperature for 15 min, then take out the chamber and wash the inside of the chamber with cotton swabs and PBS.Finally, the chambers will be placed to take pictures of the counts under the microscope in order to finally calculate the mobility of the cells in each group( 160).
2.12 Scratch Experiment
UMUC3 cells were digested and centrifuged, 5 mL of medium was added to resuspend and counted, 2×105cells per well were spread into 6-well plates, and after the cells were cultured to a density of about 60%~70%, transfection experiments were carried out, and a control group and an experimental group were set up respectively.When the cell density reaches more than 90% in each well, scratch vertically using a 10 μL pipette tip and take pictures under the microscope to record the scratch area at 0 h, 24 h, and 48 h( 64).
2.13 Statistical Processing
Statistical analysis was performed using Graphpad prism 8.0 software and SPSS25.0 statistical software, measured using mean± standard deviation, t-test was used for comparison between two groups, and correlation test was performed using spearman, and P<0.05 was statistically significant for the difference.
3.Results
3.1 Expression and prognosis of ADAM12 in bladder cancer
Based on the TCGA database, paired and unpaired analyses showed that the expression level of ADAM12 in bladder cancer tissues was significantly higher than that in normal tissues (P<0.05, Fig.1A-B),and the high expression was negatively correlated with prognosis(P=0.007, Fig.1C).The qRT-PCR results showed that the expression of ADAM12 in J82 and UMUC3 cell lines was higher than that of normal bladder cells (t-values were 15.01 and 19.91, both P<0.05,Fig.1D).Pan-cancer analysis showed that ADAM12 was generally expressed in pan-cancer, and the expression level in bladder cancer tissues was higher than that in normal tissues (P<0.05, Fig.1E).
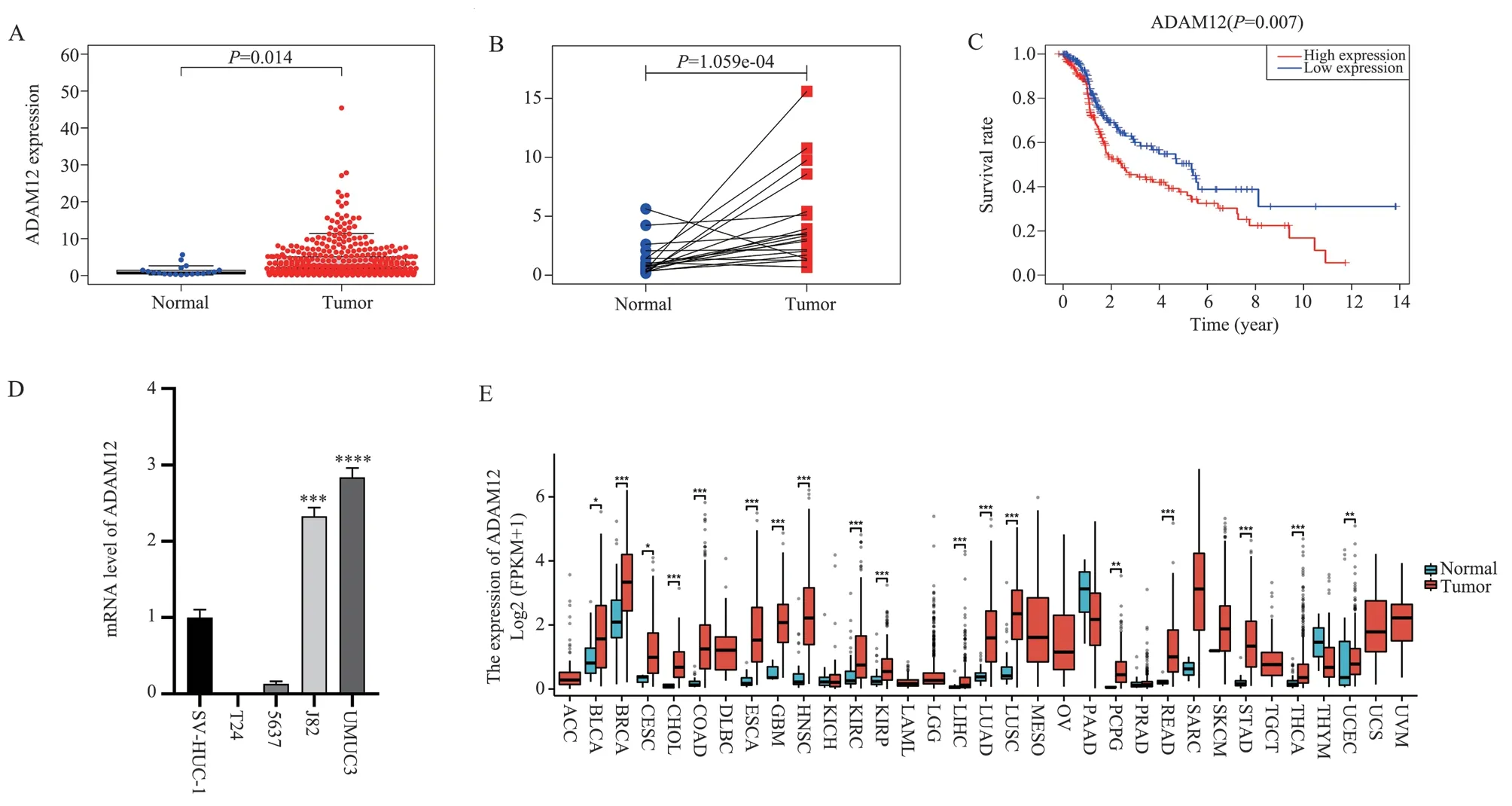
Fig 1 Expression and prognosis of ADAM12 in bladder cancer
3.2 Immunohistochemistry assays
Immunohistochemistry (IHC) staining images show that ADAM12 is located in the cytoplasm and plasma membrane.The scoring results showed that the expression level of ADAM12 in bladder cancer tissues was significantly higher than that in neighboring tissues(t=8.308, P<0.05,Fig.2)
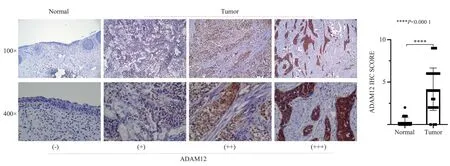
Fig 2 ADAM12 expression in bladder cancer and adjacent tissues detected by IHC
3.3 Relationship between ADAM12 and clinicopathological features
Based on TCGA clinical data, the expression of ADAM12 in bladder cancer was related to age, Grade, Stage, T and N stages(P<0.05), but not to gender and M stage (P>0.05, Fig.3).In univariate COX regression analysis, age, stage, T stage, N stage,and ADAM12 were all potential prognostic factors for bladder cancer (P<0.05).The results of Cox multivariate regression analysis suggested that ADAM12 could be used as an independent prognostic variable for bladder cancer (P<0.05).Table 1.
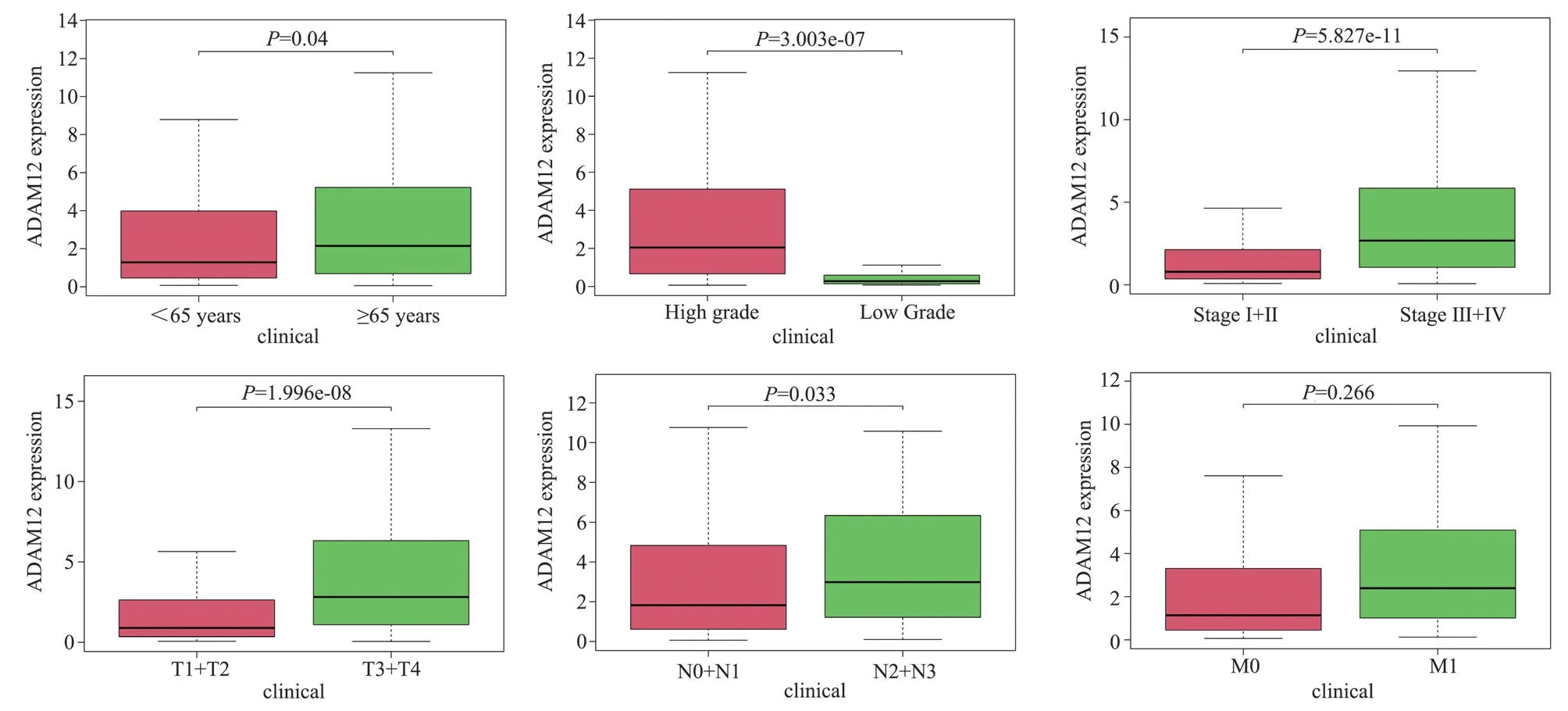
Fig 3 Relationship between ADAM12 and clinicopathological features of bladder cancer
3.4 GESA enrichment analysis
GSEA was used to identify relevant genes enriched in response to high expression of ADAM12.In the ADAM12 high expression group, ADAM12 was significantly associated with tumor pathways,cell adhesion molecules, and ECM receptors related to invasion and transformation (P<0.05, Fig.4A).In addition, ADAM12 is also involved in chemokine signaling pathways, tumorigenesis, and cancer-related pathways (P<0.05, Fig.4B).
3.5 Knockdown of ADAM12 inhibits bladder cancer cell biological behavior
Compared with the NC group, the results of qRT-PCR showed that the knockdown efficiency of the siADAM12-2 group (referred to as si2) was the best (t=8.676, P<0.05, Fig.5A).Western blot detection further confirms interference efficiency (Fig.5B).Compared with the NC group, the results of CCK8 detection showed that cell viability in the siADAM12-2 group decreased (t=11.64, P<0.05, fig.5C).Transwell results showed that the capacity of cells migrating in the siADAM12-2 group was significantly reduced (t=7.852, P<0.05,Fig.5D).Scratches the results showed that the healing ability of cells in the siADAM12-2 group was significantly reduced in scratches detection (t=8.49, P<0.05, Fig.5E).

Fig 4 GSEA exploring the potential mechanism of ADAM12 in bladder cancer
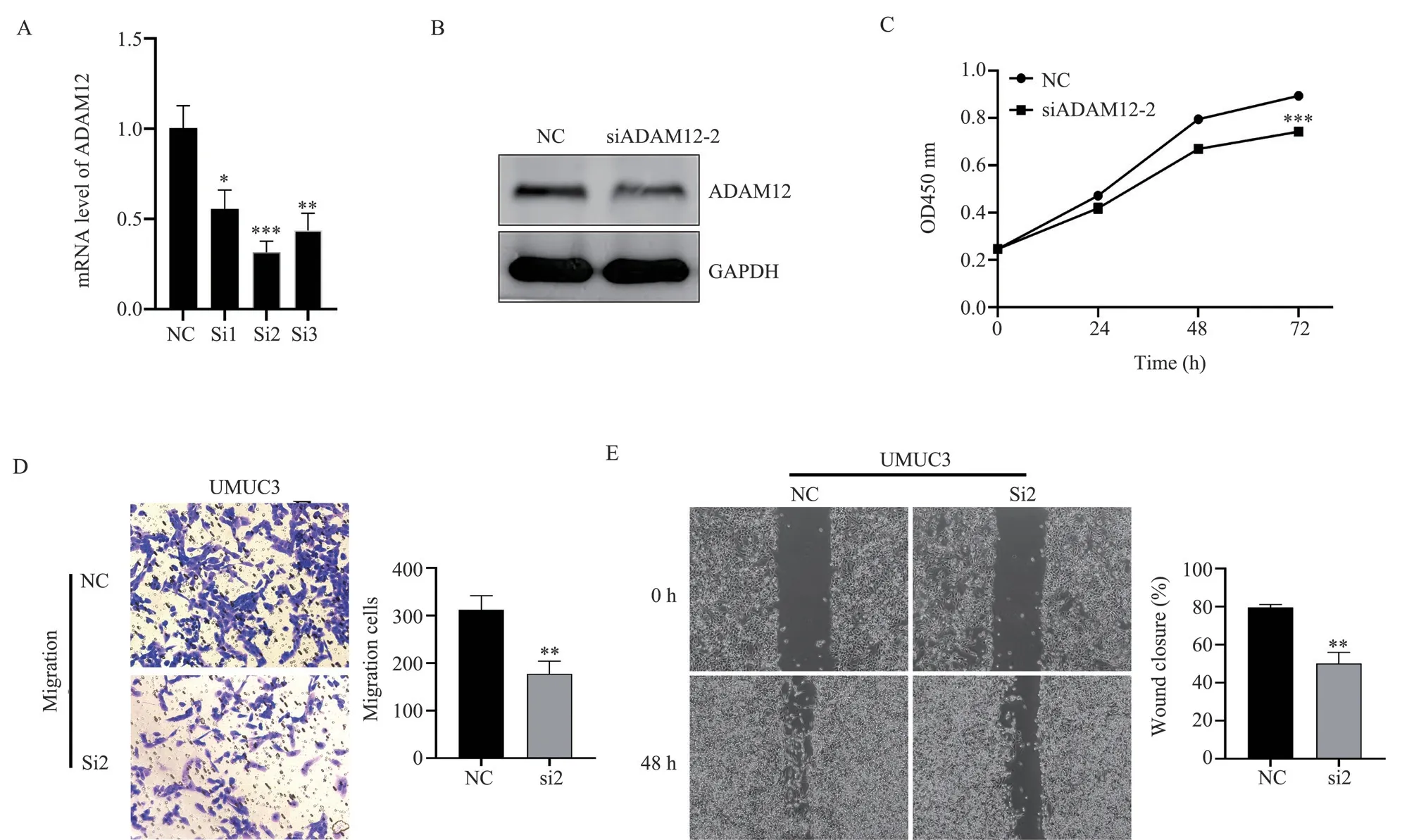
Fig 5 ADAM12 affecting the biological behavior of bladder cancer cells
3.6 ADAM12 promotes bladder cancer cell migration by triggering EMT
TCGA database analysis showed that ADAM12 was negatively correlated with EMT-related marker E-cadherin (r=-0.144, P=0.003).It was positively correlated with N-cadherin (r=0.646, P<0.001),vimentin (r=0.754, P<0.001), Snail (r=0.605, P<0.001), MMP2(r=0.784, P<0.001) and MMP9 (r=0.596, P< 0.001, Fig.6A).The results of Western blot analysis showed that compared with the NC group, the protein expression of E-cadherin in the knockdown group was increased, and the expression of N-cadherin and vimentin was decreased (Fig.6B).In over-expression, the results are reversed(Fig.6C).
3.7 Correlation between ADAM12 and bladder TICS
CIBERSORT counted 19 normal samples and 412 tumor samples,and the filtered results were shown in a histogram (P<0.05, Fig.7A).According to the resultes of the CIBERSORT method, ADAM12 was positively correlated with M1 macrophages (r=0.19, P=0.01),M2 macrophages (r=0.46, P<0.000 1), CD4+memory quiescent T cells (r=0.16, P=0.03), and negatively correlated with dendritic cells (r= 0.4, P<0.000 1), follicular helper T cells were negatively correlated (r= 0.2, P=0.0076), and regulatory T cells were negatively correlated (r= 0.16, P=0.032, Fig.7B).Correlation analysis showed the relationship between ADAM12 and immune checkpoints.Its results showed that ADAM12 was associated with CTLA4 (r=0.15,P=0.002 7), PDCD1 (r=0.13, P=0.007 9), and LAG3 (r=0.1,P=0.038) (Fig.7C).

Tab1 Univariate and multivariate regression analysis of the role of ADAM12 in bladder cancer
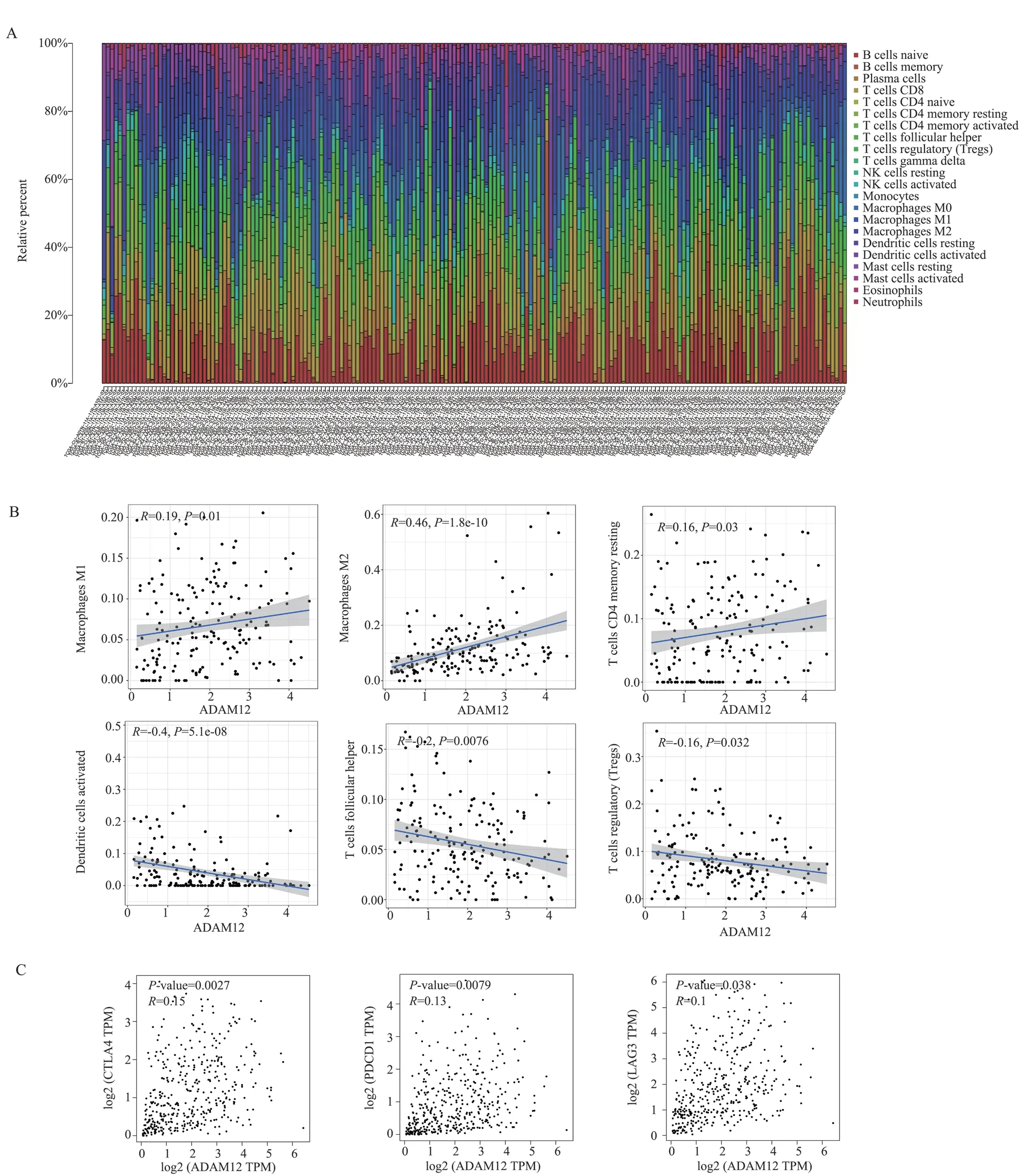
Fig 7 ADAM12 expression affecting the immune microenvironment of bladder tumors
4.Discussion
At present, more and more studies have shown that epithelialmesenchymal transition (EMT) in cancer plays an important role in progression and metastasis[16].Again, EMT is the a key process of bladder cancer metastasis.Previous studies have shown that EMT is an important biological process that facilitates NMINC to MIBC[17].Therefore, it is important to actively explore molecules that can predict the prognosis of bladder cancer and become a target molecule that can regulate the metastasis of bladder cancer.
In this study, TCGA database analysis showed that ADAM12 was highly expressed in bladder cancer and was associated with poor prognosis, and in vitro cell assays and immunohistochemical assays further confirmed that ADAM12 was highly expressed in bladder cancer.The results of clinicopathological feature analysis showed that ADAM12 was significantly associated with poor staging and grading.Multivariate cox regression analysis suggests that ADAM12 can be used as a solitary for bladder cancer prognostic factors.Previous studies have also reported that ADAM12 has a promising prognostic marker for bladder cancer[18].These findings suggest that ADAM12 may serve as a strong predictor of bladder cancer prognosis.
Secondly, in order to further explore whether ADAM12 affects the occurrence of EMT in bladder cancer, GSEA enrichment analysis showed that it was mainly associated with “cell adhesion”,“ECM receptor” and “multiple signaling pathway”.It indicating that ADAM12 has a potential role in tumor metastasis and cancer signaling pathway activation.Related publications report that ADAM12 is involved in cell adhesion and can degrade various extracellular matrices(ECM) proteins.Gelatin, type IV collagen, and fibronectin play an important role in the extracellular matrix (ECM)remodeling process, which is a feature of neoplastic diseases[19-20].In addition, ADAM12 is closely related to matrix metalloproteinases(MMPs) and is involved in extracellular matrix (ECM) remodeling and cell signaling through ECM cleavage and release of growth factors[21].ECM changes are known to be closely related to the development of EMT, and EMT in cancer is associated with tumor invasion and migration[22-24].Similarly, in the results of this study,ADAM12 was negatively correlated with E-cadherin and positively correlated with N-cadherin, vimentin, Snail, MMP2 and MMP9.The experiment results showed that the expression of E-cadherin was increased, the expression of N-cadherin and vimentin was decreased when ADAM12 was knocked down.When ADAM12 was over-expressed, the expression of E-cadherin was decreased,the expression of N-cadherin and vimentin was increased.In cell experiments, knockdown of ADAM12 inhibited bladder cancer cell migration.All results suggest that ADAM12 is an important regulator of EMT in bladder cancer cells.In related studies, it has been reported that ADAM12 is closely related to the occurrence of EMT, and it promotes tumor invasion and migration by regulating EMT in gastric cancer cells[25]; In breast cancer, ADAM12 has even been established as a new EMT marker[26].This is consistent with the results of this study, where ADAM12 promotes bladder cancer cell migration by stimulating EMT.
Finally, whether ADAM12 affects the bladder cancer immune microenvironment was further explored.Relevant literature suggests that ADAM12 is involved in a variety of tumor progression and may affect tumor initiation and progression through immune invasion[21,27,28].In this study, ADAM12 was a significant correlation between levels and the infiltration of a variety of immune cells in bladder cancer patients.The expression of ADAM12 was significantly positively correlated with M2 macrophages.Macrophages are essential for tumor metastasis and mmunosuppression, and macrophages have both M1 and M2 phenotypes, which can promote tumor immune response or stimulate tumor development, respectively[29].Tumor-associated macrophages predominantly exhibit an M2-like phenotype that stimulates tumor growth by promoting tumor immunosuppression[30].M2 polarization of TAM by causing macrophages to produce growth factors and cytokines, in regulating tumor growth, migration, and angiogenesis plays an important role[31].Finally, we further analyzed the relationship between ADAM12 and immune checkpoints, and found that it was positively correlated with the expression of CTLA4,PDCD1, and LAG3.These results suggest that ADAM12 may affect bladder cancer progression by regulating immune cell infiltration.
In summary, studies have confirmed that ADAM12 is highly expressed in bladder cancer and is associated with poor prognosis.Bioinformatics and a series of experimental studies have shown that ADAM12 promotes the migration of bladder cancer cells by regulating the occurrence of EMT.In addition, ADAM12 may affect the tumor immune microenvironment.However, in this study, there are still shortcomings and high expression of ADAM12 in patients is associated with poor prognosis based on database data.It lacks clinical follow-up data to further support it.In addition, the specific mechanism of ADAM12 affecting EMT and M2 macrophage polarization has not been studied in depth, and further exploration is needed in future studies.In conclusion, ADAM12 may serve as a poor prognostic factor and therapeutic target for bladder cancer.
Authors’ Contribution
Zhang Xiaolin: Complete the conception and experiment of this paper, and unify the data of the article computer analysis; Wang Sheng guided the experiment and revised the paper.
All authors declare no conflicts of interest.
杂志排行
Journal of Hainan Medical College的其它文章
- Systematic review and meta-analysis of endometrial thickening during endocrine therapy combined with Chinese herbal medicine intervention after breast cancer surgery
- Clinical observation on the efficacy and safety of different doses of alfentanil for painless gastroscopy
- Effects of intravenous infusion of esketamine on analgesia and postpartum antidepressant after cesarean section
- The expression of TUSC3 in Preeclampsia and the function in trophoblast cell
- Effect of hepatocyte growth factor on inflammatory factors associated with CCL4-induced hepatocyte injury
- Inhibitory effect of water soluble propolis on oxidative damage in rats with ulcerative colitis
