CircROBO1 promotes retinal Y79 cell tumor invasion by targeting KLF5
2023-12-23LiuYuefengZhangZhiweiLiXinLuoWeimin
Liu Yue-feng, Zhang Zhi-wei, Li Xin, Luo Wei-min
1.Department of Ophthalmology, Taihe Hospital (Affiliated Hospital of Hubei University of Medicine), Shiyan 442000, China
2.Cancer Research Institute of Hengyang Medical College, University of South China, Hengyang 421001, China
3.Hubei University of Medicine, Shiyan 442000, China
Keywords:
ABSTRACT Objective: To explore the role of circROBO1 in promoting the invasion of retinal Y79 cells by targeting KLF5 and its possible regulatory mechanism.Methods: RNase R enzyme digestion and qRT-PCR experiments were used to detect the structural stability of circular circROBO1 in retinal Y79 cells; cytoplasmic and nuclear RNAs of retinal Y79 cells were extracted for localization analysis of circROBO1; The expression of circROBO1 in retinal Y79 cells were silenced by siRNA.The effect of circROBO1 on the migration and invasion ability of HT-29 cells was detected by scratch assay, Transwell cell invasion and migration assay.The target binding sites of circROBO1 and its downstream miRNA and that of miRNA and its downstream target gene KLF5 were predicted by CircInteractome and TargetScan online software respectively, and the target regulation relationship between them was verified by double luciferase reporter gene experiment.Western blot was used to detect the effect of siRNA silencing the expression of circROBO1 in Y79 cells on the expression of KLF5.Results: Compared with the control group without RNase R enzyme treatment, relative circROBO1 levels did not change significantly after treatment, while relative linear ROBO1 levels decreased significantly after treatment (t=16.18, P<0.05); the content of circROBO1 in the cytoplasm was significantly higher than that in the nucleus (P<0.05); compared with sicontrol group, the migration rate and the invasion and migration abilities of Transwell cells were all lower in the si-circROBO1 group (t=22.54, P<0.05); circROBO1 can adsorb miR-885-5p, and there is a target binding site between miR-885-5p and KLF5(t=11.39, P<0.05);compared with the si-control group, the KLF5 protein expression in the si-circROBO1 group was significantly decreased (t=17.26, P<0.05).Conclusions: circROBO1 promotes retinal Y79 cell tumor invasion by targeting KLF5.
1.Introduction
Retinoblastoma (RB) is a commonly malignant intraocular tumor in infants, which is more common in children under the age of three.The eyes clinical characteristics are mainly red, pain, low vision or white pupil disease,etc.In severe cases, it even imparis the visual function and life of children[1].At present, the early diagnosis of RB is still more difficult, most of the children are already in the advanced stage of the disease when they are diagnosed, so exploring the pathogenesis and development of RB and finding the key molecules regulating the process may be the key to improve the early diagnosis of RB and reduce recurrence and metastasis[2-3].
circRNAs are covalently closed single-stranded non-coding RNA molecules, and their expression levels are significantly abnormal in a variety of malignant tumors (such as breast cancer, lung cancer, RB, etc.)[4-6].circRNAs are closely related to the malignant progression of a variety of cancers.circRNAs have the potential to become a target for early diagnosis or clinical treatment of various malignant tumors.For example, circAGFG1 can increase the proliferation and migration ability of breast cancer cells, and it is a potential therapeutic target for triple-negative breast cancer[7].Studies have shown that circROBO1 can promote the carcinogenesis and metastasis of breast cancer, and this process is realized by regulating KLF5[8].Our team’s previous studies[9] found that microRNA miR-222 can promote the growth and invasion ability of RB by targeting RB1, and accumulated a certain basis for RB research.At the same time, we also found that circROBO1 can promote the proliferation of RB cells by targeting and regulating miR-217, and confirmed the promotion effect of circROBO1on RB growth, but its invasion and migration ability has not been further studied.Therefore, this study aims to explore the effect of circROBO1 on RB invasion and its regulatory mechanism, in order to find potential therapeutic targets for RB treatment.
2.Materials and methods
2.1 Main materials
Human retinoblastoma cell Y79 (Shanghai Institute of Biochemistry and Cell Chemistry, Chinese Academy of Sciences),Lipo 3000, protein lysate and inhibitor,antibody KLF5 and β-actin(Beijing Boer, China), DMEM medium, serum and antibiotic(EsECO, China), circROBO1 interfering small RNA and its control, RNase R enzyme (Gisai Bio, China), Nuclear/cytoplasmic Component Extraction kit (BioVision, USA), TaqMan Reverse transcription and Fluorescence Quantitative PCR kit (Takara, USA),Intelligent Counter (Shanghai Mowei, China), Transwell Chamber and matrix gel (0.8 nm, Corning, USA), Nanodrop 2000 UV-VIS spectrophotometer (Thermo, USA), OLYMPUS BX51 upright microscope (Olympus, Japan).
2.2 Cell culture and grouping
According to the instructions provided by the supplier, Y79 cells were cultured in DMEM medium containing FBS, penicillin and streptomycin at concentrations of 10%,102U/mL and 0.1 g/L, respectively.The experiment was divided into si-control group,blank group and si-circROBO1 group.si-circROBO1 group was transfected with circROBO1 interfering small RNA(5'-GAAAACACAAGAUAUGAAA-3') sequence,and sicontrol group was transfected with no control sequence (5'-UUCUCCGAACGUGUCACGU-3').The blank group was transfected with no sequence.
2.3 Cell transfection
Y79 cells were collected, diluted into 104cells /ml cell suspension in DMEM medium containing 10% FBS, and inoculated into 6-well plates, in which 2 mL of Y79 cell suspension was added to each well, and incubated in a cell incubator containing 5% CO2at 37 ℃.When the confluence of Y79 cells reached about 80%, lipofectamin 3 000 and transfection reagents (si-circROBO1 and si-control) were mixed in a sterile EP tube,and the above mixture was added to the serum-free medium (without antibiotics) 20 min later.The cells were added into a 6-well plate to be transfected and cultured at 37 ℃ with 5% CO2for 7 h and 10% FBS DMEM for 48 h, which could be used for follow-up experiments.
2.4 RNA extraction and qRT-PCR detection
Total RNA of Y79 cells was extracted according to the RNA extraction kit provided by the reagent manufacturer, and detected on the UV-visible spectrophotometer.RN meeting the experimental requirements was stored in the refrigerator at -20 ℃ for future use.Using the total RNA of Y79 cells as a template, the reverse transcription kit and qSYBR-Green-containing PCR kit provided by the reagent manufacturer were operated, and relative quantitative analysis was carried out (BioRad IQTM5 Multicolor real-time fluorescence quantification System, USA).circROBO1 primer and its control primer were used for qRT-PCR experiment, in which U6 was the internal reference and the relative expression was calculated according to 2-ΔΔCT,ΔΔCT=(CTcircRNA-CTU6) target-(CTcircRNA-CTU6)control.The total reaction system was 20 μL, and each group of experiments was set up with 3 compound pores.
2.5 Scratch experiment
Use marker pen on the back of the 24-hole plate, draw a line in the center with a straight ruler, and cross the hole (all instruments have been sterilized or ultraviolet sterilized).1mL cell suspension of 5×105logarithmic growth phase was added to the 24-well plate, and the cells were cultured overnight.The next day, a 200 μL gun tip was used to mark the cells perpendicular to the transverse line of the back with the ruler,and the cells were cleaned with PBS for 3 times to remove the scratched cells.Serum-free medium was added.Put in a 5%CO2incubator at 37 ℃ and culture.24 h later,the sample was taken and photographed under the mirror.
2.6 Transwell experiment
Y79 transfected 24 h later was configured into 3×105/mL singlecell suspension(see 1.3 for transfection method ).The upper layer of Transwell was coated with matrixglue and left for 6 h for film formation (this step was required for Transwell invasion experiment and not required for Transwell migration experiment).150 μL single-cell suspension was inoculated into the upper compartment layer, and 350 μL medium containing 20% fetal bovine serum was added to the lower compartment layer,incubated in an incubator containing 5% CO2 at 37 ℃ for 24 h, and crystal indigo staining was performed.The upper compartment cells were removed, cleaned and dried,observed and counted under the microscope.
2.7 RNase R enzyme digestion experiment
After the total RNA of Y79 cells was extracted, the RNA was divided into two roups according to whether Rnase R was added:Rnase R group (digested with Rnase ) and control group (digested without Rnase R).Then, in a PCR instrument, the enzyme was reactivated at 37 ℃ for 30 min and then at 70 ℃ for 10 min, and then reverse transcription was performed.qRT-PCR detection was performed using primers designed according to circROBO1 ring junction and linear parent gene ROBO1.The expression levels of linear parent gene ROBO1 and circROBO1 in Y79 cells were compared to determine the structural stability of circROBO1 in Y79 cells.
2.8 circROBO1 subcellular localization experiment
According to the instructions of the nuclear/cytoplasmic cell component extraction kit provided by the reagent supplier,cytoplasmic and nuclear RNA were extracted from Y79 cells, and the relative expression levels of circROBO1, cytoplasmic control and cytoplasmic control 18S were detected by qRT-PCR, respectively.According to the proportion of circrobo1 in the two cells, the statistical graph was made.
2.9 Double luciferase reporter gene experiment
104 Y79 cells were inoculated into each 6-well plate and cultured in a 5%CO2incubator at 37 degrees for 24 h.According to the instructions of Lipo 3 000, circROBO1 wild type and mutant type, or KLF5 mRNA wild type and mutant type, were co-transfected with miR-885-5p or miR-NC, and subsequent operations were performed according to the instructions of the kit after transfection[13].To verify the binding relationship between circROBO1 and KLF5 and miR-885-5p, respectively.
2.10 Western blot analysis
The experiments were divided into si-circROBO1 group (same as 1.2), si-control group (same as 1.2) and blank group.Total cell protein was isolated from Y79 cells using RIPA lysate and PMSF,and then separated by SDS-PAGE gel.The transmembrane condition of the protein was 300 mA 2 h.Finally, PVDF membranes were transferred to a refrigerator at 4 ℃, treated with KLF5 and β-actin antibodies overnight (concentration 1:1 000), incubated with specific secondary antibodies at room temperature the next day, and Image J was used for analysis of results after development.
2.11 Data analysis
SPSS 20.0 was used to process the data, the results were recorded as “mean ±standard deviation”, and the T-test was used for intergroup analysis.P<0.05, which was statistically significant.
3.Results
3.1 Identification of structural stability of circROBO1 in retinal Y79 cells
The results of qRT-PCR after digestion by RNase R showed that the relative expression level of circROBO1 in circular RNA was not significantly changed after treatment with RNase R compared with the control group.However, the relative expression level of linear ROBO1 was significantly decreased after digestion by RNase R(t=16.18,P<0.05,FIG.1)
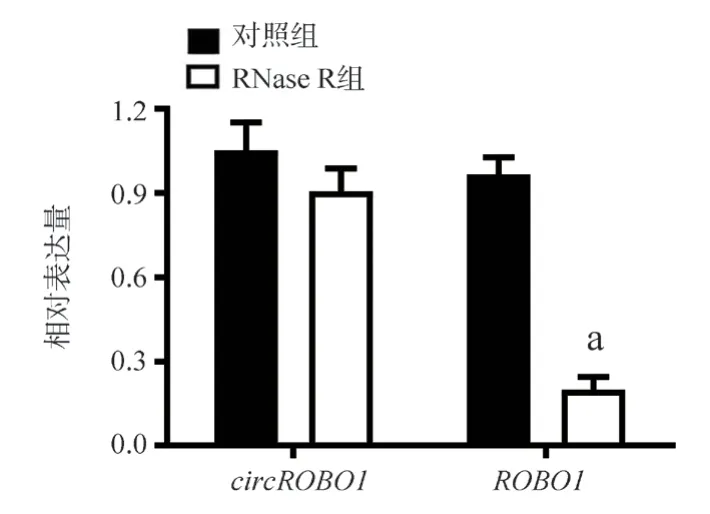
Fig 1 The relative expression levels of circROBO1 and linear ROBO1 after digestion by RNase R
3.2 Localization of circROBO1 in retinal Y79 cells
RNA from Y79 cytoplasm and nuclear were extracted to analyze the location of circROBO1 in cytoplasm and nucleus.qPCR results showed that according to the ratio of circROBO1 in cytoplasm to nucleus, the content of Circrobo1 in cytoplasm was higher than that in nucleus (t=13.04, P < 0.05, FIG.2).That is, the subcellular localization of circROBO1 was mainly in the cytoplasm (Figure 2),while β-actin was mainly located in the cytoplasm and 18S was mainly located in the nucleus.

Fig 2 Subcellular localization of circROBO1 in retinal Y79 cells
3.3 Effect of circROBO1 on the migration ability of retinal Y79 cells
Scratch test results showed that: compared with the si-control group, the mobility of retinal Y79 cells in the si-circROBO1 group was significantly reduced (t=22.54, P<0.05, FIG.3A, B), that is,after exogenous silencing of circROBO1, the migration ability of retinal Y79 cells was significantly reduced.

Fig 3 Effect of circROBO1 silencing on the migration ability of retinal Y79 cells
3.4 Effect of circROBO1 on invasion ability of retinal Y79 cells
Transwell results showed that compared with the si-control group,the invasion and migration of retinal Y79 cells in the si-circROBO1 group were significantly decreased (t侵袭=11.65, t迁移=12.06, P<0.05,FIG.4A, B).In other words, exogenous silencing of circROBO1 significantly reduced the invasion ability of retinal Y79 cells.

Fig 4 Effect of circROBO1 silencing on Transwell invasion and migration of retinal Y79 cells
3.5 Prediction and verification of binding sites of circROBO1 and miR-885-5p
According to the prediction by the bioinformatics software CircInteractome, the results showed that there might be one miR-885-5p binding site in the circROBO1 sequence.The results of double luciferase reporter gene experiment showed that when cotransfected with wild type circROBO1, compared with miR-NC,the luciferase activity in miR-885-5p co-transfection group was significantly decreased (0.42±0.05) compared with (1.00±0.02)(t=11.39, P<0.05, FIG.5B).When co-transfected with mutant circROBO1, compared with miR-NC, the activity of miR-885-5p co- transfection group (0.96±0.06) had no significant difference compared with (1.09±0.12) (t=1.112,P > 0.05, FIG.5B).In other words, circROBO1 can interact with miR-885-5p.
3.6 Prediction and validation of downstream target genes of miR-885-5p
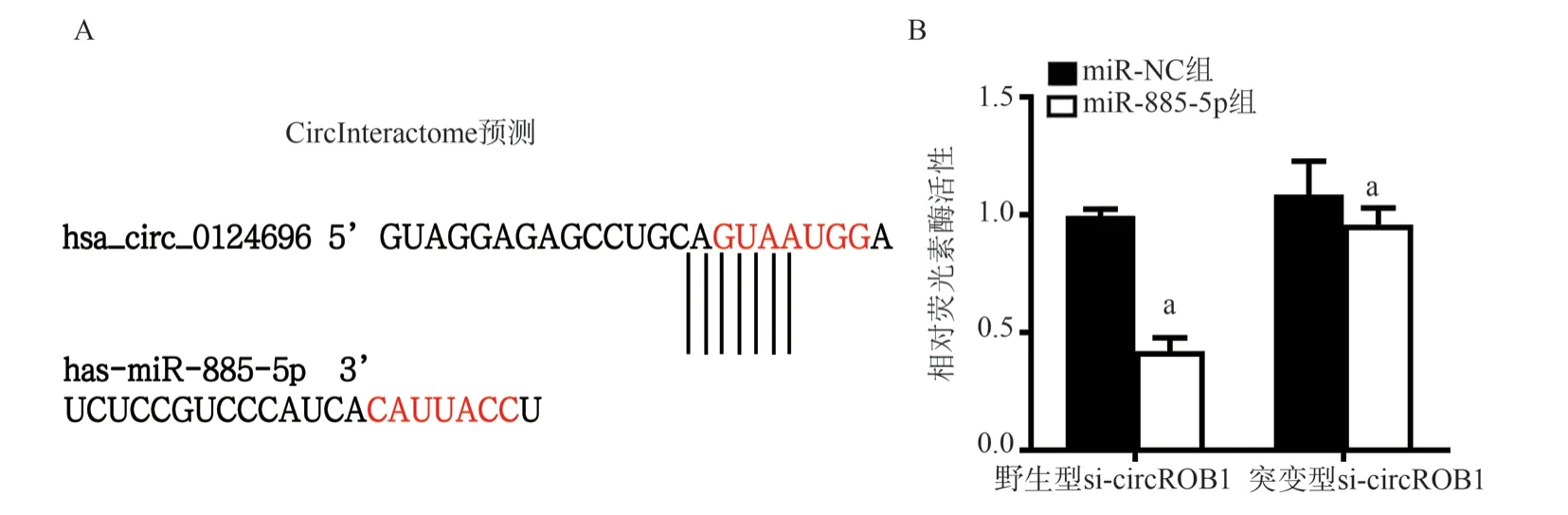
Fig 5 Prediction and verification of binding sites between circROBO1 and miR-885-5p
According to the prediction of TargetScan software, the results showed that there may be a binding site for KLF5 mRNA in the miR-885-5p sequence (Figure 6A).The results of double luciferase reporter gene experiment showed: Compared with the control group “miR-NC+KLF5 wild-type” co-transfection group,the cytoluciferase activity of “miR-885-5p+KLF5 wild-type” cotransfection group was significantly decreased (0.45±0.03) compared with (0.98±0.02) (t=17.26, P<0.05, FIG.6B).Compared with the control “miR-NC+KLF5 mutant” co-transfection group, the activity of “miR-885-5p+KLF5 mutant” co-transfection group (0.97±0.05)had no significant difference compared with (1.02±0.04) (t=1.684,P > 0.05, FIG.6B).In other words, miR-885-5p can interact with KLF5.
3.7 circROBO1 promoted retinal Y79 cell tumor invasion through KLF5
Western blot assay after circROBO1 expression was silenced by siRNA in retinal Y79 cells showed that the protein expression of KLF5 in the si-circROBO1 transfection group was significantly decreased (FIG.7A, B).In other words, exogenous silencing of circROBO1 could significantly reduce KLF5 protein expression.
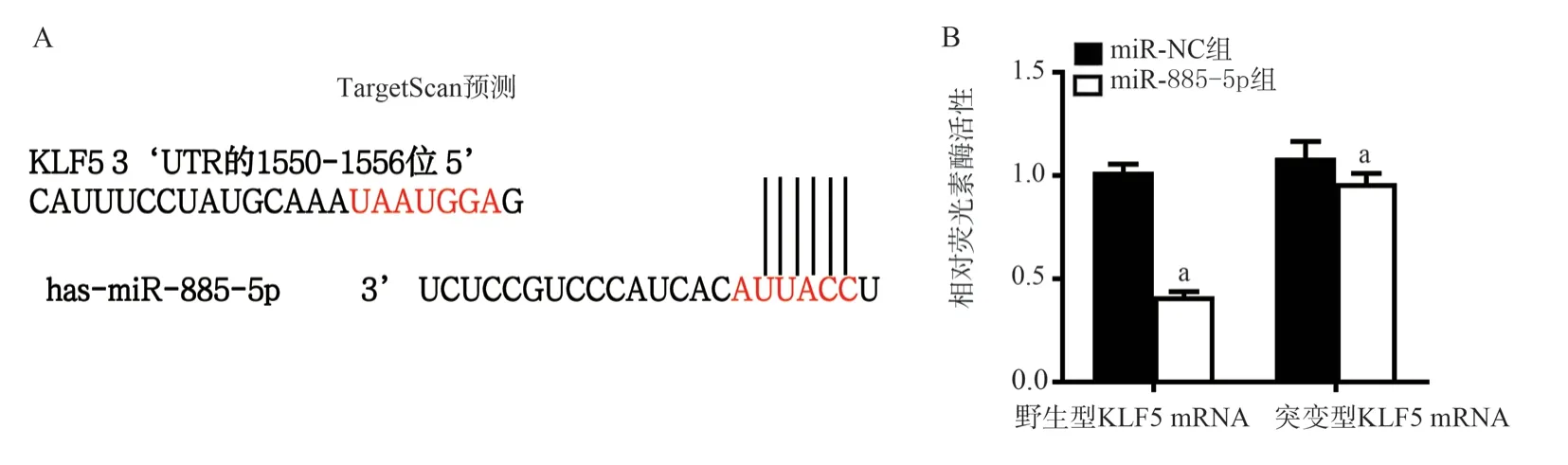
Fig 6 Prediction and verification of binding sites of miR-885-5p and KLF5
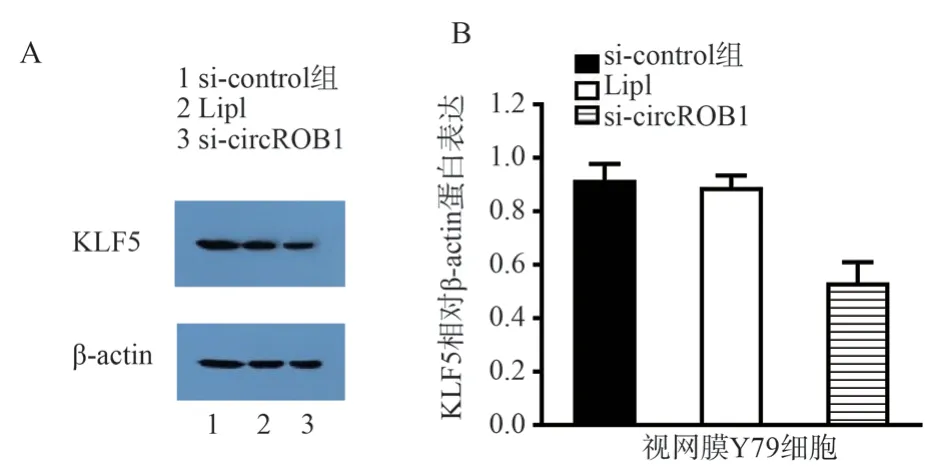
Fig 7 circROBO1 promotes retinal Y79 cell tumor invasion through KLF5
4.Discussion
circRNA is a newly discovered subclass of non-coding RNA, which is produced by reverse splicing mechanism with covalent closedloop structure.Circrna not only acts as a sponge for microRNA(miRNA) and proteins, but also regulates gene expression, carries out epigenetic modification, transforms into peptides, and generates pseudogenes[10,11].Dysregulation of circRNA expression can lead to the occurrence and rogression of a variety of human diseases,including malignant tumors and eye diseases[12].In recent years,more and more studies have found that circRNA playsan important role in the pathogenesis of various eye diseases, indicating that circRNA has the potential to be used as a biomarker for the diagnosis, progression evaluation and prognosis of eye diseases.
Previous studies have shown that circ_0000527 is highly expressed in RB tissues and cells, circ_0000527 silencing can inhibit the proliferation, migration and invasion of RB cells and promote cell apoptosis, and circ_0000527 knockdown can inhibit tumor growth in RB cells.This process is mainly achieved through circ_0000527 sponging miR-98-5p, miR-98-5p targeting XIAP and mediating the malignant progression of RB cells[13].It has been reported that circMKLN1 is closely associated with a better prognosis in RB patients.As a circular RNA, circMKLN1 is resistant to RNase R digestion in vitro, and its enrichment mainly plays a role in cytoplasm.In addition, overexpression of circMKLN1 can inhibit proliferation, migration and invasion of RB cells.Overexpression of circMKLN1 can inhibit the growth of tumors in vivo, which is realized through the regulation of miR-425-5p and its target gene PDCD4 by circMKLN1[14].These studies suggest that circRNAs can indeed promote or inhibit the malignant progression of RB through sponge adsorption of miRNAs.
In this study, qRT-PCR after digestion by RNase R enzyme showed that circROBO1 was structurally stable in RB cell Y79.This result is consistent with the results confirmed by previous studies that“circrobo1 is structurally stable and resistant to RNA exonuclease due to its ring structure”[15].Enrichment analysis of circROBO1 in the nucleus and cytoplasm showed that circROBO1 was mainly located in the cytoplasm, and silencing Circrobo1 significantly inhibited the migration and invasion ability of RB cells.Further western blot results showed that silencing circROBO1 significantly inhibited KLF5 protein expression.In breast cancer, cytoplasmic circROBO1 has been shown to promote tumor liver metastasis[8]via the miR-217-5p/KLF5 axis.These results are consistent with“circRNA, which is widely distributed in the cytoplasm, can act as a miRNA sponge to regulate the expression of downstream key genes”[16].
Kruppel-like factor 5 (KLFs) is a member of the KLF family of transcription factors and is involved in a variety of biological processes and different types of diseases, especially cancer.KLF5 regulates the expression of multiple target genes, such as Cyclin D1, Nanog and Slug[17-20].Studies have shown that KLF5 can be involved in a variety of biological functions, such as cell dryness, proliferation, apoptosis, autophagy and migration[21-24].Moreover, KLF5 can promote the metastasis and progression of a variety of malignant tumors.For example, BAP1 antagonizes the ubiquitination of WWP1-mediated transcription factor KLF5 and inhibits autophagy, thus promoting the malignant progression of melanoma[25].KLF5 deubiquitination can promote the proliferation and metastasis of breast cancer cells[26].Long non-coding RNA LncRNA PVT1 can regulate the malignant progression of triplenegative breast cancer through KLF5/β-catenin signaling pathway[27].As a transcription factor, KLF5 is overexpressed in breast and colorectal cancer and is a potential biomarker for poor prognosis[28-30].Studies have shown that circROBO1 can adsorb downstream miR-217-5p through bioinformatics prediction and dual luciferase reporter gene.KLF5 is the direct target gene of miR-217-5p, and miR-217-5p can directly regulate KLF5 expression.The positive feedback loop established by the circROBO1/KLF5/FUS axis can promote liver metastasis of breast cancer[8].In this study, we found that circROBO1 silencing could significantly inhibit KLF5 protein expression in retinal Y79 cells.Through prediction and luciferase reporter gene verification, we found binding sites between circROBO1 and miR-885-5p, and between miR-885-5 and KLF5.The results of this study are similar to those in breast cancer,suggesting that circROBO1 in RB also promotes the invasion of retinal Y79 cell tumors by targeting KLF5, which is realized through the interaction between circROBO1 and miR-885-5p.At present,the exploration of this mechanism is not perfect, and we will include more RB cell lines for verification in the future, and conduct RIP and other experiments for further verification.
This study preliminarily confirmed that circROBO1 can promote the invasion of retinal Y79 cell tumor by upregating KLF5, and this result can provide a new idea for the development of clinical RB targeting drugs.
Statement of conflict of interest for all authors
Liu Yuefeng, Zhang Zhiwei, Li Xin and Luo Weimin declare that there is no conflict of interest related to this article
Statement of contribution for all authors
Molecular Biology Experiments and data Processing: Yuefeng Liu; Data Processing and Mapping: Zhang Zhiwei; Administrative support: Li Xin, research ideas and article review: Luo Weimin.
杂志排行
Journal of Hainan Medical College的其它文章
- Cigarette induced release of exo-miR-34a from 16HBE vesicles targeting CASP2 promoted the proliferation of COPD MRC-5 cell
- To investigate the effect of Shenqi Tiaoshen Formula on CSE induced inflammatory response of MH-S cells based on TLR4/NF-kB/NLRP3 pathway
- Effect of estrogen pretreatment in GnRH antagonist protocol-Metaanalysis
- Effect of Astragalus-hawthorn on ovarian reproductive function and inflammatory mechanism of action in rats with polycystic ovary syndrome
- Inhibitory effect of thymoquinone on neuroinflammation in Parkinson's disease model by regulating NLRP3 inflammatory bodies
- Abnormal expression and significance of circ-CBLB/miR-486-5p in patients with rheumatoid arthritis of spleen deficiency and dampness excess type
