Biosensors for waterborne virus detection: Challenges and strategies
2023-12-14XixiSongZinaFredjYuqiaoZhengHongyongZhangGuoguangRongSuminBianMohamadSawan
Xixi Song,Zina Fredj,Yuqiao Zheng,Hongyong Zhang,Guoguang Rong,Sumin Bian ,Mohamad Sawan
CenBRAIN Neurotech, School of Engineering, Westlake University, Hangzhou, 310030, China
Keywords:Waterborne viruses Biosensors Optical Electrochemical Human samples Wastewater
ABSTRACT Waterborne viruses that can be harmful to human health pose significant challenges globally,affecting health care systems and the economy.Identifying these waterborne pathogens is essential for preventing diseases and protecting public health.However,handling complex samples such as human and wastewater can be challenging due to their dynamic and complex composition and the ultralow concentration of target analytes.This review presents a comprehensive overview of the latest breakthroughs in waterborne virus biosensors.It begins by highlighting several promising strategies that enhance the sensing performance of optical and electrochemical biosensors in human samples.These strategies include optimizing bioreceptor selection,transduction elements,signal amplification,and integrated sensing systems.Furthermore,the insights gained from biosensing waterborne viruses in human samples are applied to improve biosensing in wastewater,with a particular focus on sampling and sample pretreatment due to the dispersion characteristics of waterborne viruses in wastewater.This review suggests that implementing a comprehensive system that integrates the entire waterborne virus detection process with high-accuracy analysis could enhance virus monitoring.These findings provide valuable insights for improving the effectiveness of waterborne virus detection,which could have significant implications for public health and environmental management.
1.Introduction
Waterborne viral infections,such as diarrhea,encephalitis,hepatitis,and even cancer,remain a major cause of mortality worldwide [1,2].These viruses are primarily transmitted through the fecal-oral route,posing significant challenges for preventing waterborne diseases[3].They greatly threaten human health due to their discrete distribution,lasting activity,strong tolerance to chemical disinfectants,and low infection dose [4].Continuous monitoring and effective detection methods to ensure the safety of water resources and protect public health are essential,given the ability of some infectious viruses to survive in wastewater for extended periods [5].According to World Health Organization(WHO)data,improving water quality can reduce the global burden of diseases by approximately 4% [2].Detection and analysis of viruses introduced into wastewater via human excreta can provide dynamic public health information by detecting the presence of infectious diseases early on while assessing the spatial distribution and trends in the occurrence of viruses [6].Monitoring activities typically concentrate on identifying potential human exposure points in the water cycle,including wastewater from residential areas,public facilities in urban areas,surface runoff,and treated water after large-scale biological events.Thus,systematic monitoring and detection protocols facilitate selecting appropriate treatment processes and crisis response strategies.
Despite the importance of monitoring waterborne viruses,several challenges remain.First,virus tracking methods based solely on population symptoms are limited since symptoms caused by waterborne viruses are not specific;second,due to the diversity of viruses and the complexity of human activities,it is often difficult to identify the transmission chain of waterborne viruses;and third,there is a lack of institutions providing comprehensive surveillance data on virus occurrence,concentration,and removal efficiency[7].Therefore,developing advanced monitoring tools that effectively detect waterborne viruses is critical for managing and predicting the rapid spread of these viruses (Fig.1A).
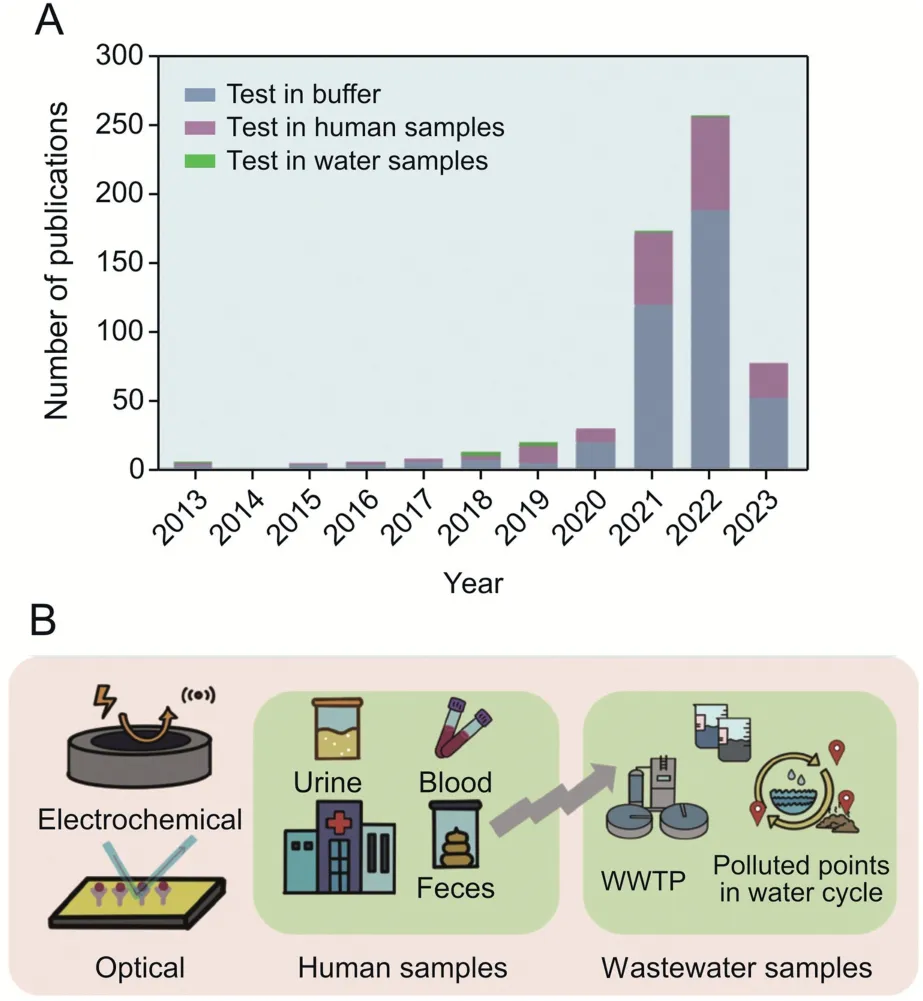
Fig.1.Trends and evolution in biosensors for waterborne virus detection.(A) Publications regarding biosensors for waterborne virus detection in the past two decades(Web of Science).(B) Developed biosensors (electrochemical,optical) applied in human samples and wastewater samples.The insights gained from the biosensing waterborne viruses in human samples can be adapted to improve biosensing in wastewater.WWTP: wastewater treatment plant.
Traditional methods such as nucleic acid amplification techniques and immunology-based methods such as real-time quantitative polymerase chain reaction (RT-qPCR) and enzyme-linked immunosorbent assay (ELISA)can be time-consuming and require well-trained personnel and sophisticated equipment,preventing their broad implementation [8].In regard to low-and middleincome countries (LMICs) with limited resources,the need for accessible and affordable virus detection methods is becoming even more urgent[9].Biosensors are powerful analytical tools with relatively low cost,fast,and even near real-time response compared to the aforementioned conventional analytical methods[10].A typical biosensor identifies analytes in samples through specific biological receptors (nucleic acids,antibodies,proteins,molecularly imprinted polymer (MIP),cells,microorganisms,etc.).It evaluates readable signals (electrical,optical,thermal,mass,frequency changes,etc.) based on the transducer [11].Several recent studies have been carried out to investigate biosensors for detecting different markers in wastewater,such as heavy metals,organic materials,drugs,and microorganisms [12,13].The developed approaches can be classified into two main categories:optical and electrochemical according to the input and response signals[11] or even a stand-alone device (lab-on-chip/fiber platforms) to pretreat samples,capture pathogens,and output signals [14].Complex wastewater matrices can be addressed by optimizing fabrication parameters to enhance the performance of biosensors,typically by selecting biological elements that interact efficiently with the analytical target while ensuring fast and reliable output[15,16].Overall,these diagnostic assays can be versatile and adaptable,making them valuable tools for detecting various viral targets in different contexts.
In fact,the development of biosensing for wastewater samples has lagged the urgent demand for diagnostic tools in health care settings due to increased detection challenges and scarce funding in LMICs.Moreover,water samples,particularly those sourced from natural water bodies or wastewater,often lack standardized protocols and regulatory guidelines for effective biosensor applications[6,17,18].
Several reviews have previously summarized the state-of-theart biosensors for preventing waterborne pathogens to date.For example,Kotsiri et al.[11]emphasized the detection of bacteria in water and food matrices,while Kadadou et al.[19] demonstrated the screening of bacteria and viruses in clinical and water samples.Pilevar et al.[8]focused on detecting multiple viruses in water and wastewater,and Bhardwaj et al.[20] described the approaches to developing nanomaterials-assisted optical biosensors for waterborne pathogens.However,there remains a significant gap in providing a comprehensive overview of the latest breakthroughs in waterborne virus biosensors,encompassing both clinical and wastewater samples present in real-life settings.
In light of this gap,this review aims to not only provide a comprehensive overview but also summarize various promising strategies to enhance the sensing performance of optical and electrochemical biosensors when used with human samples.Furthermore,we leverage the insights gained from biosensing waterborne viruses in human samples to specially improve the biosensing performance in wastewater.Lastly,we propose future perspectives in this direction to further advance the field(Fig.1B).
2.Waterborne viruses and their detection barriers in humans and wastewater
2.1.Typical waterborne viruses and their threat to the public
The WHO highlights the nine most common species of enteric viruses with evidence of transmission in drinking water:adenovirus(AdV),astrovirus (AV),norovirus (NoV),sapovirus (SV),hepatitis E virus (HEV),enterovirus (EV),pararechovirus,hepatitis A virus(HAV),and rotavirus(RV)[2].NoV and RV infection is often associated with exposure to contaminated water or shellfish and is considered the most significant cause of diarrhea deaths,especially in developing countries [7,21].Both HAVs and HEVs cause viral hepatitis epidemics in developing countries,resulting in serious health problems [22].Research has shown that enteric viruses pollute surface water and groundwater to varying degrees [23].Predictably,incomplete disinfection of focally contaminated water is a potential source of related infectious disease outbreaks.Humans can be infected by ingesting or contacting polluted domestic sewage,groundwater,or surface water.For instance,it was reported that human AdVs have high resistance to different tertiary and advanced treatments of wastewater treatment plant(WWTP)[24].
In recent decades,multiple viral pandemics have raised concern about enveloped viruses,although the waterborne transmission route for these viruses has not been definitively proven.Evidence suggests that not all enveloped viruses lose their infectivity quickly.For example,infectious severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) and avian influenza virus particles have been detected in feces[5,25].It has also been suggested that SARSCoV-2 may be transmitted via water in the community [26].Additionally,water supplies may eventually be contaminated with SARS-CoV-2 through urine,sputum,vomit,or blood [27].
2.2.Direct detection of waterborne viruses in human and wastewater samples: challenges and barriers
Multiple factors should be further evaluated and controlled before proceeding with large-scale waterborne virus biosensing.First,it is essential to note that viral detection is more complicated than bacterial detection.Unlike bacteria,it is challenging to cultivate large numbers of waterborne viruses in vitro,making their analysis more complex[28].Concurrently,the generic materials of some waterborne viruses,such as AV,NoV,SV,EV,HAV,HEV,and RV,are RNA that is more dynamic and variable than DNA,which poses a significant challenge to molecular detection techniques[4,8].Moreover,low-copy viruses hinder detection activities,especially in wastewater samples.To accurately detect waterborne viruses(e.g.,AdV,HAV,and RV),varying from 1 to 100,000 genome copies per liter in concentration,it is necessary to collect tens or even thousands of liters of water samples and concentrate them prior to measurement [29].
Complex matrices can significantly impact the efficiency of waterborne virus detection in several ways.For instance,human samples,such as blood and urine,contain a diverse range of cells,proteins,and biomolecules that may hinder virus binding to biosensor surfaces or antibodies,leading to decreased sensitivity and specificity.Fecal samples may contain multiple microorganisms,metabolites,and dietary components that can interfere with biosensor performance.Similarly,wastewater is often contaminated with organic substances,humic acids,heavy metals,and microorganisms due to natural or anthropogenic causes [30,31].Also,untreated wastewater is typically characterized by high levels of turbidity,suspended solids,and organic matter concentrations,which can cause fouling of interfering components on sensor surfaces[24],resulting in reduced sensitivity and prolonged response time.Additionally,samples mixed with sludge can be challenging to handle as the flocs in the sludge can adsorb a considerable amount of virus,making it difficult to detect[32].
3.Current biosensors for waterborne virus detection in human samples
Biosensors are typically designed to detect waterborne viruses in feces,urine,serum,and saliva,providing important reference information for detecting excreta-contaminated water.
3.1.Recognition element: classification
The recognition element,alternatively called a bioreceptor,is responsible for the specialized recognition of the target analyte.Choosing a bioreceptor is critical to biosensor development,as it determines the sensor's specificity,sensitivity,and stability [33].Antibodies have been widely used as bioreceptors due to their high specificity toward target analytes.However,antibodies require stringent environmental conditions to maintain their stability and functionality,which can limit their practical applications in biosensors [34].This brings the concern of biosensor stability under transportation,storage,and operation conditions.To address this limitation,peptides,aptamers,DNA probes,and MIPs have been alternatively explored.These bioreceptors offer several advantages over antibodies,such as improved stability,lower cost,and ease of production,as explained in detail in Table 1 [34-36].
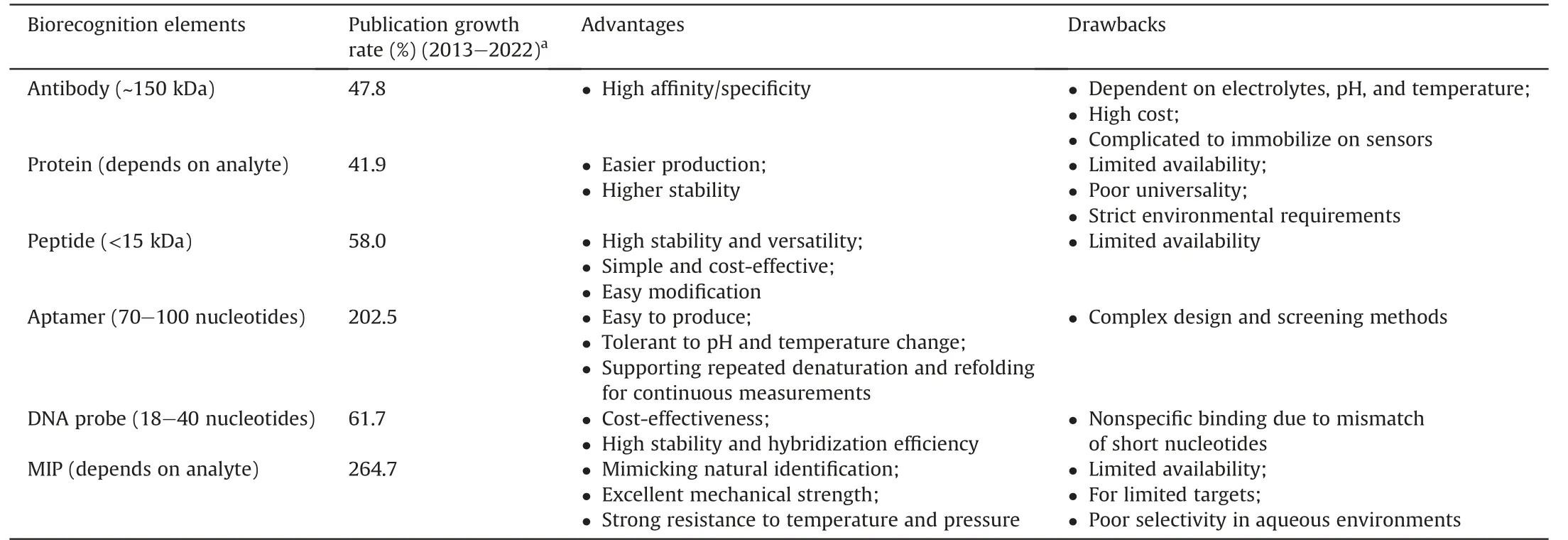
Table 1 Emerging biorecognition elements and their features [34-36].
Specific peptides,composed of amino acid polymers,possess structural units identical to those of proteins and can be further modified to maintain high stability and affinity [37].In this framework,Baek et al.[38]introduced an anti-fouling peptide and flexible linkers into the NoV-specific peptide,which improved its nonfouling properties and binding interaction with NoVs.When detecting NoVs from human excreta using gold electrodes as biotransformers,the optimal modified peptide exhibited a lower limit of detection (LOD) of 1.78 copies/mL.Designing novel peptide receptors with high affinity is still challenging due to the limited knowledge of molecular recognition,resulting in limited peptide options for viral recognition [39].
Aptamers are degradation-resistant single-stranded DNA(ssDNA)and RNA molecules that mimic monoclonal antibodies.Precise design and chemical modifications can regulate their binding properties.Recently,researchers have focused on developing aptamers that target NoV/enterovirus type 71 (EV71) for detection and diagnostic purposes[40-43].Researchers have also reported that pairing an aptamer with other recognition factors can lead to more sensitive target binding and stronger biosensing signals[40,42].
In DNA probe-based biosensors,a specific complementary DNA sequence in the sample can excite an electrical or optical signal.This method can support on-site pathogen detection without requiring professional operators.Nevertheless,it is necessary to overcome the interference of complex target genomes during the design phase and the mismatch problem of short nucleotide chains as recognition molecules.To date,DNA sensors have been reported to detect HAV,HEV [44,45],and NoV [46] with the use of nanomaterials such as silver(Ag)/gold(Au)/magnetic nanoparticles or quantum dots(QDs).
MIPs are also biomimetic bioreceptors made from synthetic materials with a shape and functionality complementary to those of the target molecule,allowing for selective capture from a complex mixture [47].Compared to antibodies,MIPs are more stable,costeffective,and easier to produce in large quantities.They also have the potential for higher batch-to-batch consistency to ensure assay reproducibility[48].However,the impact of larger and more flexible three-dimensional targets (such as viruses) on the imprinting ofMIPs needs to be considered due to technical limitations[49].
3.2.Recognition element: surface immobilization
The immobilization strategy of biorecognition elements is a critical factor in ensuring the stability of sensors and maintaining the integrity of their binding sites.Various immobilization methods are available,including physical adsorption,covalent binding,affinity binding,and encapsulation,among others.The choice of immobilization method depends on several factors,such as the nature of the biorecognition element,the type of assay or detection system,the surface chemistry of the solid support,and the desired application.Immobilization strategies include random or oriented forms,and maximum functionality can be obtained by enhancing the capacity of biosensors or the orientation of antibody binding sites.Random immobilization is generally conducive to higher surface coverage of bioreceptors,while oriented immobilization provides better sensitivity for analyte detection [50].A discussion on random and oriented immobilization strategies has been reported in previous studies [51,52] and is illustrated in Fig.2.To produce uniform bioreceptor adhesion,it is essential to emphasize the need for oriented immobilization of functional groups on nonfunctional sites of bioreceptors to overcome the allosteric effect of biomolecules.
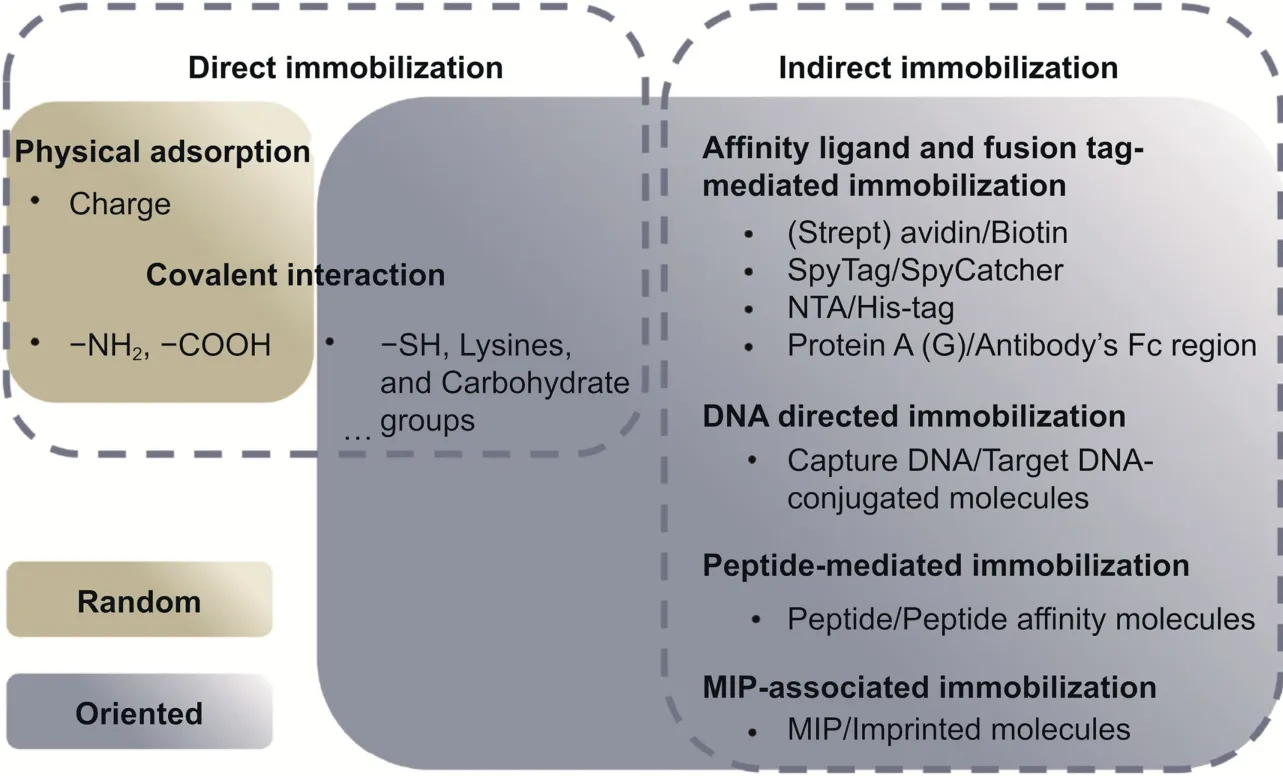
Fig.2.Simplified representation of currently available immobilization strategies.NTA: nitrilotriacetic acid;MIP: molecularly imprinted polymer.
Site-directed immobilization is always ideal but challenging,as it requires specific functional groups or chemical modifications for effective immobilization.Peptide-based biosensors are typically modified with the thiol group of cysteine to enable their immobilization onto the gold surface.Spacers are frequently introduced between the thiol group and the peptide chain to make the peptide probes more flexible and accessible[53].It is easy to synthesize and modify aptamer/DNA probes that incorporate nucleic acid structures to carry thiol or biotin groups,which help control the direction of immobilized aptamers[54].
3.3.Electrochemical biosensors for waterborne virus detection in human samples
Electrochemical biosensors have become increasingly active in detecting waterborne viruses due to their high sensitivity and fast response.The working electrode(WE),serving as the transduction element,is responsible for recognizing the analyte of interest.Gold electrodes are commonly used owing to their cost-effectiveness,high conductivity and compatibility [38,55].Other types,such as indium tin oxide (ITO) electrodes [56] and carbon electrodes[57-59],share similar advantages.Common electrochemical techniques include electrochemical impedance spectroscopy (EIS),differential pulse voltammetry(DPV),cyclic voltammetry(CV),and square wave voltammetry(SWV)[60].Various electrode materials and nanomaterials can be integrated into electrode manufacturing in different forms to enhance the transduction capacity.
3.3.1.Electrode modification:nanomaterials
Electrode modification is a well-established method for enhancing sensing performance,which involves surface modification and structure optimization.For example,gold nanoparticles(AuNPs) can improve electrode sensing and the binding of biomolecules to the surface due to their high surface energy [61].A multiplexed simultaneous detection system for five types of hepatitis viruses (HAV-HAE) based on this pattern was fabricated by Tang et al.[62].In addition,graphene has favorable chemical and physical properties,such as a high surface area,strong mechanical strength,fast electron transport,good thermal performance,and optical transparency [63-65].Field-effect transistor (FET)-based graphene as the active region has achieved highly responsive detection of RV [66] and SARS-CoV-2 [64].
Multiple nanomaterial modifications can be even more exciting.Chand et al.[41] introduced graphene-gold nanocomposites as a conductive film on the surface of a screen-printed carbon electrode(SPCE).Through the high-affinity effect of biotin and streptavidin,ferrocene redox probe-labeled aptamers were site-directed immobilized on electrochemical substrates to capture the target NoVs.A microfluidic chip based on polydimethylsiloxane (PDMS)was used to integrate a graphene-gold nanocomposite-modified electrochemical sensing structure with a microfiltration zone.This filtration zone was utilized to filter and enrich NoV-infected clinical samples before subsequent sensitive detection.The entire test process requires only 35 min with an LOD of approximately 100 pM(Fig.3A) [41].
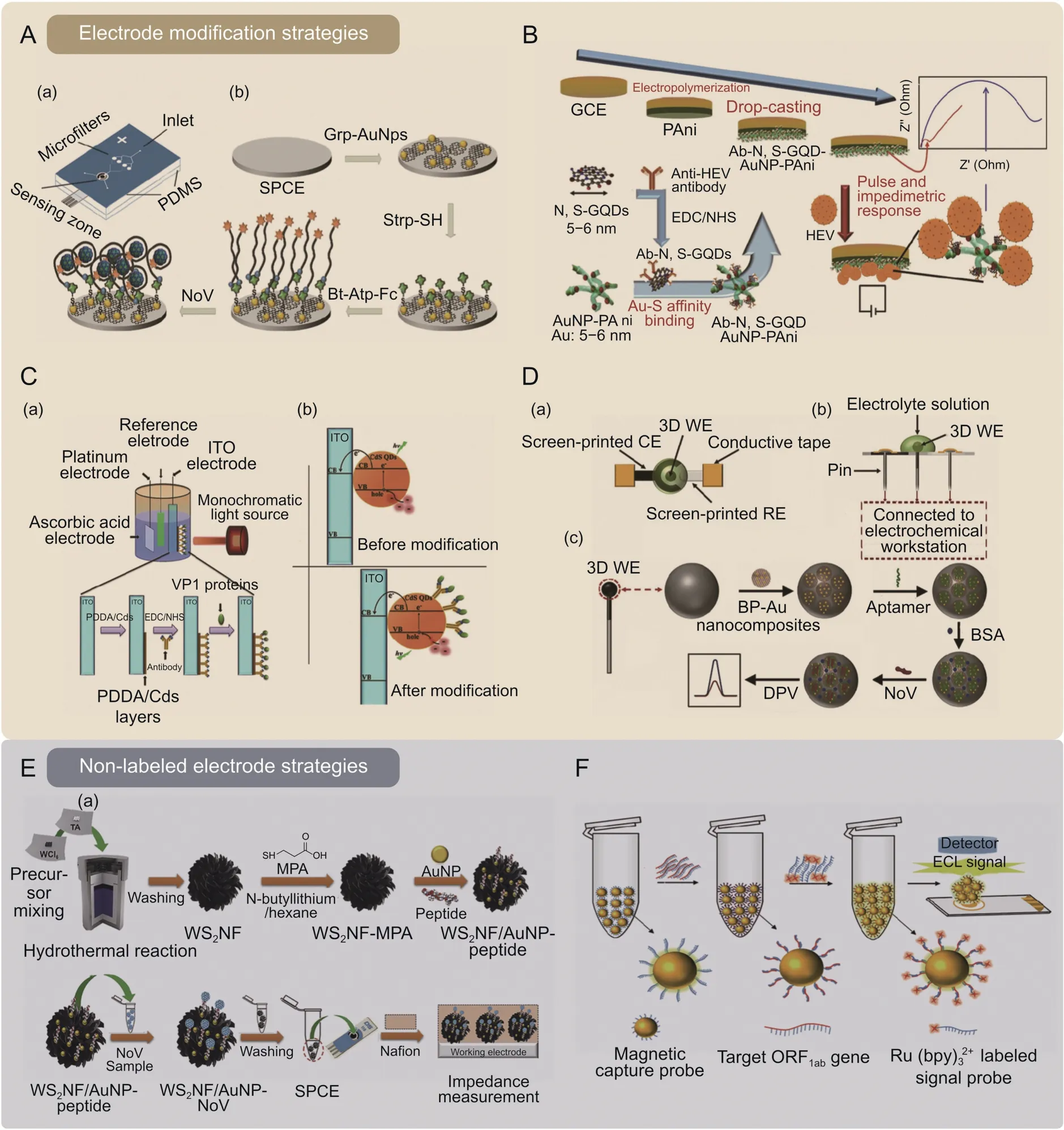
Fig.3.Schematic of the device fabrication and target detection in electrochemical biosensors.(A)A microfluidic electrochemical aptasensor for norovirus(NoV)detection in clinical samples.(a) Graphene-gold nanoparticles (Grp-AuNPs) modified electrochemical sensing zone integrated with a microfiltration zone on a polydimethylsiloxane (PDMS) microfluidic chip;(b) process of electrode functionalization and NoV aptasensing[41].(B) An impedimetric biosensor for hepatitis E virus (HEV) gene detection in human serum,using specific anti-HEV antibody-conjugated to nitrogen-and sulfur-codoped graphene quantum dots (Ab-N,S-GQDs) and gold-embedded polyaniline nanowires (AuNP-PAni) nanocomposite to modify the electrode [57].(C) A photoelectrochemical (PEC) biosensor for NoV detection.(a) The design and mechanism of CdS quantum dots (Cds QDs) combined indium tin oxide (ITO) electrode;(b) The energy transformation before and after Cds QDs modification [56].(D) A three-dimensional (3D) electrochemical aptasensor for NoV detection.(a-b)Top-view and side-view of the NoV aptasensor;(c)Workflow diagram for electrode functionalization and NoV aptasensing[68].(E)Detecting NoVs using peptides functionalized AuNPs decorated tungsten disulfide nanoflower (WS2NF) on electrochemical biosensor.(a) The preparation of WS2NF/AuNP-peptide;(b) the workflow diagram for NoV sensing [58].(F) Monitoring open reading frame 1 ab (ORF1ab) gene through dual-probe hybridization mediated electrochemiluminescence (ECL) biosensor for SARS-CoV-2[71].Strp-SH: thiolated streptavidin;Bt-Atp-Fc: biotin and ferrocene tagged aptamer;SPCE: screen-printed carbon electrode;GCE: glassy carbon electrode;PDDA: poly (diallyldimethylammonium chloride);EDC: 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide;NHS: N-Hydroxysuccinimide;VP1 protein: NoV GII.12 capsid protein VP1;BP-Au nanocomposite: phosphorene-gold nanocomposites;WE: working electrode;CE: counter electrode;RE: reference electrode;DPV: differential pulse voltammetry;BSA: bovine serum albumin;MPA: 3-mercaptopropionic acid.Reprinted from Refs.[41,56-58,68,71] with permission.
Recently,Alafeef et al.[67] fabricated a graphene-based paper platform that utilizes specific ssDNA probes attached to AuNPs and showed a high sensitivity for electrochemical sensing of the genetic material of SARS-COV-2.In addition,Chowdhury et al.[57] combined gold-embedded polyaniline nanowires (Au-PAni) and antibodies conjugated to nitrogen-and sulfur-codoped graphene quantum dots (N,S-GQDs) for HEV quantification.This biosensor performed well in detecting HEV in serum,cell culture supernatant,and monkey fecal specimen samples (Fig.3B) [57].
Furthermore,Guo et al.[56] developed a photoelectrochemical(PEC) biosensor system based on ITO electrodes and QDs.QDs enhanced the system's light absorption properties and electron transfer capabilities.For this approach,the photocurrent generated by the QD-coupled PEC system is reduced when the fixed antibodies on the ITO electrodes are bound to the virus targets,resulting in spatial steric hindrance.This reduction in photocurrent is used to detect the presence of NoVs of 46 copies/μL in inactivated samples within 30 min,suggesting its potential as a low-cost pointof-care (POC) tool(Fig.3C) [56].
Other studies focused on three-dimensional (3D) structures to increase the conductivity of sensing platforms and anchor sites for biorecognition molecules.Jiang et al.[68] proposed an electrochemical aptasensor with multiple phosphorene-gold nanocomposite-modified 3D WE layers.The novel composite amplifies the electron transfer process at the electrode interface,enhancing the electrochemical signal.This approach provided a durable supporting material for the adapters and holds promise for improving biosensor sensitivity,selectivity,and stability.The movable spherical WE has a larger surface area,facilitating NoV collection and avoiding cross-contamination (Fig.3D) [68].
3.3.2.Non-labeled electrode:free nanoprobes
Despite significant progress in electrode modification,several concerns remain: i) only limited materials are biocompatible;ii)evidence regarding the long-term stability of composite materials and processed sensors is lacking;and iii) technologies to fabricate such platforms are complex and expensive,such as microfabrication,lithography,and nanofabrication [69].Thus,electrochemical sensing without any electrode modification has garnered significant attention.
In the process of identifying the target with a free nanoprobe,the analyte-nanoprobe complex will be enriched on the WE through deposition or magnetic adsorption,which allows the complex to participate in electrical signal transmission.Baek et al.[58]proposed AuNPs decorated with tungsten disulfide nanoflowers (WS2NF/AuNPs) to provide more immobilization sites for recognition peptides and enhance electrochemical conductivity.It allows for easy capture of the insulation effect caused by NoV.The nanocomposite that completes the virus recognition reaction in solution was entrapped onto the SPCE.The performance of the biosensor was evaluated using EIS,which measures changes in electrical resistance in response to the presence of target viruses.The EIS signals indicated that the biosensor with a 3D flower-like shape structure has high specificity and sensitivity to oyster samples(Fig.3E)[58].
Moreover,modify-free electrodes have stronger expansibility.Examples include the addition of magnetic nanoparticles(MNPs)to facilitate the rapid separation and enrichment of target analytes from complex samples,thereby significantly improving the sensitivity of bioassays andeliminating matrix interference[44,70].Jianget al.[71]proved that magnetic capture probes introduced a dual-probe hybridization-mediated electrochemiluminescence(ECL)biosensor for SARS-CoV-2 sensing.Magnetic capture probes and Ru(bpy)32+-labeled signal probes underwent specific dual-probe hybridization with the open reading frame 1 ab(ORF1ab)gene of SARS-CoV-2.The ECL signal of the subsequently formed hybridization complex was measured through SPCE,whose intensity was positively correlated with the ORF1ab concentration.Such an ECL biosensor is highly sensitive,reproducible,and stable with high interference resistance,achieving an LOD of 0.1 fM in saliva and urine (Fig.3F) [71].Additionally,the modification-free system can achieve electrical signal amplification through various methods,such as sandwich detection[44,70],nucleic acid hybridization [45,72],and gene amplification[72,73],as further discussed in Section 3.5.
3.4.Optical biosensors for waterborne virus detection in human samples
Optical biosensors measure the optical response signal due to receptor-target binding,such as spectrum shift,spectrum intensity/pattern change,or liquid color difference.They can be designed onchip,on-fiber,on-paper,or in-tube with principles including surface plasmon resonance(SPR),localized surface plasmon resonance(LSPR),surface-enhanced Raman scattering (SERS),fluorescence,and colorimetric techniques.
3.4.1.Plasmonic sensing
Optical plasmonic biosensors use plasmonic active metals with high refractive index (RI) sensitivity for virus detection.Their versatile and flexible structures and dimensions suit various applications and facilities.SPR-based RI sensing is realized by introducing a plasmon-active metal(such as Au and Ag)layer on various dielectric substrates(such as glass or flexible polymers)[74,75].The sensitivity of SPR devices to environmental noise limits their ability to detect small targets,particularly at low concentrations in a complex matrix[20].Thus,it is necessary to improve the detection process and biorecognition [76],for example,by incorporating a filtering procedure[77]or a rinse step[78]into the viral detection scheme and using the sandwich principle for ultrasensitive detection[40].
Compared with SPR,LSPR is more sensitive to the RI changes caused by biomolecular binding events near the surface due to the localized nanoscale field around metal nanoparticles [75].Metal nanoparticles or nanopatterned plasmonic metasurfaces,which rely on locally enhanced electromagnetic fields,exhibit reduced evanescent tails and stronger light confining ability.This enhances the light-matter interaction in the vicinity of the sensor.In LSPR,metal structures such as gold nanocups[79]or nanoislands[80]can contribute a stronger electronic field.Combining a plasmonic metasurface with a dielectric microcavity brings a high-quality resonance mode [81,82].Qiu et al.[80] further combined the plasmonic photothermal (PPT) effect and LSPR sensing to detect SARS-CoV-2.The PPT/LSPR system comprises 2D gold nanoislands functionalized with DNA probes to target the RNA-dependent RNA polymerase (RdRp) gene of SARS-CoV-2.This system generates local PPT heat to transduce in situ hybridization under laser induction.This dual-functional biosensor reached an LOD of approximately 0.22 pM for the RdRp gene and a 96% recovery(consistent with RT-qPCR) in respiratory samples (Fig.4A) [80].
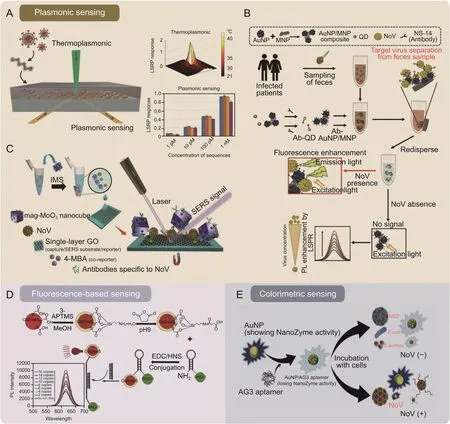
Fig.4.Typical optical biosensors for waterborne virus detection.(A) Schematic illustration of a dual-functional plasmonic photothermal (PPT) biosensor for the detection of the SARS-CoV-2 gene.The PPT effect assisted the hybridization of DNA probes to the SARS-CoV-2 gene of RNA-dependent RNA polymerase (RdRp),and the localized surface plasmon resonance(LSPR)effect acted as signal sensing[80].(B)An LSPR-based fluoroimmunoassay for the detection of noroviruses(NoVs).Antibody-modified AuNP/MNP nanocomposites simultaneously capture targets and excite fluorescent signals in fecal samples[85].(C)A sandwich surface-enhanced Raman scattering(SERS)immunoassay for NoV detection.This work showed a dual SERS nanotag/substrate platform formed by a graphene-mediated SERS capture substrate and plasmonic/magnetic molybdenum trioxide nanocubes (mag-MoO3 NCs) [88].(D) Quantum dot (QD)-molecular beacon nanobiosensor for NoVs [46].(E) Colorimetric sensing via a NoV nanozyme aptasensor [43].AuNP: gold nanoparticle;MNP: magnetic nanoparticle;Ab-QD: anti-NoV antibody conjugated quantum dot;Ab-AuNP/MNP: anti-NoV antibody conjugated AuNP/MNP nanocomposites;PL enhancement:photoluminescence enhancement;IMS: immunomagnetic separation;GO: graphene oxide;4-MBA: 4-Mercaptobenzoic acid;APTMS: (3-aminopropyl) trimethoxysilane;MeOH:methanol;EDC: 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide;NHS: N-Hydroxysuccinimide;AG3 aptamer:murine norovirus capsid-specific AG3 aptamer; E.coli: Escherichia coli;MS2: E.coli bacteriophage MS2; S.aureus: Staphylococcus aureus.Reprinted from Refs.[43,46,80,85,88] with permission.
Using analytes as triggers to alter the plasmon resonance behavior between nanoparticles also becomes a simple and highly promising biosensing modality.A frequently used couple is fluorescent QDs and AuNPs,where the LSPR energy transfer between these two types of nanoparticles leads to fluorescence enhancement or bursting of the QDs [83].The size,shape,and distance of the adjacent AuNPs modulate the fluorescence intensity [84].Takemura et al.[85] introduced MNPs to form hybrid nanocomposites with AuNPs and subsequently participate in the fluorescence interaction of QDs.The AuNP/MNP nanocomposites separate target viruses from impurities by a magnetic field,thus making the LSPR effect uninterrupted by pollutants,which is very beneficial for detecting complex matrices.Consistent with expectations,the assay system detected as low as 84 genomic copies/mL of NoVs in fecal samples (Fig.4B) [85].
SERS-activated platforms,which incorporate engineered plasmonic nanostructures,have also been developed to detect trace biological particles.However,interference in SERS detection of viruses in complex matrices arises from Raman signal impurities caused by numerous contaminants and biomolecules.It is necessary to capture the virus molecules before SERS detection in these systems to achieve a high signal-to-noise ratio.Given this,Yang et al.[86] proposed an oblique gold-nanoneedle array functionalized by specific protein catchers of angiotensin-converting enzyme-2 to capture SARS-CoV-2.Sandwich immunoassays utilizing performance-enhanced SERS substrates and SERS nanotags confirmed outstanding advantages over other SERS platforms[87,88].Achadu et al.[88] combined a graphene-mediated SERS capture substrate with plasmonic/magnetic molybdenum trioxide nanocubes(mag-MoO3NCs)to form a dual SERS nanotag/substrate platform.The antibody-conjugated mag-MoO3NCs separated the NoVs from human feces.NoVs-mag-MoO3NCs deposited on the capture substrate under magnetic influence for the SERS immunoassay,achieving a signal amplification of up to 109-fold and an LOD of 60 RNA copies/mL (Fig.4C) [88].
3.4.2.Fluorescence-based sensing
Fluorescence-based sensing strategies have been applied to various matrices,such as wastewater,soil,and serum.Typically,they do not require expensive,complex signal receivers and are ideal for POC devices.Fluorescent nanomaterials,such as QDs,are often functionalized with biorecognition elements for specific detection of pathogens [89].
Some fluorescence-based biosensors take advantage of the interaction of fluorescent tags and quenching agents in various subtle forms.For instance,a unique DNA probe named“molecular beacon” acts as a sensing “inducer” to which CdZnSeS QDs and quenchers(black hole quencher 1,BHQ-1)are attached at each end.The molecular beacon increases the fluorescence signal when it binds to the target viral RNA,spatially separating the CdZnSeS QDs from BHQ1.Adegoke et al.[46] screened CdZnSeS QD-conjugated molecular beacon bioprobes with different modifications and detected 8.2 copies/mL NoV RNA in serum (Fig.4D) [46].Alternatively,surfaces such as graphene oxide (GO) substrates behave as quenchers onto which fluorescent label-labeled antibodies/aptamers are adsorbed when in the unbound state.It has been reported that Chen et al.[90] used different colored QDs in GO solution to fabricate a dual-color immunosensor targeting human EV71 and coxsackievirus B3 (CVB3).
3.4.3.Colorimetric sensing
Colorimetric sensing strategies are often based on the visual detection of a color change that occurs in response to the interaction between the target molecule and a sensing material,which can be a variety of substances such as dyes,nanoparticles,or other molecular probes.AuNPs are one of the most frequently used.When the target molecule binds to the functionalized AuNP,it can cause a change in the size,shape,or aggregation state of the particles,which in turn affects their optical properties and leads to a detectable color change [91-93].The strategy mentioned above must avoid nonspecific aggregation in complex matrices,which can lead to nonspecificity and false-positives.
For other cases,converting colorless substrates to colored products using the enzyme-mimicking catalytic activity of nanoparticles has become an alternative color development strategy.However,this approach requires attention to reduce the occurrence of broad-spectrum catalytic activity [94].Accordingly,Weerathunge et al.[43] controlled the enzyme-mimicking catalytic activity of AuNPs by DNA aptamers to achieve ultraspecific detection of infective murine norovirus (MNV).Considering the extensive dynamic range that is equally popular with low detection limits,Weerathunge and coauthors investigated the possibility of actively modulating the dynamic operating range of the biosensor by varying the concentration of AuNPs.When the AuNPs changed from an initial concentration of 75 μM-100 μM and 150 μM,the linear range could be adjusted from 20 to 1,000 viruses/mL initially to 132-1,980 and 330-3,300 viruses/mL,respectively,giving the biosensor more design flexibility for future practical applications (Fig.4E) [43].Furthermore,we should also be aware that biomolecule-modified AuNPs exhibit low catalytic activity and sensitivity.This drawback can be overcome by doping the AuNPs with silver ions,as demonstrated in the immunoassay of NoV-like particles (NoV-LPs) and NoV subtypes by Khoris et al.[95].
3.5.Signal enhancement of the two main biosensing systems
The sensitivity of biosensors usually depends on the relationship between the analyte concentration and the signal.However,the target virus concentration in water media is too low to be distinguished by conventional sensing methods.To date,many signal amplification strategies have been used for biosensors,focusing on enhancing signal sensing mechanisms [96],output signal-to-noise ratio [97],and aggregate size of target-probenanomaterial complexes [91,97].
3.5.1.Nanomaterial-assisted amplification
To achieve optimal sensitivity,various factors need to be considered during nanomaterial design,such as manipulating the physical characteristics of nanomaterials(material,size,geometry/shape,dimensions,etc.).The size and shape of nanoparticles can be optimized to increase their surface area and enhance their binding capacity with viral targets [98].Furthermore,MNPs help to guide viral enrichment and signaling.
Zhao et al.[42] fabricated a novel magnetic covalent organic framework/pillararene heterosupramolecular nanocomposite to serve as a bioconjugate with multiple identified sites.The composite was combined into a peptide-target virus-aptamer sandwich strategy on an electrode with high electrical conductivity for sensing NoVs.The developed platform could detect NoVs without extraction and amplification of viral nucleic acids.Additionally,it had a short turnaround time (~1.5 h) and exhibited high accuracy and selectivity(Fig.5A)[42].The same group further demonstrated a hypersensitive response to SARS-CoV-2 RNA with a LOD as low as 200 copies/mL in clinical samples.Here,the signal-amplifying moleculep-sulfocalix[8]arene-functionalized graphene forms long concatamers via multiple probe hybridizations.The resulting supersandwich complex enriches the electrochemical mediators(toluidine blue) to produce an ultrasensitive DPV signal on the modified SPCE.The detectable ratios of the proposed method for clinical samples are higher than those obtained using RT-qPCR[99].
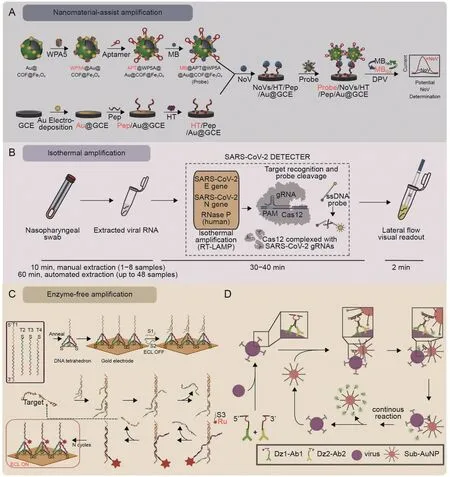
Fig.5.Schematic representation of three types of signal amplification.(A)Human norovirus(NoV)detection using the electrochemical biosensor integrates the magnetic covalent organic framework/pillararene heterosupramolecular nanocomposites [42].(B) The LAMP-CRISPR-Cas12a lateral flow system for SARS-CoV-2 detection [103].(C) The DNA tetrahedron-based entropy-driven electrochemiluminescence (ECL) biosensor for the detection of SARS-CoV2 RNA-dependent RNA polymerase (RdRp) gene [112].(D) DNAzyme walker for homogeneous detection of enterovirus type 71(EV71)and coxsackievirus B3(CVB3)[108].COF:covalent organic framework;WP5A:water-soluble pillar[5]arene;APT:NoV-specific aptamer;Pep: NoV-specific pepteide;MB: methylene blue;GCE: glassy carbon electrode;HT: 1-hexanethio;DPV: differential pulse voltammetry;RT-LAMP: reverse transcription-loop-mediated isothermal amplification;PAM:protospacer adjacent motif;gRNA:guide RNA;Cas 12:CRISPR-Cas 12;S1/S2/S3:DNA probes;Ru:Ru(bpy)32+;Dz1-Ab1:EV71 capsid protein VP1 antibody conjugated DNA1;Dz2-Ab2:EV71 capsid protein VP2 antibody conjugated DNA2;AuNP:gold nanoparticle.Reprinted from Refs.[42,103,108,112]with permission.
3.5.2.Isothermal amplification
Isothermal amplification is a much-advanced technique over traditional amplification techniques(such as PCR).It involves DNA polymerization at a constant temperature over several minutes,eliminating the need for costly instrumentation and complex handling procedures[100].This technique is widely used in clinical diagnostics,genetic analysis,and environmental monitoring.Common strategies include recombinase polymerase amplification(RPA),loop-mediated isothermal amplification(LAMP),and rollingcircle amplification (RCA) [101].
The RPA reaction in the microenvironment has been integrated with the POC fluorescent device to complete the AdV DNA isolation,detection,and signal amplification process in less than 1 h [102].LAMP-based logic gate strategies are prevalent in the nucleic acid detection of viruses.In the LAMP-CRISPR-Cas12a lateral flow system reported by Broughton et al.[103],the LAMP reaction is responsible for identifying the RNA sequences of SARS-CoV-2 extracted from nasopharyngeal or oropharyngeal swabs,and Cas12a then releases the signal by cleaving the FAM-biotin reporter molecule.The lateral flow test paper visualizes the final result.The whole system showed remarkable advantages,including easy-touse reporting format,accuracy (10 copies/μL of LOD,95% positive predictive agreement),fast run times (<40 min),and laboratoryfree (Fig.5B) [103].For another logic element,RCA,Chaibun et al.[104] described a multiplex RCA-based electrochemical biosensor for rapid detection of the N (nucleocapsid) and S (spike) genes of SARS-CoV-2 in clinical samples.The system integrates RCA amplicons and probe hybridization into a one-step sandwich hybridization procedure followed by electrochemical testing for redox labeling,allowing the amplification of N/S genes down to 1 copy/μL into a detectable signal in less than 2 h.The assay results on clinical samples are consistent with the RT-qPCR results.
3.5.3.Enzyme-free amplification
As mentioned above,enzyme-assisted signal amplification strategies have shown effective signal enhancement capabilities.Nevertheless,biological enzymes are not exempt from the drawbacks caused by harsh storage and working conditions.Therefore,enzyme-free signal amplifications,also called DNA programming tools,are gaining popularity due to technological advancements and their potential to overcome some of the limitations associated with enzyme-assisted signal amplification methods.The specific DNA programming tools include DNA enzymes,entropy-driven amplification (EDA),hybridization chain reaction (HCR),catalytic hairpin assembly (CHA),and DNA walkers[105].
Enzyme-like elements,such as nanomaterials [106] and DNAzymes [107,108],have greatly expanded the possibilities for generating highly sensitive biosensing signals.As described in 3.4.3,the peroxidase-like activity of nanomaterials such as AuNPs enhances the extinction coefficient of the solution,allowing colorimetric sensing of trace targets.Additionally,peroxidase-like catalytic activity has been demonstrated in a special ssDNA structure,the G-quadruplex,which has unique folding properties due to its enrichment in guanine[109]and folds in the presence of hemin[110] to form a nonclassical nucleic acid secondary structure.Cui et al.[111] designed logical reactions in a microfluidic platform to reduce nonspecific color.The hemin/G-quadruplex-conjugated DNA probe is the chromogenic generator,while GO-coated microbeads assist in MNV identification and signal amplification.
Enzyme-free EDA is attractive as a tool for DNA-based competitive hybridization.Fan et al.[112] presented an EDA cascade amplifier circuit with an ECL biosensor targeting the SARSCoV-2 RdRp gene.The tetrahedral DNA capture probes coupled to a gold electrode serve as a backbone for EDA.A set of hybridization probes bind competitively to RdRp DNA at the DNA tetrahedron apex,prompting enrichment of Ru(bpy)32+and thus turning on the ECL signal.This DNA tetrahedron-coupled EDA cascade achieved femtomolar detection levels(less than 2.67 fM)with good stability and fouling resistance,and spiked serum recoveries ranged from 98.21 to 102.3% (Fig.5C) [112].
Three other DNA tools (HCR,CHA,and DNA walker) utilize toehold-mediated strand displacement.The main principle is based on the kinetic capture of target sequences in DNA substable hairpins [113].Zhang et al.[106] introduced a low background interference and high sensitivity sensor for ECL employing the HCR,which adopted three-stranded Y-type DNA probes as vital reaction tools.Based on this strategy,detecting SARS-CoV-2 nucleic acid in pharyngeal swabs was highly feasible,with an LOD of 59 aM.DNA walkers are a series of toehold-mediated DNA nanocomponents that mimic the directional movement of biological protein motors(e.g.,myosins and kinesins)and can move along an elaborate 2D/3D DNA track.Hybridization or hydrolysis with the DNA track provides the driving force [114].Recently,an advanced study focused on using DNA walkers and proximity ligation assay (PLA)-derived methods to enhance the sensitivity of detection systems for EV71 and CVB3 viruses [108].In this study,hybridization of the target virus with the probe resulted in the assembly of DNA walkers with DNAase activity,which cut the fluorescent markers one by one along the AuNP tracks.The fluorescence intensity released was proportional to the number of target viruses in the sample(Fig.5D)[108].This approach is commonly used in molecular diagnostics because it allows for the rapid and sensitive detection of a wide range of pathogens.
3.5.4.Sandwich-type-based signal enhancement
The sandwich method is a prominent technique in biosensor design and assay development.It uses multiple recognition elements to create a “sandwich” structure around the target analyte.This method forms stable and specific complexes by capturing the analyte between these recognition elements,resulting in benefits like increased signal amplification,improved selectivity,and enhanced detection limits.It surpasses traditional direct detection methods and enables highly sensitive and reliable biosensor signals.Kim and his coworkers [115] have demonstrated that implementing the sandwich strategy resulted in a sensitivity increase of approximately 15 times compared to the case without the sandwich assay.Zhang et al.[116]proposed a supersandwich ECL assay to achieve remarkable LOD of viral gene (0.022 pM) based on multiple DNA probes.Huang et al.[79] utilized paired antibodies coupled to the LSPR sensor and AuNP to capture two distinct epitopes of the SARS-CoV-2 spike protein.This approach improved the assay's specificity and reduced the LOD to 370 viral particles/mL,which is significantly lower than the viral load in nasopharyngeal swabs and saliva (104-1010viral particles/mL).There is also a research team developed a sandwich assay using aptamers to detect NoV capsid protein.They utilized gold nanorods as signal enhancement molecules for SPR detection,effectively reducing the LOD to attomolar concentrations [40].
Nevertheless,sandwich assays still have certain limitations.These include cost and complexity,as the use of two antibodies and labeling chemistry increases expenses and skilled personnel are required for assay optimization.Availability and specificity of antibodies pose a challenge,requiring suitable antibodies without crossreactivity.The stability and storage conditions of immobilized and labeled antibodies are critical.Additionally,the multi-step workflow can be time-consuming,hindering real-time applications.Moreover,complex sample matrices can introduce interference,leading to false results.The dynamic range of sandwich assays may be limited,necessitating dilutions for accurate quantification of highconcentration samples.To overcome these limitations,ongoing research focuses on various approaches.Antibody engineering improves availability,specificity,and affinity.Assay optimization strategies simplify workflows and reduce time requirements.Integration with advanced platforms like microfluidic systems or nanotechnology enables more efficient and sensitive detection.
3.6.Integration of complementary transduction elements
Several nanoscale integrated structures based on optical and electronic systems capable of detecting waterborne viruses with high sensitivity have been developed,particularly in the wake of the COVID pandemic.Multimodal sensing typically combines complementary transduction elements in an integrated system for virus detection.Data from multiple sensors can be rapidly processed and analyzed to provide a well-rounded situation of virus presence and concentration.
Examples include AuNP-mediated colorimetric and low-cost electrochemical assays [45,92].Ngamdee et al.[45] developed a one-pot assay strategy for detecting HEV viruses.DNA strand replacement reactions and magnetic separation are combined to eliminate the negative effects of residual contamination and pipetting errors.The HEV-DNA spontaneously displaces with the AuNP-labeled probe,and the resulting red color can be observed visually and measured by absorbance.The disposable SPCE provides a linked reusable electrochemical pathway to assess free AuNPs.In summary,the system achieves detection limits of 10 pM(visual),10 pM (spectrophotometric),and 1 fM (electrochemical).However,some potential drawbacks,such as the high loss rate due to robust nucleic acid extraction and the impact of the pH of the extraction buffer on the DNA strand replacement efficiency,need to be considered (Fig.6A) [45].
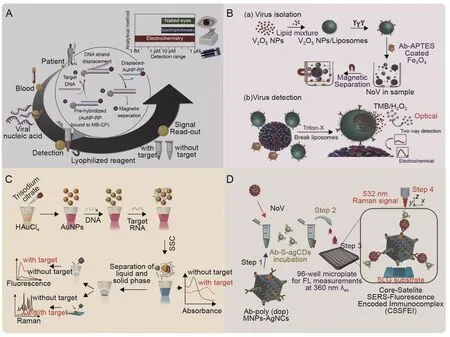
Fig.6.Schematic representation illustrating the detection principle of multimode biosensors.(A) One-pot detection of hepatitis E virus (HEV) with three ways of signal interpretation[45].(B)Liposome-based dual modality sensor including virus isolation via V2O5 nanoparticle(V2O5 NP)-encapsulated liposomes and dual modality of virus detection[70].(C)Triple-mode biosensors for virus RNA detection[93].(D)Sulfur-doped carbon dots(CDs)@polydopamine-functionalized magnetic silver nanocubes for dual-modality detection[117].AuNPs:gold nanoparticles;AuNP-RP:AuNP-labeled reporter probes;MB-CP:magnetic bead-labeled capture probes;APTES:(3-aminopropyl)triethoxysilane;NoV:norovirus;TMB: 3,3′,5,5′-Tetramethylbenzidine;SSC: saline sodium citrate;Ab: NoV-specific Antibody;S-agCDs: sulfur-doped agar-derived carbon dots;Poly (dop)-MNPs-Ag NCs:polydopamine-functionalized magnetic silver nanocubes;FL: fluorescence signal;SLG: single-layer graphene.Reprinted from Refs.[45,70,93,117] with permission.
Another study by Ganganboina et al.[70] presented a redoxcatalyzed dual-mode sensing platform for detecting lowconcentration targets,such as NoV,by integrating the advantages of magnetic separation,encapsulation of liposomes,and artificial enzyme amplification.The V2O5nanozymes are encapsulated into liposomes functionalized with NoV-specific antibodies and MNPs,eliminating the oxidase's nonspecific signal.Only after the NoV-LP recognition process can the V2O5nanozyme be artificially released to generate a colorimetric/electrical signal.The quantitative analysis performed in this study revealed that the dual-modal sensing platform achieved LODs of 4.1 fg/mL (DVP) and 0.34 pg/mL(colorimetric)for this bimodality,with linear ranges of 10 fg/mL to 10 pg/mL (DPV) and 1 pg/mL to 100 ng/mL (colorimetric) (Fig.6B)[70].Nevertheless,the high LOD of the optical signal is a limiting factor for the performance of this dual-modal sensor.Replacing the colorimetric elements with fluorescence materials may be an improvement direction to enhance the sensor performance of dual confirmation.
Fluorescence and SERS spectroscopy are complementary optical modalities used as practical analytical tools for biosensing and bioimaging applications.Gao et al.'s SARS-CoV-2 project [93]implemented a fluorescently labeled DNA probe and SERS detection in addition to colorimetry,running three modes of signal:absorbance,fluorescence,and SERS,reaching ultralow detection limits of 160 fM,259 fM,and 395 fM,respectively.This scheme significantly reduced false-negatives (Fig.6C) [93].Achadu et al.[117] then used fluorescent carbon dots (CDs) and a sandwichcomplex-enhanced SERS system to improve bioassay sensitivity and eliminate matrix interference.The immunocomplex formed by CDs,NoV-LPs/NoVs,and magnetic silver nanocubes can be rapidly detected with the fluorescence signal after deposition using fluorescence confocal imaging.It can further be magnetically enriched to the SERS platform for high-precision detection.Although the fluorescence modality remains tricky for most cases in natural specimens,in this system,CDs bring the fluorescence LOD down to 80 copies/mL of NoVs,close to the SERS signal (10 copies/mL).Overall,this design successfully tackles difficult-to-detect targets in fecal samples (Fig.6D) [117].
4.Current and future biosensors for waterborne virus detection in wastewater
Wastewater from different sources varies greatly in physical appearance,chemical parameters,and biological substance abundance.It is a potential resource to provide much dynamic population information.Nonetheless,this also makes subsequent analysis of specific targets difficult because it is necessary to overcome variable and complex components and aim for target analytes with unpredictable concentrations [118].Different factors must be considered to successfully implement biosensors for infectious disease/epidemic surveillance,including sampling,sample pretreatment,and target monitoring [6].
4.1.Current biosensors for waterborne virus detection in wastewater
To date,biosensors for waterborne virus detection in wastewater are limited compared to those for detection in human samples.The available examples of biosensors for detecting waterborne viruses in wastewater are now summarized in Table 2[44,59,73,119,120].Although these biosensors are not yet fully developed for controlling wastewater disease,they provide important insights into the potential of dealing with wastewater samples.Examples involve creating high-precision identification events without complex pretreatment,e.g.,a SARS-CoV-2 MIPmodified electrode [59].On the other hand,biosensing platforms integrating signal enhancement strategies and target capture/enrichment have also proven effective in spiked wastewater,such as the external force-assisted near-field illumination (EFA-NI)biosensor based on AuNPs and MNPs[119]and the electrochemical detection assisted by magnetic beads and horseradish peroxidase(HRP)-labeled HEV probes[44].Cost-effectiveness,portability,and user-friendliness were also taken into account for the biosensors,as evidenced by the use of low-cost disposable electrodes (printed circuit board electrodes[73]and screen printed electrodes[44,59])and paper-based devices (microfluidic paper-based analytical devices,μPADs[120]).However,most of these studies have validated their feasibility in artificial samples but not in real-world scenarios.
4.2.Future biosensors for waterborne virus detection in wastewater
Despite the limited studies on the biosensing of waterborne viruses in wastewater,the lessons we learned from biosensing in human samples do provide good insights.When designing future biosensors for detection in wastewater,several challenges need to be considered,including 1) limitation of sample preparation;2)poor stability of recognition elements;3)low detection sensitivity;4)false-negatives/positives;and 5)imperfect quantitative analysis.A high sensitivity at the picomolar or even attomolar level[121]is essential,as we need to monitor waterborne viruses down to pathogenic amounts (1-100 virus particles) [8].
4.2.1.Sampling and pretreatment of wastewater

Implementing sufficient sample collection ensures successful application of waterborne virus monitoring.Urban and rural wastewater systems reflect the public health status of densely populated areas.Attention must be paid to the disposal and fate of wastewater in different regions.The urban sewage system facilitates the collection of wastewater samples representing the entire community,as all wastewater eventually flows into WWTPs.In contrast,due to nonstandard disposal,more sampling points must be set in rural areas or LMICs,such as open spaces,toilets,or septic tanks,and even downstream of rivers [122].In addition,viral sample data (concentration,vitality,integrity,and diffusion rate)are variable in different areas of the wastewater system [6,123].Well-designed spatial sampling for the possible starting point and possible transmission path of infection outbreaks is crucial to control the epidemic.
The complex environment consisting of abundant contaminants,interfering chemicals,and inhibitors poses a more significant challenge to designing highly sensitive biosensors,which highlights the importance of sample pretreatment and concentration steps.Current virus detection has involved filtration and preconcentration technologies that combine flocculation and precipitation strategies [124],such as ultrafiltration,electronegative filtration,polyethylene glycol precipitation,skimmed-milk flocculation,and other virus recovery methods [125].However,a low recovery rate and poor repeatability are their shortcomings.The storage of samples and the impact on viruses are also among the challenging factors [6].Moreover,there is still no consensus on a standard procedure for preprocessing,making it difficult to obtain reliable and comparable results [126].Simultaneously,integrating immediate wastewater treatment with performance-enhanced biosensors will serve as an alternative approach,indicating the current development direction of wastewater biosensors.
In the biosensing system,pretreatment of samples is influenced by factors such as the purity and volume of the samples,the biosensing elements' performance,and the biosensing system's running mode.For wastewater samples with high impurity content and low viral load,the virus concentration may be lower than the biosensor's LOD,in which case a prefiltration centrifugal separation or a preconcentration step is required [41].In pathogen detection applications,isolating the target using a magnetic system and superimposing a washing step is a popular method to reduce background inhibitory material and concentrate the target[85].For DNA/RNA-based assays,extraction of genetic material and reverse transcription (RNA scope) may be desirable(Fig.7A).
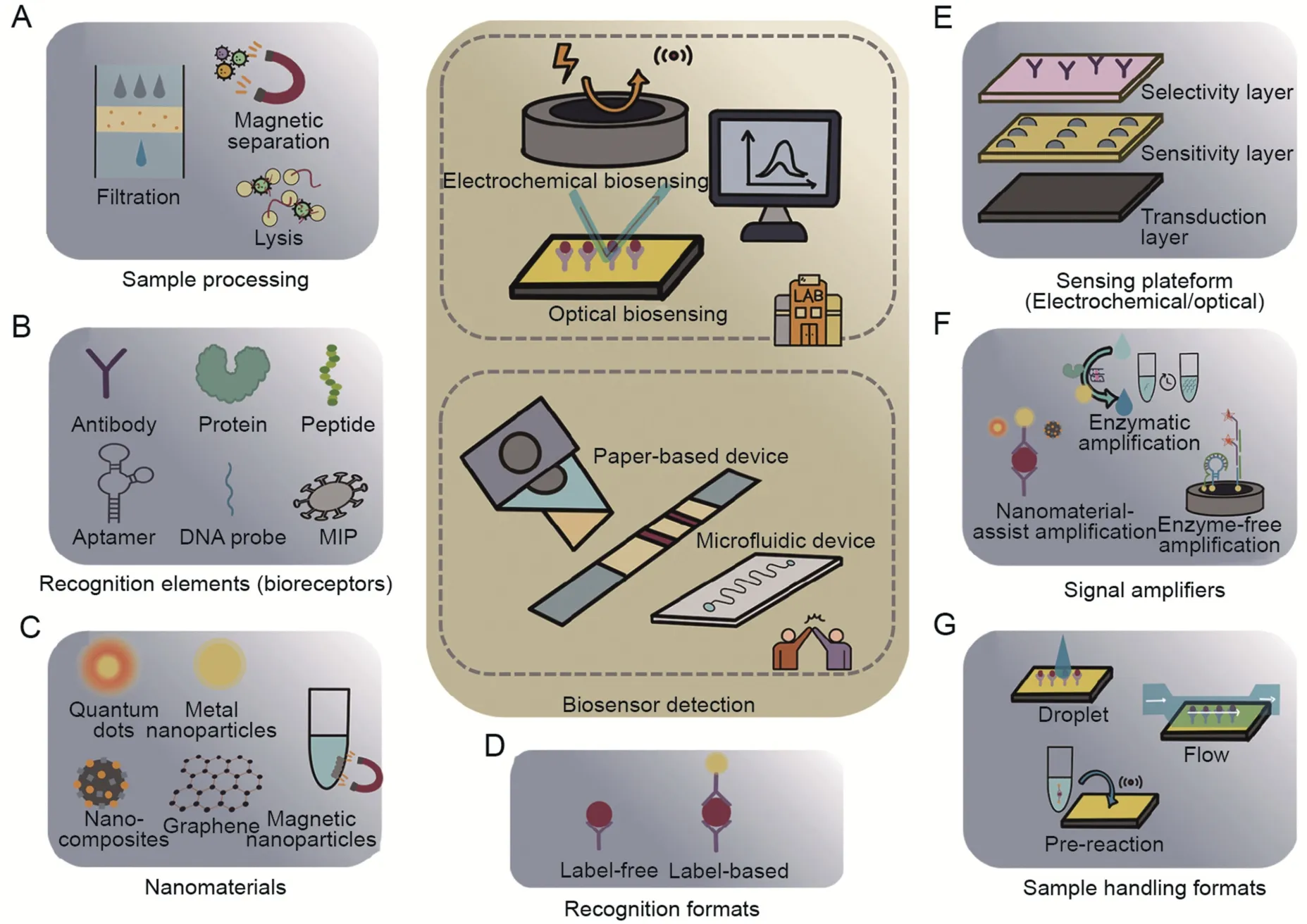
Fig.7.Summary of the components involved in waterborne virus biosensing in wastewater.(A)Sample preprocessing to reduce interferents.(B)Target recognition elements with high specificity and stability.Selection of (C) nanomaterial types,(D) recognition formats,(E) sensing platform,(F) signal amplifying methods,and (G) corresponding sample handling formats.
4.2.2.Loading of wastewater on the biosensors
Typically,three formats can be considered to handle wastewater over a biosensor: flow,droplet/immersion,and preincubation/prereaction.Flow formats are compatible with large sample volumes(from hundreds of microliters to several liters),allowing the sample to be exposed to the sensor surface in a controlled and reproducible manner.The most common flow formats are flow cells [40,77] and microfluidic channels [41,102,111],both requiring consideration of parameters such as flow rate,measurement time,and template identification efficiency.Droplet/immersion is a simple sample handling method that reads the signal by adding a drop of the sample directly onto the functionalized transducer or immersing the functionalized transducer in the sample.Preincubation/prereaction formats are often coupled with a simple detection device,as they allow efficient and sensitive target identification in a small-volume reaction system.Biosensors based on signal-enhanced labels and enzymatic amplification have been reported in combination with such formats.Label-based sandwich detection,for example,typically involves preincubating the sample with an identification probe and signal probes,followed by an appropriate processing step before detection with a transducer[44,127].
4.2.3.Biosensing platforms for waterborne virus in wastewater
To improve environmental wastewater monitoring,the ready-touse solution is used to introduce nanotechnology for well-designed biorecognition events and various transduction strategies.
The highly stable recognition elements mentioned in Section 3.1 are user-friendly for long-distance transport and detection conditions [128] (Fig.7B).The use of biomimetic bioreceptors,such as aptamers and MIPs,offers several advantages over their natural counterparts,including improved stability,selectivity,and tunability,making them promising alternatives for biosensing and other applications.Furthermore,the optimized sensing modalities consisting of multifunctional nanocomposites have greater robustness and signal reproducibility within the error range.High surface area,unique shape and dimensions,catalytic activity,and electrochemical and physicochemical properties are the prominent features of nanoparticles,making them widely used in bioimmobilization,signal generation,and amplification.The combination of emerging nanomaterials such as nanoparticles,nanorods,carbon nanotubes,and nanocomposites with biosensors has enlightened the possibility of performing heterogeneous structures and improved the sensitivity and analytical properties[129,130],as discussed in Sections 3.3 and 3.4 (Fig.7C-E).However,there are concerns that need to be considered.Metallic nanomaterials,such as gold,silver,and platinum nanoparticles,face challenges related to limited surface area and biocompatibility issues.Additionally,QDs,like CdSe QDs,can pose toxicity concerns.In contrast,carbonbased nanomaterials,like graphene and carbon nanotubes,offer desirable properties such as high sensitivity,biocompatibility,and flexibility.However,their production costs are relatively high due to the complex manufacturing processes involved.Alternatively,composite nanomaterials can leverage synergistic effects between different materials,leading to improved sensing and catalytic performances while also being cost-effective.The selection of nanomaterials depends on specific biosensor requirements,target biomolecules,and intended applications.
Signal amplification methods are needed to generate measurable signals [96,97,131],including 1) Nanomaterial-assisted amplification (nanoparticles,nanorods,nanocomposites,etc.)[40,42],2) enzymatic amplification: enzyme catalysis (enzymes and nanozymes,etc.) [43],isothermal nucleic acid amplification(LAMP,RPA,RCA,etc.)[102,104],and cascade amplification strategy integrating multiple amplification strategies (LAMP-CRISPR-Cas system) [103],3) enzyme-free amplification (DNA walker,HCR,CHA,etc.) [106,108] (Table 3) [40,42,43,102-104,106,108] (Fig.7F).Nanomaterial-assisted sandwich assays employ a dual-binding mechanism that enhances sensitivity and specificity by amplifying signals but rely on precise nanofabrication and a multi-step workflow.Isothermal nucleic acid amplification is a rapid and robust method for signal amplification that does not require thermal cycling.However,it requires skilled personnel and strict transport conditions.Enzyme-free amplification,based on DNA programming,offers a simple and cost-effective approach through strand displacement reactions and self-assembly for signal amplification without enzymes.Meanwhile,challenges in DNA element design,potential non-specific adsorption interference,spatial site hindrance during biorecognition,and sensitivity to environmental conditions must be addressed by carefully optimizing the process for reliable performance.
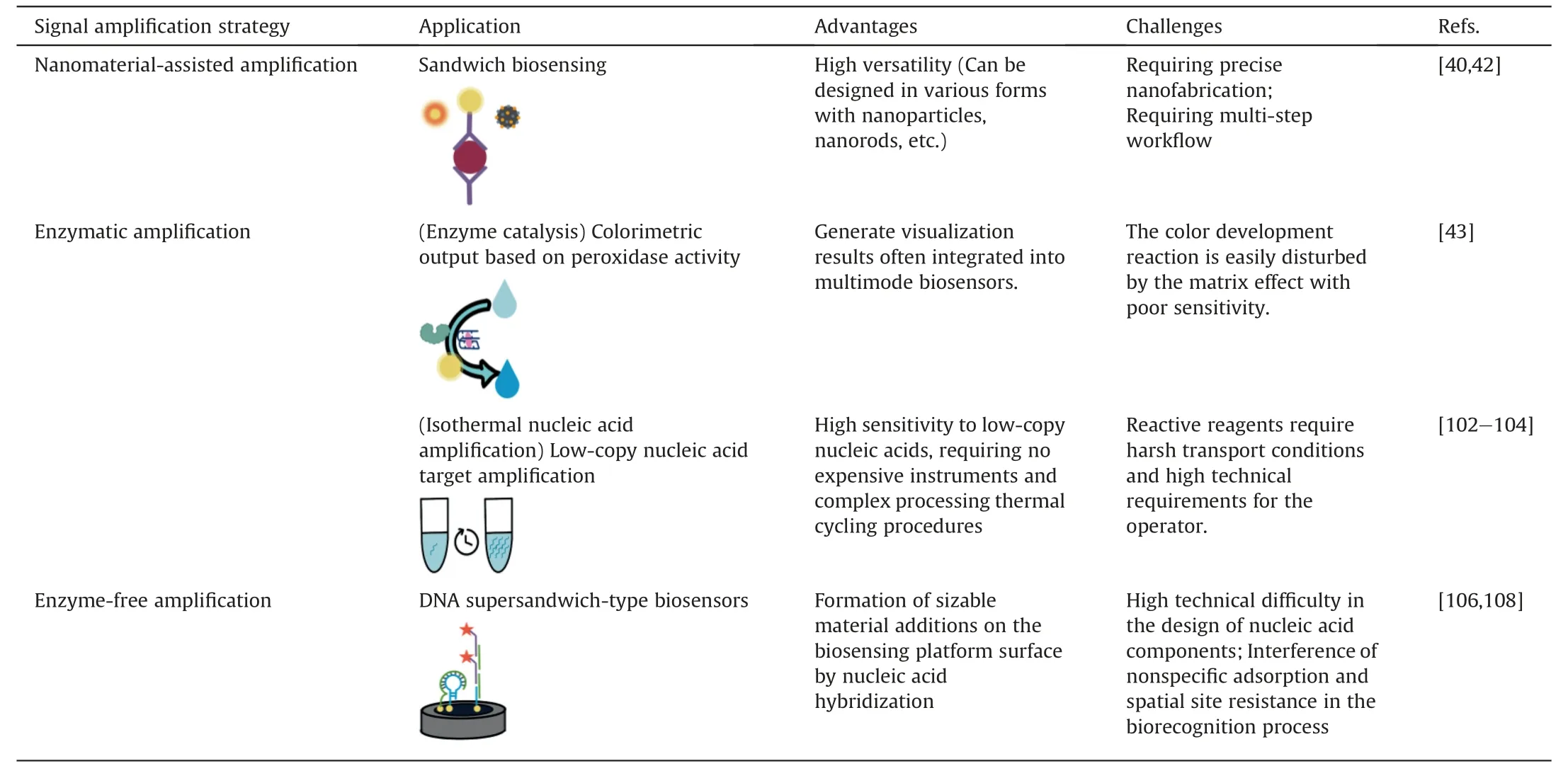
Table 3 Comparison of signal amplification strategies in virus detection.
5.Future perspectives
For waterborne virus detection in complex samples,integrating advanced biosensors with automated and portable systems while achieving desirable analytical precision and reproducibility in batch processing with minimal interference is highly desired.
First,the biosensors are expected to detect the target viruses directly from real samples,where lowering background noise and condensing the waterborne virus are necessary.Several studies have built biosensing systems that achieve higher input-to-output responses employing advanced hybrid nanocomposite materials,ultrasensitive recognition events,and logic programs[58,70,85,88,106,117].Colorimetric and visual assays are helpful in resource-limited areas or laboratories because of their simplicity and ease of use,and the current trend is to combine them with other advanced electrochemical/optical technologies into multifunctional detection devices to reach reliable results through multiple signal cross-valuation.On the other hand,trapping analytes at controlled translocation rates at biosensing interfaces improves precise sensing for accurate measurements.In addition to conventional methods of controlling analyte flow rates in flow cells and microfluidic channels,a recent study using nanopore technology has shown that single molecules can be manipulated for assays under precise control of both space and time [132].This highlights the potential of using advanced techniques for precise analyte manipulation in biosensing applications.
Second,a combination of experimental methods and mathematical modeling can be interesting to characterize and evaluate biosensor performance.Experimental methods could include testing the biosensor with a range of virus concentrations and sample volumes,as well as testing the biosensor's selectivity against other viruses and molecules.Mathematical modeling can predict biosensor performance under different conditions and optimize biosensor design and operation.In addition,machine learning (ML) holds the potential to further advance biosensors.With the ability to discern low-resolution sensing signals from high noise through big data analysis,ML can optimize virus feature selection and concentration prediction by analyzing complex datasets and training models.This allows for rapid predictive modeling of virus behavior.Moreover,ML can be integrated into real-time biosensing platforms to enable continuous monitoring of viral contamination in various settings,such as water sources and healthcare facilities.An example is a label-free platform that combines SERS and ML for rapid and accurate detection of SARSCoV-2 [133].
Third,the aging of biosensors over time needs to be considered.In addition to overcoming the stability barrier of biological recognition elements,it is critical to consider the“scaling”effect caused by nonspecific adsorption from other biomolecules.Possible solutions include introducing various antifouling layers on the electrode surface in electrochemical systems [134].
To continue,developing multiplex detection systems with multiple modular functional areas should be considered to form an on-site detection instrument that integrates sample processing,multiple virus strain identification,and reliable signal output[135].Microfluidic systems are expected to enable small portable devices with integrated modular functional areas[136],as in the case of the proposed one-step microfluidic NoV sensing platform [41].Additionally,incorporating smartphones could facilitate portability,easy-to-understand results,and broad practical applicability.
Furthermore,the currently available biosensors have demonstrated excellent scalability,allowing for the integration of diverse biorecognition elements with nanostructures and sensing platforms for multiplexed biosensing.These platforms can be updated with next-generation algorithms to accurately distinguish between different virus strains [67].The identification and monitoring of multiple viral pathogens are crucial for safeguarding human health and ensuring water safety.Multiplexed biosensing plays a vital role in this regard by enabling the simultaneous detection of multiple viruses in a single sample.Its advantages are evident:multiplexing significantly reduces the time and cost required for individual virus detection,making it highly efficient for large-scale monitoring and surveillance efforts.By combining multiple detection targets,it not only enhances sensitivity and specificity but also reduces the likelihood of false positives or false negatives.This forward-looking approach to virus biosensing holds great promise in combating virus transmission and protecting public health.
6.Conclusions
Waterborne viral pandemics have been a significant threat to public health for several decades,making the development of sensitive and reliable detection methods crucial for early diagnosis and timely intervention.Biosensing platforms have emerged as promising tools for waterborne virus detection owing to their high sensitivity and specificity.This work focuses on optical and electrochemical biosensing platforms for waterborne virus detection,highlighting the mechanism,transducer,biointerface design,signal amplification strategies,and sensing platform design.
Obtaining a higher input-to-output response is essential,and this can be achieved by adopting a multifunctional transducer.Additionally,reducing nonspecific signals and enhancing the property change brought about by receptor-target binding and sufficient signal enhancement is worth further study from the perspective of the biointerface.For platforms,highly integrated and compact devices are expected for POC testing and on-site diagnosis.Future perspectives on waterborne virus biosensing to improve the accuracy and sensitivity of biosensors are in various aspects,including innovations and improvements in materials science,engineering,and biology,as well as the exploration of ML techniques.
Overall,the development of biosensing platforms for waterborne virus detection is a challenging yet rewarding task,and it holds tremendous potential for improving public health and safeguarding our environment.
CRediT author statement
Xixi Song: Conceptualization,Writing -Original draft preparation,Reviewing and Editing;Zina Fredj: Validation,Writing -Reviewing and Editing;Yuqiao Zheng: Writing -Reviewing and Editing;Hongyong Zhang: Writing -Reviewing and Editing;Guoguang Rong: Writing -Reviewing and Editing;Sumin Bian:Supervision,Validation,and Writing -Reviewing and Editing;Mohamad Sawan:Conceptualization,Supervision,Validation,and Writing -Reviewing and Editing,Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This research was supported by the Research Center for Industries of the Future of Westlake University,China (Grant No.:210230006022219/001),the National Natural Science Foundation of China(Grant No.:82104122),Westlake University,China(Grant No.:10318A992001),and the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang,China(Grant No.:2020R01005).
杂志排行
Journal of Pharmaceutical Analysis的其它文章
- Stage-specific treatment of colorectal cancer:A microRNA-nanocomposite approach
- Development status of novel spectral imaging techniques and application to traditional Chinese medicine
- Oridonin restores hepatic lipid homeostasis in an LXRα-ATGL/EPT1 axis-dependent manner
- Ginsenoside Rg5 enhances the radiosensitivity of lung adenocarcinoma via reducing HSP90-CDC37 interaction and promoting client protein degradation
- Canonical transient receptor potential channel 1 aggravates myocardial ischemia-and-reperfusion injury by upregulating reactive oxygen species
- Nanoscale coordination polymer Fe-DMY downregulatingPoldip2-Nox4-H2O2 pathway and alleviating diabetic retinopathy
