miR-483-5p regulates osteoclast generation by targeting Timp2
2023-12-02NIUTianqiLIUCaixiaXIONGJunJIAHaoWANGHuaLIShuangDENGHuimingZENGXiangzhou
NIU Tian-qi, LIU Cai-xia, XIONG Jun, JIA Hao, WANG Hua, LI Shuang, DENG Hui-ming,ZENG Xiang-zhou
Hainan Medical University, Haikou 571199, China
Keywords:
ABSTRACT Objective: To investigate whether miR-483-5p regulates osteoclast generation by targeting Timp2.miR-483-5p can promote osteoclast differentiation and bone destruction.Methods:Target genes of miR-483-5p were predicted by miRNAs target gene prediction software TargetScan8.0, and wild type and mutant 3' UTR plasmids were constructed.Dual luciferase reporter genes were used to verify whether target genes had a targeted regulatory relationship with miR-483-5p.Western blotting was used to detect the corresponding changes in the expression level of target protein after adjusting the level of miR-483-5p in cells.Cells were transfected or infected with target gene siRNA or target protein lentivirus, and TRAP staining and q-PCR assays were performed.In addition, for osteoclast induction experiment,RAW264.7 cells were co-transfected with ago-miR-483-5p and target protein-overexpressed lentiviruses q-PCR and TRAP staining were performed respectively.Results: Bioinformatics software was used to predict the target gene of miR-483-5p, and the Timp2 gene was found to regulate osteoclasts, and the dual luciferase reporter detection system found that miR-483-5p could be associated with the 3-UTR of the predicted target gene Timp2 gene.There are complementary loci and targeted regulatory relationship between them.Subsequently, we upregulated miR-483-5p in RAW264.7 cells to reduce the expression of Timp2.Compared with the normal group, the number of osteoclasts and the expression of osteoclast-specific genes increased significantly after the induction of Timp2 in knockdown cells.After co-transfection of target gene and miR-483-5p into cells, the number of osteoclasts and the expression of specific genes decreased significantly compared with the normal group.Conclusion: Timp2 is a downstream target gene of miR-483-5p and is involved in and inhibits osteoclast generation.
1.Introduction
As the only cell type responsible for bone resorption in the human body, Osteoclasts (OCs) are multi-nucleated bone resorption cells from the mononuclear macrophage spectrum[1].As osteoclasts are crucial for normal bone development and bone balance in vivo,over-activation of osteoclasts will lead to imbalance of intraosseous environmental balance[2], thus leading to a variety of osteolytic bone diseases.Such as osteoporosis, bone Paget’s disease and erosive arthritis[3].Therefore, osteoclasts have become an important target for the prevention and treatment of osteolytic diseases.MicroRNAs are a class of highly conserved single-stranded small non-coding RNA that regulate over 30% of genes in eukaryotes[4].miRNA can specifically bind to the 3’-untranslated region (3’-UTR) of the target and inhibit mRNA expression by interfering with mRNA stability and/or blocking protein translation[5].A large number of studies have shown that miRNA affects various signaling pathways by regulating the above factors and hormones, leading to differentiation and functional changes of OCs[6-8].miR-483 is a kind of miRNA found in human fetal liver reported by Landgraf in 2007.miR-483-5p and miR-483-3p are formed after enzyme digestion[9].Most of the literature on miR-483-5p has been associated with tumors.However, the regulation of miR-483-5p on OCs generation and its mechanism through Timp2 has not been reported.In our previous study,Timp2, the target gene of miR-483-5p, was discovered through biological information software, and some gene sequences of the two were compared, and complementary sequences were found.In addition, the target was verified by luciferin analysisinvitro, and RANKL was used to induce RAW264.7 cells to differentiate into OCs system for targeting verification.To clarify the mechanism of miR-483-5p targeting to Timp2 affecting OCs differentiation and activity.
2.Materials and methods
2.1 Materials
RAW264.7 cells and human embryonic kidney 293T cells were purchased from the cell bank of the Chinese Academy of Sciences.Gp-transfection reagents were transfected from Suzhou Transfection company transfected with transfection reagents GP-transfect-Mate.Luciferase reporter gene detection kit from Annolun Beijing Biotechnology Co., LTD.The Lipo 2 000 transfection reagent was derived from Thermo Fly.Leukocyte acid phosphatase (TRAP) kit from Sigma; Protein extraction kit and SDS-PAGE gel preparation kit were from Shanghai Biyuntian Biotechnology Co., LTD.Prime Script TM RT Master Mix (Perfect Real Time) reverse transcription kit, TB Green® Premix Ex TaqTⅡ (TⅡ RNase H) fluorescent quantitative detection kit Plus) from TaKaRa, Japan; TRIzol reagent from Invitrogen (USA); Fetal bovine serum (FBS), DMEM culture-medium, penicillin and streptomycin were obtained from Gibco.Synergy HTX, from Biotek, USA; Real-time fluorescence quantitative PCR instrument (Model: Roche LightCycler480Ⅱ)from Germany Roche Diagnostics.
2.2 Biological information software prediction
With TargetScan8.0 database website https://www.targetscan.org/vert_80/ may predict and miR-483-5p targeted target genes, the predicted target genes and miR-483-5p gene sequence comparison,to find whether there is a complementary sequence.
2.3 Double luciferase activity detection
The construction ofTimp2wild type and mutant 3’-UTR plasmid was obtained from Suzhou Jima Gene Co., LTD.Transfection: 293T cells were inoculated into a 12-well plate at a concentration of 5×105 cells/well.Each group was set up with three multiple Wells and cultured with DMEM containing 10% FBS, 100 U/mL penicillin G,and 100 μg/mL streptomycin at 37 ℃ and 5% CO2for 24 h to fully adhere to the wall.The transfection groups were as follows: mimic NC+Timp2wild, miR483 mimic+Timp2wild, mimic NC+Timp2-miR-483-mutant, and miR-483 mimic+ Timp2-miR-483 mutant.Transfection was performed according to GP-transfect-Mate instructions.The transfection solution was incubated in the incubator for 5 h, and 500 μL of DMEM containing 10% FBS was added.The culture was continued at 37 ℃ with 5% CO2for 48 h.Double luciferase system test: Discard the culture medium in the 12-well plate, wash the cells twice with 500 μL PBS, add 300 μL 1×PLB into the culture hole, gently shake the culture plate at room temperature for 15 minutes, and transfer the lysate to the test plate with 100 μL per well.Firefly luciferase activity and sea kidney luciferase activity were detected using Synergy HTX’s double luciferase assay system with selected 1-2 second delay and 5-10 readings.
2.4 Cell transfection
miR-483-5p mimic, miR-483-5p inhibitor andTimp2siRNA were transfected into RAW264.7 cells, respectively, for subsequent experiments.Transfection methods: RAW264.7 cells were taken from a 24-well plate with a density of 5×104/ well, and cultured with DMEM medium containing 10% FBS, 100 U/mL penicillin G and 100 μg/mL streptomycin for 24 h at 500 μL well, so that the cells were fully adherent to the wall.Transfection was performed according to lipo 2 000 instructions.After 5 h culture at 37 ℃ in 5%CO2incubator, discard the old transfer dye solution in the plate, add DMEM medium containing 10% FBS 500 μL/well, except for Blank group, add RANKL with final concentration of 100 ng/mL, and continue to culture.
2.5 Co-transfection experiment
Firstly, lectin virusTimp2-LV constructed from Suzhou Jima Gene Company was infected into RAW264.7 cells according to the instructions.After the cells were cultured to more than 90% in 6-well plates, the cells were then incubated into 24-well plates with a density of 5×104/ well, and subsequent transfection of miR-483-5p mimic was performed.
2.6 Western blot assay
Whole cell proteins were extracted from RAW264.7 cells cultured for 2days after transfection using RIPA buffer containing PMSF.Whole cell protein concentrations were measured with a BCA protein assay kit.Equal amounts of whole cell extracts were isolated by 10% SDS-PAGE and electrophoresis and transferred to polyvinylidene fluoride membrane.After blocking TBST with 5%skim milk for 1h, the bands were incubated with Timp2 and β-actin primary antibody at 4 ℃ overnight, and then with HRP coupled secondary antibody for 1h.Finally, the bands were detected with ECL reagent using protein development instrument.The signal obtained by western blotting was quantified using ImageJ software.Normalization was performed with β-actin.
2.7 Trap staining
RAW264.7 cells were transfected and cultured for 4days before TRAP staining.Specific experimental procedures were carried out according to the kit instructions.The purplish red part observed in the visual field was the cytoplasm of OCs, and the blue-stained part in the cytoplasm was the nucleus of OCs.Osteoclasts were counted according to TRAP staining positive and the number of nuclei was more than 3.
2.8 RT-PCR
Total RNA was isolated from RAW264.7 cells cultured for 2 d after transfection with TRIzol reagent.cDNA was synthesized from 500 ng total RNA by reverse transcription kit Prime Script TM RT Master Mix(Perfect Real Time).The reaction system was prepared according to the instructions of TB Green® Premix Ex TaqTⅡ (TⅡ RNaseH Plus) kit.The reaction was amplified by Roche LightCycler480 realtime quantitative PCR instrument.The amplification parameters were as follows :95 ℃30 s, 95 ℃ 5 s, 60 ℃20 s, a total of 40 cycles.The primers of experimental mice were as follows: upstream primer of TRAP: 5 ‘-GTGGAAGCCTCTGGAAAATC-3’, downstream primer: 5 ‘-CTCCTCCCTCACACCCGTTA-3’; NFATc1 upstream primer: 5 ‘-CCGTTGCTTCCAGAAAATAACA-3’, downstream primer: 5 ‘-TGTGGGATGTGAAACTCGGAA-3’; cathepsin K upstream primer: 5 ‘-CttccaatacgtgCagaga-3’, downstream primer:5 ‘-TCTTCAGGGCTTTCTCGTTC-3’; MMP-9 upstream primer:5 ‘-CAAAGACCTGAAAACCTCCAA-3’; Downstream primer: 5‘-GGTACAAGTATGCCTCTGCCA-3’; β-actin upstream primer:5 ‘-CTGTCCCTGTATGCCTCTG-3’; Downstream primer: 5‘-TGATGTCACGCACGATTT-3’.The expression of related genes was evaluated by 2-ΔΔCT, and all values were normalized by β-actin.
2.9 Statistical processing
SPSS 26.0 software was used for statistical analysis of the data.One-way analysis of variance and LSD post hoc test were used to analyze the statistical significance of the measurement data.P<0.05 was considered to be statistically significant.
3.Results
3.1 microRNA target prediction
The target gene of mmu-miR-483-5p was predicted by Target Scan 8.0 software, and the following luciferase reporter gene found that Timp2 might be the target of miR-483-5p affecting OCs differentiation, and the following verification was conducted on Timp2.
3.2 Luciferase reporter gene detection to verify whether Timp2 is the target of mmu-miR-483-5p
By comparing the complementary regions ofTimp2’s 3’-UTR and mmu-miR-483-5p, we found complete complementarity, as shown in Figure 1(A).In order to verify the targeting relationship betweenTimp2and mmu-miR-483-5p, we successfully clonedTimp23’-UTR and constructed recombinant vectors of wild-typeTimp2WT and mutantTimp2MUT for miRNA target detection.Compared with the mimics NC group, the fluorescence activity of the mmumiR-483-5p mimics group was significantly decreased after 2d transfection of HEK 293T cells with wild-type recombinant vector Timp2 WT (F=19.401, P<0.05), as shown in Figure 1(B); However,there was no significant difference in luciferase activity between the mimics NC group and the mmu-miR-483-5p mimics group when the mutant recombinant vectorTimp2MUT was co-transfected (F=0.05,P<0.05), as shown in Figure 1(C).These results indicated that mmumiR-483-5p could complement the predicted 3’-UTR site of Timp2 gene, and there was a targeted regulatory relationship between the two.These results preliminarily suggest thatTimp2gene may be the target gene of mmu-miR-483-5p regulating osteoclast generation.
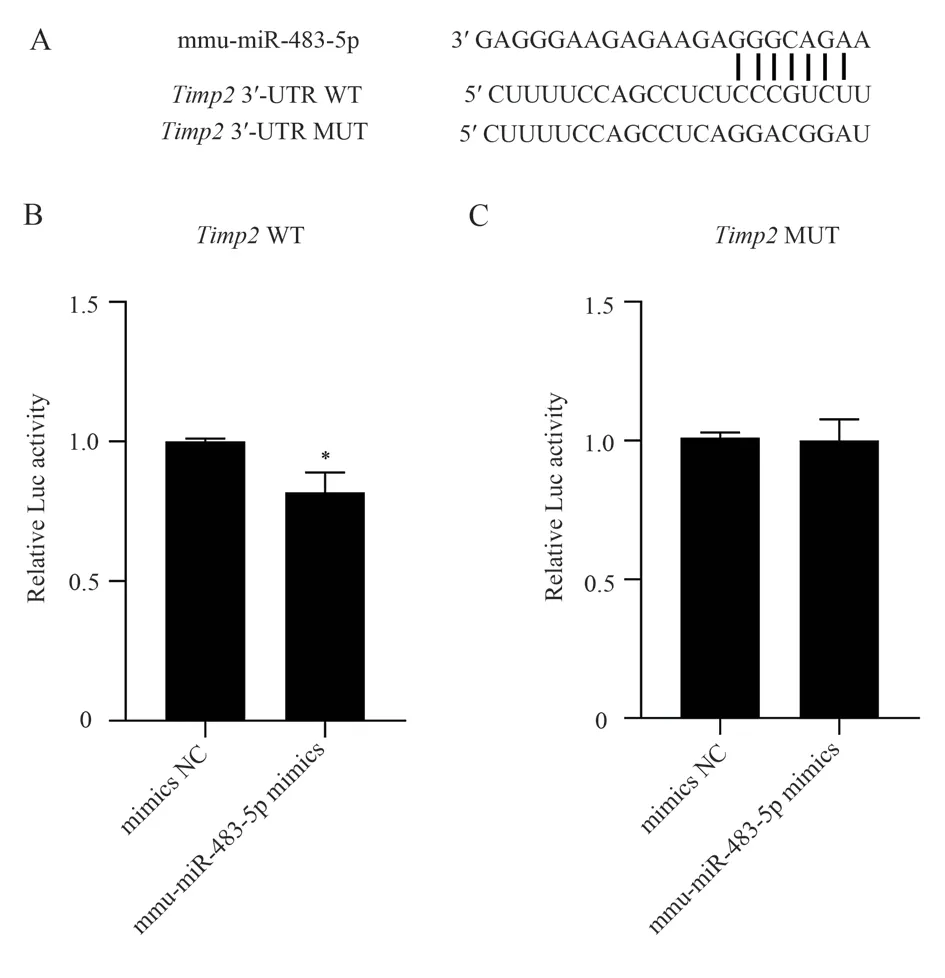
Fig 1 Luciferase reporter assays
3.3 Influence of transfected miR-483-5p content on Timp2 in RAW264.7 cells
After transfecting miR-483-5p mimic and miR-483-5p inhibitor into RAW264.7 cells, the protein content of Timp2 was significantly decreased after increasing the expression of miR-483-5p, compared with that of NC group (F=13.218,P<0.05), as shown in Figure 2.On the contrary, after inhibiting the expression of miR-483-5p,compared with the in NC group, the protein content of Timp2 was increased (F=25.693, P<0.01), as shown in Figure 2, indicating that miR-483-5p is at least negatively correlated with Timp2 protein in vitro.
3.4 Effect of silencing Timp2 on OCs differentiation in RAW264.7 cells
In order to clarify the relationship betweenTimp2and OCs, in this study,Timp2siRNA was used to silenceTimp2in RAW264.7 cells.After inducing OCs, TRAP staining showed that Timp2 in silenced cells resulted in more OCs generation than in Rankl-induced positive control group (F=118.021,P<0.05), as shown in Figure 3(A).The RT-PCR results showed that the specific gene TRAP (F=7358.715,P<0.05), NFATc1 (F=1741.066, P<0.05), Cathepsin K (F=239.981,P<0.05) and MMP-9 (F=4456.654,P<0.05), as shown in Figure 3(B), suggesting that Timp2, as the target of miR-483-5p in RAW264.7 cells, can affect the differentiation and activity of OCs.
3.5 Effects of miR-483-5p and Timp2 on OCs differentiation of RAW264.7 cells
TRAP staining results showed that transfecting only miR-483-5p mimic withTimp2-LV and miR-483-5p mimic could promote OCs differentiation and increase the volume of the differentiated OCs compared with that of RANKL group.However, after transfecting miR-483-5p mimic, overexpression ofTimp2could significantly reverse the excessive OCs activation induced by miR-483-5p mimic (F=108.721, P<0.05), as shown in Figure 4(A).The results of RT-PCR were consistent with those of TRAP staining (TRAP:F=323.829, P<0.05; NFATc1: F=197.148, P<0.05; Cathepsin K:F=431.888, P<0.05; MMP-9: F=448.821, P<0.05), as shown in Figure 4(B).Therefore, miR-483-5p andTimp2have targeted and antagonistic effects in vitro, and miR-483-5p can lead to overdifferentiation of OCs by decreasing the expression of Timp2.
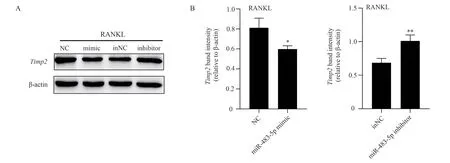
Fig 2 Western blot analysis

Fig 3 TRAP staining and RT-PCR analysis
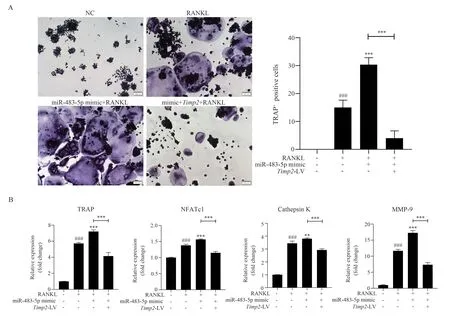
Fig 4 TRAP staining and RT-PCR analysis after co-transfection
4.Discussion
More and more studies have proved that the abnormal formation and activation of OCs under pathological conditions have a critical effect on bone destruction in various diseases[10].The differentiation process of OCs is affected by a variety of factors and hormones,including RANKL, M-CSF, sex hormones,etc.These factors as signaling molecules lead to the activation of multiple downstream signaling pathways, such as NFATc1 (main), MAPK, MITF, PI3K/AKT and NF-κB,etc.Thus regulating the differentiation and activity of OCs[11,12].In addition to these osteoclast promoting factors, there are some inhibitory regulatory factors, such as the soluble binding of osteoprotectin (OPG) to RANKL, resulting in the silencing of the activation of RANK/RANKL signaling pathway.Estrogen can indirectly inhibit bone resorption by stimulating OPG production[13].In addition, some pro-inflammatory cytokines can also promote the formation of OCs[14].
However, many studies believe that it is of great significance to interfere with the regulation of osteoclast-related miRNA during the formation of OCs.Since the discovery of the first miRNA named“lin4” in 1993, more and more studies have found many miRNAs in a variety of organisms, which can control a variety of biological processes, including organogenesis and development, hematopoietic function, apoptosis, proliferation and tumorigenesis, by negatively regulating the expression of multiple genes.miRNAs are key regulatory molecules that control cell proliferation, migration and cell differentiation[15].miRNAs are derived from RNA polymerase II, and the transcription sequence is derived from non-coding DNA sequences, introns or coding sequences in untranslated regions (UTRs)[16], and their regulatory gene functions at the posttranscriptional level[17].Many miRNAs have been identified as key regulators of lymphocytes and macrophages (e.g.miR146a or miR155)[18].
In the study on miR-483-5p, Wang et al.[19] reported that miR-483-5p could participate in the occurrence and development of osteoarthritis by regulating chondrocyte hypertrophy.Laura et al.[20]found that the expression level of miR-483-5p in cancellous bone samples from osteoporosis patients was significantly up-regulated compared with the control group through miRNA chip analysis.
In addition, studies have shown that miR-483-5p can contribute to the differentiation of monocytes into OCs[21].In this study, biological information software was first used to predict that miR-483-5p might have a targeting relationship with Timp2.Furthermore, the wild-type (WT) and mutant (MUT) dual luciferase reporter vector of Timp2 3 ‘-UTR was constructed, and the target of miR-483-5p was determined by RT-PCR detection and gene sequencing.Subsequently,target proteins were detected using miR-483-5p mimic and miR-483-5p inhibitor to artificially up-regulate or down-regulate miR-483-5p content in RAW264.7 cells.The results showed that miR-483-5p could affect the expression of Timp2 protein in RAW264.7 cells.Further, we explored the role of Timp2 in OCs differentiation.Combined with the two partial data, we preliminatively suggested that miR-483-5p could regulate the differentiation process of OCs and its osteoclast activity by targeting Timp2.Notably, subsequent co-treatment of RAW264.7 cells confirmed this idea, asTimp2significantly reversed the overactivation of OCs induced by miR-483-5p during the differentiation of RAW264.7 cells into OCs.Therefore, through the above series of in vitro experiments, we verified that miR-483-5p affects OCs differentiation and activity through regulation of Timp2.This finding is of great significance in intervening the regulation of osteoclast-related miRNA to affect the formation of OCs, and provides new potential ideas and strategies for the treatment of bone destruction diseases caused by excessive differentiation of OCs.
Author’s contribution:
Zeng Xiangzhou: Host of the project, responsible for experimental design, experimental guidance and paper revision and correction;Niu Tianqi: Responsible for experiment implementation, data processing and paper writing; Liu Caixia: Responsible for some data processing; Xiong Jun, Li Shuang, Deng Huiming: responsible for sample collection and sorting; Wang Hua, Jia Hao: Provide technical guidance for target gene prediction.
All authors declare no conflict of interest.
杂志排行
Journal of Hainan Medical College的其它文章
- Tapinarof inhibits proliferation and induces apoptosis of non‑small cell lung cancer A549 cell
- Protective effect of lycopene on Parkinson's disease cell model based on endoplasmic reticulum stress
- Chemical components and mechanism of Jiuwei Huaban pill in treating psoriasis
- Expression of miR-9-5p and RHOA in aluminum-induced rat cognitive dysfunction
- Intervention of Xuduan Zhongzi Formula on spermatogenesis epididymal morphological changes in a mice model of oligospermia
- Exploration of the molecular mechanism of Qishen decoction in regulating miR-495/FTO pathway mediated macrophage polarization to improve insulin resistance therapy of type 2 diabetes
