Tapinarof inhibits proliferation and induces apoptosis of non‑small cell lung cancer A549 cell
2023-12-02GAOErkeQIANFengJINQili
GAO Er‑ke, QIAN Feng, JIN Qi‑li
1.Bengbu Medical College, Bengbu 233004, China
2.The Second Affiliated Hospital of Bengbu Medical College, Bengbu 233004, China
Keywords:
ABSTRACT Objective: To explore the mechanism of tapinarof inhibiting proliferation and promoting apoptosis of non‑small cell lung cancer A549 cell.Methods: The proliferative ability of non‑small cell lung cancer cells A549 and H1299 was detected by clonal formation assay,and the cell viability of non‑small cell lung cancer cells A549 was detected by CCK‑8 assay.A549 cells were treated with different concentrations of tapinarof (0, 5, 10, 20) μmol/L for 24 h.The effects of tapinarof on the cell cycle of A549 were detected by flow cytometry using PI single dye method.Western blot assay was used to detect the effect of benzene‑moder on the expression of cycle‑associated protein P21 and CDK2.A549 cells were treated with different concentrations of tapinarof (0, 10, 20, 40) μmol/L for 48 h.Annexin V‑FITC and PI double staining were used to detect the effect of tapinarof on apoptosis of A549 cells.Western blot assay was used to detect the effects of different concentrations of tapinarof on expression of apoptosis‑related proteins, cleaved‑caspase 3 and cleaved‑PARP.Western blot analysis was performed to determine the expression of p‑AKT and FOXO1 in A549 cells at different concentrations (0, 5, 10, 20) μmol/L.Results: The results of clonal formation experiment showed that 40 μmol/L tapinarof could completely inhibit the proliferation of A549 and H1299 cells.CCK‑8 assay showed that compared with the control group (0 μmol/L), A549 cell activity decreased gradually with the increase of tapinarof concentration.A549 cells were stained with Annexin‑FITC/PI.Flow cytometry showed that the apoptosis rate of A549 cells increased with the increase of tapinarof treatment concentration (40 μmol/L tapinarof treatment, P<0.001).After treatment with tapinarof for 48 h, the protein expression of cleaved‑caspase 3 and cleaved‑PARP in A549 cells was increased, compared with control group (0 μmol/L).Flow cytometry after PI staining of A549 cells showed that tapinarof induced A549 cell arrest in G1 phase.In A549 cells treated with tapinarof for 24 h, the expression of P21 protein was increased and CDK2 decreased.The phosphorylation level of AKT was significantly inhibited and the expression level of FOXO1 was increased in A549 cells treated with phenylenmode at different concentrations.Conclusion: Tapinarof inhibits the proliferation of A549 cells and induces apoptosis, which may be related to the AKT/FOXO1 signaling pathway.
1.Introduction
Lung cancer is the cancer with the highest incidence and mortality among malignant tumors[1].Among the diagnosed lung cancer patients in our country, non‑small cell lung cancer accounts for about 80%[2].Unfortunately, the prognosis of NSCLC is poor.Many patients have developed to advanced stage or metastases when the disease is discovered, and the five‑year survival rate is only about 15%, which seriously affects the health and safety of human beings[3, 4].Chemotherapy is a common clinical treatment for non‑small cell lung cancer, but the sensitivity of chemotherapy drugs is low,the therapeutic effect is not ideal.With the emergence of molecular targeted drugs, the therapeutic effect of non‑small cell lung cancer has been significantly improved.However, gene mutation occurs in most patients after using targeted drugs, which leads to drug resistance.Therefore, new targeted drugs need to be developed to solve the corresponding treatment problems.
Aromatic hydrocarbon receptor (AhR) exists in cytoplasm and is dependent on ligand activation for its function.bHLH‑PAS is a member of the basic helix‑loop‑helix‑Per/Arnt/Sim (BHLH‑PAS)protein family.The bHLH domain is a common component that binds to DNA for transcription function, while the other two PAS domains bind to AhR receptors and form dimers with AhR nuclear translocation (ARNT), respectively[5].At rest, AhR forms complexes in the cytoplasm with two heat shock protein 90 (HSP90), hepatitis B X‑associated protein 2 (XAP2), and prostaglandin E synthetase 3 (P23)[6].After ligand binding and activation, AhR shuttles in the nucleus, translocation into the nucleus, and HSP90 is isolated during the shuttling process, thus exposing the PAS domain binding to AhR to regulate the expression of different heterologous biological reaction elements.In addition, AhR has been shown to be involved in the transcription of genes involved in proliferation, development,and immune regulation[7].Studies have confirmed that the activation of AhR inhibits the proliferation of hepatoma cells by increasing the expression of CDK inhibitor P27kip1[8].Studies have also shown that treatment with the AhR agonist Tetrachlorodibenzo‑p‑dioxin(TCDD) (0.1‑100 nM) can significantly reduce the colony formation and proliferation of human colon cancer cells[9].As an AhR agonist,canine has also been reported to inhibit tumor cell growth by activating AhR.Therefore, AhR may be one of the future targets for cancer treatment.
Benzomod is a novel AhR agonist derived locally from Gram‑negative bacilli with luminescent properties[10].The current Benzomod 1% Cream (VTAMA®) was approved by the FDA in May 2022 for the topical treatment of plaque psoriasis in adults.It specifically binds to and activates AhR in a variety of cell types,including CD4+T cells, HaCaT cells and human skin grafts[11, 12].Current studies have shown that benzomod regulates the expression of cytokines and skin barrier proteins as well as antioxidant activity by activating AhR, promoting the normalization of skin barrier[13,14].However, at present, the mechanism studies of benenmod at home and abroad are limited to its role in activating AhR, regulating immunity, anti‑inflammatory and maintaining the integrity of the skin barrier, and whether it has anti‑tumor effect on tumors remains unknown.This study elucidates the effects of benzomod on the proliferation and apoptosis of non‑small cell lung cancer A549 cells,and further explores its mechanism.
2.Materials and methods
2.1 Reagent
Tapinarof (CAS: 79338‑84‑4) was purchased from MedChemExpress, Shanghai, China; RPMI‑1640 medium, trypsin(containing EDTA and phenol red) purchased from Gibco, USA;Fetal bovine serum was purchased from Ex Cell Biologic.Penicillin‑streptomycin double antibody (100×), CCK‑8 reagent was purchased from Biosharp Corporation in China; Dimethyl Sulfoxide (DMSO)was purchased from Solarbio, China; 4% tissue cell fixative solution, crystal violet reagent, RIPA lysis solution, PMSF (100 mM),Western primary antibody diluent,Western secondary antibody diluent, BCA protein quantitative kit, HRP biotin labeled secondary antibody Rabbit antibody were purchased from Shanghai Biyuntian Company, China.PAGE Gel rapid preparation kit was purchased from Shanghai Yaenzyme Biological Company; PVDF transfer film(0.22 μm) was purchased from Millipore, USA.The rabbit primary antibodies GAPDH, β‑actin, cleaved‑caspase3, cleaved‑PARP,CDK2, and P21 were purchased from Cell Signaling Technology,Inc.AnnexinV‑FITC Apoptosis Kit, Cell Cycle Kit purchased from Lianke Biology, Hangzhou, China; Hifair® III 1st Strand cDNA Synthesis Kit (gDNA digester plus) was purchased from Yishengbio,Shanghai; Fluorescence quantitative PCR kit was purchased from Biosharp Corporation, China.Human non‑small cell lung cancer cell line A549 (US strain Preservation Center).
2.2 Cell Culture
Human non‑small cell cancer cells A549 cells were cultured in DMEM complete medium containing 10% fetal bovine serum and 1% penicillin‑streptomycin double antibody, and placed in an incubator containing 5% CO2at 37 ℃ for routine culture.The cells were changed or transcultured every 2 to 3 d.
2.3 Cell clonal formation experiment
A549 cells with good state were selected, digested and centrifuged,and the suspension concentration of fully cultured keynote cells was 2×103cells /mL.Each well of the 6‑well plate was inoculated with 900 cells, which were mixed and put back into the cell incubator for culture.After cell adhesion, A549 cells were treated with different concentrations (0, 2.5, 5, 10, 20, 40) μmol/L of benenmod,respectively, and then continued to culture.After the cells were observed to grow into clones with a microscope (about 9 d), the supernatant was discarded, the cells were lightly washed twice with PBS, and the residual medium was removed.600 μL of 4%paraformaldehyde was added to each well, and the fixed time was 20 min.The fixing solution was discarded and gently rinsed with PBS.550 μL crystal violet solution was added to each hole, and the staining time was 20 min.After rinsing the dye solution, dry it in a drying oven at 42 ℃.Photos were taken in bright light and the number of cell clones was counted.Each group had 5 multiple Wells and the experiment was repeated 3 times.
2.4 CCK-8 experiment
A549 cells with good state were selected, digested and centrifuged,and the cell concentration was adjusted to 5×105cells /mL.Each 96‑well plate was inoculated with 100 μL cell suspension, and PBS was added around to prevent liquid volatilization in cell plate holes.The culture was continued overnight or for more than 6 h.A549 cells were treated with 8 groups of benzomod (0, 5, 10, 20, 40, 60,80, 100) μmol/L, respectively, and cultured for 24 h.A mixture containing 10% CCK‑8 reagent was prepared away from light.The drug‑containing culture medium of the supernatant was discarded,and 100 μL mixed solution was added to each well and returned to the incubator for further culture for about 40 min.Set the single wavelength of the enzyme marker to 450 nm, accurately measure and record OD values of each hole one by one.Cell viability of each group was calculated.The experiment was repeated three times.(Cell viability =[(experimental hole‑blank hole) /(control hole‑blank hole)×100%).
2.5 Cell cycle experiment
A549 cells in a good state were selected, digested and centrifuged,and the concentration of cell suspension was adjusted to 1×106cells /mL.Each 6‑well plate was inoculated with 400 μL, and then filled with complete medium to 2 mL and placed in an incubator for further culture.A549 cells were treated with different concentrations of benzomod (0,5,10,20) μmol/L and cultured for 24 h.The cells were collected by digestion centrifugation.The precooled fixative(PBS: anhydrous ethanol =4:3) was added to the cells at the bottom of the tube and carefully blown away.Put in the ‑20 ℃ refrigerator overnight (can be stored for one month before serving).Centrifuge the fixed cells and discard the upper fixed solution before loading the machine.Add 2 mL PBS to hydrate the cells again and centrifuge the supernatant.300 μL DNA staining solution was added, blown well,incubated for half an hour under 4 ℃ dark staining, and detected by upflow cytometry.
2.6 Cell apoptosis experiment
A549 cells in a good state were selected, digested and centrifuged,and the concentration of cell suspension was adjusted to 1×106cells/ml.Each 6‑well plate was inoculated with 300 μL, and then filled with complete medium to 2 mL, and incubated overnight or for more than 6 h in an incubator.A549 cells were treated with different concentrations of benzomod (0,10,20,40) μmol/L and cultured for 48 h.Digestive centrifugation collects cells.Set the centrifuge speed at 800 r/min, centrifuge for 5 min, discard the supernatant to collect precipitation.Wash 2 times using pre‑cooled PBS.Cells were mixed with 500 μL 1× binding buffer, then 10 μL Annexin V‑FITC and 8 μL propyl iodide (PI) fluorescent dye were added into the cell suspension of each group, and mixed gently.Incubate at room temperature and away from light.5 min later, flow cytometry was performed.PI negative and PITC positive were early marcescent cells (upper right quadrant) and double positive were late marcescent cells (lower right quadrant).The apoptosis rate was the sum of the two.
2.7 Real‑time fluorescence quantitative PCR experiment
2.7.1 Cell RNA extraction and reverse transcription
Select the cells of better growth state, digestion centrifugation,planking.After adherent cells were added with 40 μmol/L benzomod,the culture was continued for 48 h.Discard supernatant, add PBS moistening wash twice.Add 1 mL Trizol to each well, gently blow ten times to fully lysate the cells, collect in EP tube, and leave at room temperature for 5 min.Add 200 μL chloroform, shake until pink and cloudy, and leave at room temperature for 2~3 min.After centrifugation at 4 ℃ 12 000 g for 15 min.Water absorption phase into new EP tube.Add 500 μL isopropyl alcohol, shake well and put on ice for 5‑10 min.After centrifugation at 4 ℃ 12 000 g for 10 min.The supernatant is emptied and the RNA is left at the bottom of the tube (white sediment is visible to the naked eye).75% ethanol was added and the EP tube was gently shaken to suspend the RNA.After that, the centrifuge was centrifuged at 7 500 g at 4 ℃ for 5 min.Repeat twice.Dry the ethanol at room temperature and add DEPC water.RNA concentration and purity were measured.Store in ‑80 ℃refrigerator.The cDNA was obtained according to the instructions of Shanghai Yi Sheng Biological reverse transcription Kit and stored at‑20 ℃.
2.7.2 Fluorescence quantitative PCR experiment
Primers included Shanghai Biosynthesis, upstream GAPDH sequence: TCAAGAAGGTGGTGAAGAG, downstream GAPDH sequence: AGGTGG AAG AAT GGG AGT TG; Bcl‑2 upstream sequence: TCG CCC TGT GGA TGA CTG AGT AC, Bcl‑2 downstream sequence: ACA GCC AGG AGA AAT CAA ACA GAG G; Bax upstream sequence: AGC GAC TGA TGT CCC TGT CTC C, Bax downstream sequence: AGA TGG TGA GTG AGG CGG TGA G; FasL upstream sequence: TTC ATG GTT CTG GTT GCC TTG GTA G, FasL Upstream sequence: GCT GTG TGC ATC TGG CTG GTA G.A reaction system of 20 μL was prepared, and the whole process was operated on the ice away from light (3 multiple holes were set).The cDNA template was diluted at 1:20, and the reaction system of each tube sample was added as follows: Add 10 μL to SYBR Green qPCR Mix, 0.4 μL to each upstream and downstream primer (10 mM), 2 μL to cDNA template,and 7.2 μL to double steaming water.After adding, oscillate on the vortex oscillator for 20 s to mix and transient.It was put into the fluorescence quantitative PCR instrument, and the relevant parameters were adjusted using the three‑step reaction procedure, as follows: predenaturation at 95 ℃, time 3 min, denaturation at 95 ℃for 10 s, annealing at 60 ℃ for 30 s, extension at 72 ℃ for 30 s, and cycling 40 times.Finally, the dissolution curve was set according to the recommended procedure of Roche instrument.The ct values of each sample were derived, and the relative expression levels of target genes were calculated by 2‑△△ctmethod, and normalized for statistical analysis.
2.8 Western Blot Protein Western blotting
2.8.1 Cell protein extraction
Discard the medium in the upper layer of the 6‑well plate and wash the cells with pre‑cooled PBS twice.An appropriate amount of RIPA lysate containing 1% PMSF was added, about 80~120 μL per well.Crack in ice for 2 min.Scrape the cells clockwise and backward with the cell scraper, and absorb the cell mucus into the EP tube.Leave on the ice for 20 to 30 min.Centrifuge at 12 000 rpm at 4 ℃ for 30 min.Absorb supernatant into EP tube to obtain protein solution.Can be temporarily stored in ‑80 ℃ refrigerator.In the follow‑up experiment, the protein concentration was determined by BCA protein kit.
2.8.2 SDS‑PAGE Gel electrophoresis
Gel was prepared according to the instructions of the gel kit on PAGE.5×protein loading buffer was evenly mixed into the protein sample and diluted into 1×protein loading buffer.Boil sample in metal bath at 100 ℃ for 10 min.Denatured can be directly subjected to SDS‑PAGE electrophoresis, or stored at ‑20 ℃.Add 20~50 μg to each well according to protein expression content.Fill the electrophoresis tank with electrophoresis solution.Run concentrated glue at constant pressure of 80 V for 30 min.After increasing the voltage to 110 V, run the separation glue for 1 h.PVDF membrane was cut into a certain size and activated in methanol for more than 10 seconds.Then carefully clip into the pre‑cooled transfer liquid foam.Open the transfer clip, put the black side on the bottom,spread the sponge and filter paper in turn, transfer the cut gel to the filter paper, and then carefully clip PVDF film (no creases), spread on the gel surface, with the stripper plate carefully remove bubbles,finally spread the filter paper and sponge in turn, clip up the film clip, put into the transfer slot.The black side of the clip faces the negative terminal and the white side faces the positive terminal.The bottom of the foam box is covered with thin ice, and then put into the transfer tank, and then pour the pre‑cooled 1× transfer liquid into the tank.Put the lid on, fill the foam box with ice cubes, adjust the constant current 200 mA, and turn the film for 2 hours.At the end of the transmembrane, the transmembrane clip was opened and the PVDF membrane was extracted with tweezers and marked.Closed at room temperature for 1‑2 h.After closure, make corresponding marks and incubate primary antibody in 4 ℃ refrigerator overnight.After applying primary antibody, the film was washed 3 times with 1×TBST shock, once 10 min.PVDF membrane was placed in secondary antibody prepared according to 1:400, and the secondary antibody was incubated at room temperature for 1 ~2 h.After applying the second antibody, the film was washed 3 times with 1×TBST shock, once 10 min.Under the condition of avoiding light,PVDF membrane was put into the prepared developer solution (ECL A liquid: B liquid =1:1).The developer was evenly coated with the membrane with pipetting gun, and each strip was displayed with Tanon‑5200 exposure instrument.Image j software was used to analyze the gray value of each sample.
2.9 Statistical processing
All experiments in this study were repeated for more than 3 times,and the experimental data were represented by mean ± standard error(SEM).If the measurement data conform to the normal distribution test, the difference between the two groups is compared by the t test of two independent samples.One‑way analysis of variance was performed for the two groups.If the normal test was not met,the rank sum test was used to compare the differences among the groups.Statistical analysis was performed using GraphPad Prism 8.0 software.P<0.05 was considered statistically significant, P<0.05 was not considered statistically significant.
3.Results
3.1 Benzomod inhibits proliferation of non‑small cell lung cancer cells
The chemical formula of benzenmod is shown in FIG.1A.Cell lines A549 and H1299 were treated with different concentrations of benzenmod for about 8 d.The number of clones formed by A549 and H1299 cells was significantly inhibited (P<0.01).As the concentration of benzomod gradually increased, the inhibition effect became more obvious, in a concentration‑dependent manner, as shown in Figure 1B‑D.CCK‑8 results showed that the cell viability of A549 and H1299 cells was significantly inhibited after 24 h treatment with different concentrations of benenmod (P<0.01) in a concentration‑dependent manner.Among them, high concentration of benzomod could significantly inhibit the activity of A549 cells(P<0.01), as shown in Figure 1E‑F.
3.2 Apoptosis of A549 cells induced by benzomod
A549 cells were treated with different concentrations of benzomod(0, 10, 20, 40) μmol/L for 48 h, and the apoptosis rate was analyzed by flow cytometry with Annexin V‑FITC double staining, as shown in Figure 2A.The results showed that compared with the control group (0 μmol/L), the apoptosis rate of A549 cells was increased by 20 and 40 μmol/L concentrations of benenmod (P<0.01) in a concentration‑dependent manner, as shown in Figure 2B‑C.
3.3 Benzomod regulates the expression of apoptosis-related proteins and gene mRNA in A549 cells

Fig 1 Tapinarof inhibits the proliferation of non‑small cell lung cancer cells
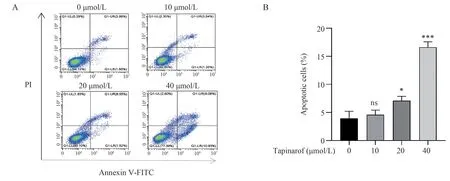
Fig 2 Tapinarof induces apoptosis of A549 cells
A549 cells were treated with different concentrations of benzomod(0,10,20,40) μmol/L for 48 h.Compared with the 0 μmol/L group,benzomod at 20 and 40 μmol/L concentrations significantly increased expression levels of cleaved‑caspase 3 and cleaved‑PARP(P<0.01) in a dose‑dependent manner, as shown in Figure 3A‑C.Fluorescence quantitative PCR results showed that compared with 0 μmol/L group, 40 μmol/L concentration of benenmod treatment group decreased the expression of anti‑apoptotic gene Bcl‑2 mRNA,and increased the expression of pro‑apoptotic genes Bax, Bad and Fasl mRNA (P<0.01), as shown in Figure 3D.
2.4 Benzomod induced A549 cell arrest in G1 phase
A549 cells were treated with different concentrations of benenmoder (0,5,10,20) μmol/L for 24 h.The results showed that the ratio of G1 phase cells in the 0 μmol/L group was 61.2%, that in the 10 μmol/L group was 66.88%, and that in the 20 μmol/L group was 75.72%, as shown in Figure 4A.Statistically, compared with 0 μmol/L group, 10 μmol/L group and 20 μmol/L group, the percentage of G1 phase cells was significantly increased (P<0.01),the cell ratio of S phase and G2 phase decreased, as shown in Figure 4B.These results suggest that benzenmodde induces A549 cell line to be blocked in G1 phase.After 24 h treatment of A549 cells with different concentrations of benzomod, we detected the expression changes of cycl‑dependent protein P21 and CDK2 by Western Blot assay.The results showed that benzomod could up‑regulate the expression of P21 and down‑regulate the expression of CDK2(P<0.01).Figure 5A‑C.
3.5 Benzomod inhibited AKT phosphorylation and increased FOXO1 levels in A549 cells
In addition, AKT plays an important role in cell proliferation,so Western Blot analysis was performed to detect the expression of total AKT, p‑AKT, and FOXO1 protein.The results showed that benzomod decreased the expression of p‑AKT in A549 cells compared with the 0 μmol/L group (P<0.01), increased FOXO1 protein expression (P<0.01), while total AKT did not change significantly, as shown in Figure 6A‑C.

Fig 3 Tapinarof regulates the expression of apoptosis related proteins and genes in A549 cells

Fig 4 Tapinarof induces cell cycle arrest in A549

Fig 5 Expression of P21 and CDK2 proteins associated with G1 phase of the cell cycle
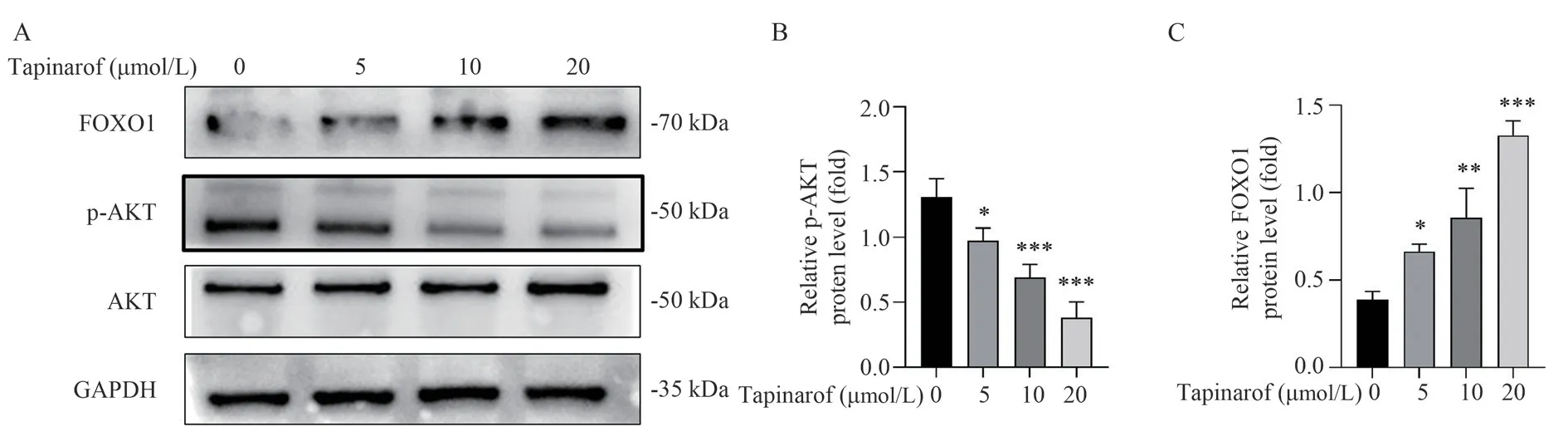
Fig 6 Effects of tapinarof on expression of p‑AKT and FOXO1 protein.
4.Discussion
In this study, benzomod significantly inhibited clonal formation and cell viability of non‑small cell lung cancer cells, induced G1 phase arrest of cell cycle, and promoted apoptosis.In non‑small cell lung cancer cells treated with benenmod, protein expressions were up‑regulated, cleaved caspase 3 and cleaved PARP, mRNA expressions of anti‑apoptotic genes Bcl‑2 were decreased, and mRNA expressions of pro‑apoptotic genes Bax, Bad, and FasL were increased.In terms of cell cycle phase progression, benenmode induced cell cycle arrest in G1 phase and upregulated the expression of P21 protein while inhibiting the expression of CDK2 protein.In a further mechanistic study, Western Blot results showed that p‑AKT expression decreased while FOXO1 expression increased.Therefore,the antitumor effect of benenmod may be related to the AKT/FOXO1 signaling pathway.These results suggest that benzomod has antitumor effects on A549 cells.
Benzomod is a small molecule AhR agonist that specifically binds and activates AhR[15].Studies have shown that benenmoder can directly bind to and activate a variety of cell types in vitro and in vivo, promote the expression of a variety of metabolism related genes such as CYP1A1 and CYP1B1 in cells, and induce nuclear translocation of AhR[12].However, whether it has antitumor effect in tumor treatment has not been reported.In this study, we found that benenmod can significantly inhibit the proliferation and induce apoptosis of non‑small cell lung cancer cells, which is of great significance for the use of benenmod drugs in anti‑tumor.
Caspase cascade signaling plays an important role in endogenous and exogenous cell apoptosis.Among its members, caspase 3 plays an irreplaceable role in initiating apoptosis, and its catalytic high activation is a common initiator of apoptosis resulting in cell death.PARP is mainly cleaved by caspase 3 in vivo, which is a key enzyme in detecting DNA damage.Therefore, the cleavage product of PARP is also considered to be a biological marker of apoptosis.Studies have shown that the natural product traline can effectively induce the apoptosis of gastric cancer cells by activating caspase 3 and PARP.In this study, it was found that after Tapinaorf was treated with A549 cells, the expression of cleaved‑caspase 3 and cleaved‑PARP were significantly up‑regulated.Bcl‑2, Bcl‑xl, Bax and Bad are important members of the Bcl‑2 family, which mainly mediate endogenous cell apoptosis (mitochondria‑dependent), and their balance regulates apoptosis.An increase in the ratio of Bax to Bcl‑2 would cause mitochondria to release cytochrome c, which would then lyse caspase 3 and RARP, leading to cell apoptosis[16].Studies have confirmed that PAHs 7, 12‑dimethylbenzene [a] anthracene can up‑regulate the expression of pro‑apoptotic molecule Bax by targeting AhR in oocytes, leading to oocyte apoptosis[17].On the other hand,Fas/FasL play a key role in exogenous cell apoptosis.When the death receptor is stimulated by external factors, the two combine to activate caspase 8 and caspase 3, and the activated caspase 3 cleaves proteins, leading to cell apoptosis[18].In this study, we found that the mRNA levels of proapoptotic genes Bax, Bad and FasL were up‑regulated, while the mRNA levels of anti‑apoptotic genes Bcl‑2 were down‑regulated in A549 cells after phenylenmod treatment by fluorescence quantitative PCR.These results indicate that benenmod can induce apoptosis of non‑small cell lung cancer cells through intrinsic and extrinsic pathways and inhibit cell proliferation.
Many compound strategies for tumor therapy focus on inducing cell cycle arrest[19, 20].The cell cycle has four distinct phases, including G1, S, G2, and M phases, which are usually regulated by several cyclin dependent kinases (CDKS)[21].The G1/S phase cell cycle transition is mainly regulated by cyclin /CDK complexes, cyclin‑CdK4/6 (early stage) and cyclin‑CdK2 (late stage), which have been shown to regulate cell cycle by promoting RB (retinoblastoma)phosphorylation[22].P21 protein can inhibit the expression of CDK2 protein and is a negative regulator of G1/S phase transition, inducing cell arrest in G1 phase.Studies have confirmed that morulin and antipsychotropic florin hepatocellular carcinoma HepG7 and Huh1 cells up‑regulate the expression of P21 protein while inhibit the expression of CDK2, thus blocking the cell cycle in G1 phase[23,24].It was also shown in this study that benenmode increased the expression of P21 protein and inhibited the expression of CDK2,promoting non‑small cell lung cancer cell arrest in G1 phase.
AKT plays an important role in tumor cells, including cell survival, proliferation and apoptosis.Phosphorylated AKT (p‑AKT)is involved in the regulation of cell proliferation, apoptosis and metastasis[25].Overexpression of p‑AKT is considered to be a target for tumor therapy.For example, it has been reported that high expression of p‑AKT is positively associated with cancer metastasis in patients with breast cancer, thyroid cancer, and colorectal cancer [26].Studies have also shown that adiponectin activates AKT signaling pathway to protect myocardium by preventing myocardial cell apoptosis, and curcumin can inhibit the growth of glioblastoma cells by reducing the expression of p‑AKT[27, 28].In this study, benzomod had no significant effect on the expression of total AKT, but decreased the activity of p‑AKT, suggesting that its antitumor activity against NSCLC was related to the activation of p‑AKT.During the cell cycle of cancer, FOXO1 signals can target P21 protein to inhibit cell proliferation[29].Studies have shown that overexpression of FOXO1 in PC3 and LNCaP cells of prostate cancer reduces cell proliferation[30].In the present study, the expression of p‑AKT and FOXO1 was decreased and increased after benenmoder treatment in non‑small cell lung cancer cells, suggesting that benenmoder’s inhibition of proliferation and induction of apoptosis in non‑small cell lung cancer cells may depend on the AKT/ FOXO1 signaling pathway.
In conclusion, this study found that benenmod could inhibit the proliferation of A549 and H1299 cells of non‑small cell lung cancer,and induce G1 phase arrest and apoptosis.The mechanism may be related to AKT/FOXO1 signaling pathway.This study revealed that benzomod may be a potential anti‑tumor drug, but its specific mode of action and mechanism has not been clarified in this study, and further research is needed.
Author’s Contribution:
Gao Erke, completed experiments, analyzed data, wrote papers,collected and analyzed literature.Qian Feng: Research guidance,paper modification; Jin Qili: Thesis writing supervisor
All authors declare no conflict of interest.
杂志排行
Journal of Hainan Medical College的其它文章
- Research progress on key genes of vitamin D signaling pathway
- Research progress on the influence of local hemodynamics on carotid atherosclerosis
- Copy number variation sequencing for diagnosis of cytomegalovirus infection based low‑depth whole‑genome sequencing technology in fetus: Three cases and literature review
- Exploration of the molecular mechanism of Qishen decoction in regulating miR-495/FTO pathway mediated macrophage polarization to improve insulin resistance therapy of type 2 diabetes
- Intervention of Xuduan Zhongzi Formula on spermatogenesis epididymal morphological changes in a mice model of oligospermia
- Expression of miR-9-5p and RHOA in aluminum-induced rat cognitive dysfunction
