LINC00662 affects the sensitivity of hepatocellular carcinoma cells to sorafenib drug by regulating miR‑106a‑5p/CAV1 axis
2023-11-14CHENBocenLIANGNaYANDongjingCHENTongXIAOManCAIWangwei
CHEN Bo‑cen, LIANG Na, YAN Dong‑jing, CHEN Tong, XIAO Man, CAI Wang‑wei
Key Laboratory of Biochemistry and Molecular Biology, Hainan Medical University, Haikou 571199, China
Keywords:
ABSTRACT Objective: To investigate the mechanism of long non‑coding RNA‑LINC00662 on induction of sorafenib resistance in hepatocellular carcinoma (HCC) cells.Methods: HCC cells(HepG2, HCCLM3), sorafenib‑resistant hepatocellular carcinoma cells HCC‑SR (HepG2‑SR,HCCLM3‑SR) were investigated by real‑time fluorescence quantitative polymerase chain reaction (RT‑qPCR) and Western blot was used to detect LINC00662, miR‑106a‑5p and cavitin‑1 (CAV1) expression in each group of cells.106a‑5p and cavitin‑1 (CAV1) expression levels were measured by RT‑qPCR and Western blot.The si‑LINC00662 and miR‑106a‑5p mimics were transfected with HCC‑SR cells, respectively, and cell sensitivity to sorafenib drug was detected by cell activity kit (CCK‑8).And the targeting relationship between LINC00662 and miR‑106a‑5p, miR‑106a‑5p and CAV1 was further determined by dual luciferase reporter assay, RT‑qPCR, and Western blot.Results: The relative expression of LINC00662 and CAV1 was significantly increased and miR‑106a‑5p expression was significantly decreased in HCC‑SR cells (P<0.01, P<0.001); interference with LINC00662 expression or overexpression of miR‑106a‑5p significantly increased the sensitivity of HCC‑SR cells to sorafenib drug(P<0.05, P<0.01).And LINC00662 targeted to negatively regulate miR‑106a‑5p expression and miR‑106a‑5p targeted to negatively regulate CAV1 expression (P<0.05).Conclusion:LINC00662 could act as a competitive endogenous RNA (ceRNA) of miR‑106a‑5p to promote the expression of CAV1 and mediate the resistance of sorafenib in HCC cells.Interfering with LINC00662 expression can inhibit sorafenib resistance and increase sorafenib drug sensitivity in HCC cells.✉Corresponding author: CAI Wang‑wei, Professor
1.Introduction
Liver cancer is the fifth most common cancer worldwide, with nearly one million new cases and deaths each year[1].Hepatocellular carcinoma (HCC) accounts for over 90% of primary liver cancers and is the second most common cause of cancer‑related deaths, and is currently one of the most common and deadly cancers, making it a major global public health problem[1].Significant advances have been made in the prevention, diagnosis and treatment of HCC, and with the implementation of targeted agents and immunotherapy,HCC‑related adverse outcomes are changing significantly and patient life expectancy has improved[2].In the last decade, the traditional first‑line treatment has been with sorafenib (Sorafenib)or lenvatinib[3].Of these, Sorafenib has also been the mainstay of treatment.Sorafenib is an oral multi‑targeted kinase inhibitor that inhibits malignant tumour growth by suppressing tumour cell proliferation and is the first first‑line agent to improve survival in patients with raw advanced HCC[4].70% of patients who relapse within the first 3 years of initial treatment have early stage HCC that is usually resectable, however, clinical sensitivity to development of sorafenib drug sensitivity is becoming more common and the quality of survival of HCC patients is greatly reduced[5-7].The aim of this study was to identify the factors that contribute to drug resistance and ways to mitigate it.Therefore, understanding the mechanisms that lead to drug resistance is an effective way to improve the survival of patients with HCC.
Long non‑coding RNAs (lncRNAs) have many biological functions and play a critical role in many human diseases.An increasing number of studies have shown that lncRNAs play an important role in the development of cancer, and that abnormal expression of lncRNAs is closely related to cell proliferation, differentiation,invasion, migration, epithelial‑mesenchymal transition (EMT), and angiogenesis[8].Recent studies have found that lncRNAs are involved in drug metabolism and are closely related to drug resistance[9,10].Therefore, studying the relationship between lncRNA and tumour drug resistance will help to elucidate the biological mechanism of lncRNA in the development of tumour drug resistance and provide a new basis for improving the effectiveness of drug therapy.
The long‑stranded non‑coding RNA LINC00662 is a sequence of 2 085 length encoded by human chromosome 19q11.In recent years, it has been shown that LINC00662 is an oncogenic lncRNA and an important regulator of cancer cell proliferation, migration and invasion[11].However, the role of LINC00662 in the development of drug resistance in hepatocellular carcinoma cells is not fully understood.In this study, we investigated the effect of LINC00662 on sorafenib resistance in hepatocellular carcinoma cells and the associated mechanisms.
2.Materials and methods
2.1 Materials
Human hepatocellular carcinoma HCC cell lines (HepG2,HCCLM3) were obtained from Professor Fengtian He’s research group at the Third University of Medical Sciences of the Army;DMEM basic medium and fetal bovine serum were purchased from Gibco (C11965500B, USA); miRNA reverse transcription kit,internal reference U6 validation primers, miRNA mimics (miR‑106a(QP113, HmiR‑SN0026); total RNA extraction kit (LS1040) from Eastep®Super, USA; cDNA synthesis kit and real‑time quantitative PCR amplification kit (11111ES92, 11202ES03) from Yisheng,Shanghai, China.The small interfering RNA (siRNA) was purchased from Shanghai Abbott Biotechnology Co.DOJINDO, item no.CK04; CAV1 rabbit polyclonal antibody, internal reference GAPDH rabbit monoclonal antibody and sheep anti‑rabbit IgG secondary antibody were purchased from Shanghai Able Antibiotics.
2.2 Experimental methods
2.2.1 Cell culture
HepG2 and HCCLM3 hepatocellular carcinoma cells were cultured in DMEM medium containing 10% fetal bovine serum at 37 ℃ in a cell culture incubator with 5% CO2.When the cells reached 80%fusion, 0.5 mL of trypsin was added to digest the cells until they were deformed and dislodged, and the digestion was terminated by adding complete medium.
2.2.2 CCK‑8 assay for cell viability
HCC cells were counted using a cell counter, inoculated into 96‑well plates at 2 500 cells/well and incubated in the incubator for 48 h.10 μL of CCK-8 solution was added to each well.The absorbance(OD) value at 450 nm was recorded.Cell survival rate = OD of experimental group/OD of control group × 100%
2.2.3 Cell transfection
Cells in logarithmic phase were collected and inoculated in 6‑well plates at a concentration of 1 × 106cells per well.si‑NC,si‑LINC00662, mimics‑NC, miR‑106a‑5p mimics and si‑CAV1 were transfected with Lipo8000™ transfection reagent at 70% of the bottom of the flask and the cells were collected after 48 h for subsequent analysis.
2.2.4 Dual luciferase reporter gene assay
The cell culture plates to be tested were removed from the incubator and the appropriate reagents were added for the assay.The absorbance of firefly luciferase and sea kidney luciferase was measured.
2.2.5 Real‑time fluorescence quantitative PCR ( RT‑qPCR)
Detection of LINC00662, miR‑106a‑5p and CAV1 mRNA expression.Total RNA was first extracted from the cells using the kit.Reverse transcription was performed using the miRNA Reverse Transcription Kit, cDNA Synthesis Kit and RT‑PCR was performed using qPCR SYBR Green Master Mix reagent with the following primer list.

Tab 1 Primer sequence
2.2.6 Western blotting to detect CAV1 protein expression
BCA kits were used to detect protein concentration.SDS‑PAGE gel electrophoresis was performed on 10% gels.Afterwards, the membranes were transferred at a constant flow of 200 mA for 2 h.After transfer, the PVDF membranes were immersed in the purchased closure solution for 30 min at room temperature and incubated overnight at 4 ℃ in a shaker with diluted primary antibody(primary antibody dilution ratio: CAV1, 1:1 000 and then HRP‑labelled sheep anti‑rabbit secondary antibody or sheep anti‑mouse secondary antibody).Then add HRP‑labelled sheep anti‑rabbit secondary antibody or sheep anti‑mouse secondary antibody and incubate for 2 h (both dilutions 1:5 000).The bands were scanned using a chemiluminescence system, and the Image J software was used to analyse the grey scale values of the bands, and GAPDH was used as an internal reference for quantification.
2.2.7 Statistical analysis Data were expressed as mean ± standard deviation and differences between groups were analysed after t‑test using prism 6.0 software,with P < 0.05 considered statistically significant between the two groups.
3.Results
3.1 Establishment of HepG2 and HCCLM3 sorafenib‑resistant cell lines
The IC50 of HepG2 and HCCLM3 cells against sorafenib was determined by the CCK8 method, and then the IC50of sorafenib‑resistant strain HepG2‑SR and sorafenib‑resistant strain HCCLM3‑SR against sorafenib was compared to determine the resistance index RI.The experimental results revealed that the IC50of HepG2 and HCCLM3 against sorafenib at the concentrations of 10+0.3 μmol/L and 8+0.12 μmol/L, and the IC50concentrations of sorafenib for HepG2‑SR and HCCLM3‑SR were further verified to be 40+0.22 μmol/L and 30+0.15 μmol/L, respectively, so the resistance index IR of HepG2‑SR was 4.00+1.40 t=3.19 and HCCML3 ‑SR had a resistance index IR of 3.75+0.08 t=3.09, indicating that the sorafenib‑resistant cell lines were successfully constructed, and the results are shown in Figure 1.

Fig 1 Cell survival rate of HepG, HCCLM3 and their resistant strains at different concentrations of sorafenib
3.2 LINC00662 can adsorb miR‑106a‑5p, and miR‑106a‑5p can target and regulate the expression of LINC00662 and CAV1 mRNAs
There is increasing evidence that lncRNAs can act as competing endogenous RNAs (ceRNAs) that interfere with the function of miRNAs and thus with the expression of miRNA‑targeted genes.To investigate the mechanism of action of LINC00662, the bioinformatics website LncBase Predicted v.2 was used to predict the miRNAs bound by LINC00662, and the results showed the presence of a binding site between LINC00662 and miR‑106a‑5p, suggesting that LINC00662 has a role in adsorbing miR‑106a‑5p See Fig.2a.Further targeting of miR‑106a‑5p was predicted by Targetscan, which showed a binding site between miR‑106a‑5p and CAV1-3’-UTR, see Fig.2b.Dual luciferase reporter assays showed that transfection of miR‑106a‑5p mimics compared to transfection of miR‑NC decreased WT LINC00662 and WT‑CAV1 relative luciferase activity with statistically significant differences (t=10.11,P<0.01 and t=11.21, P<0.01, respectively), whereas no significant changes were observed for MUT‑LINC00662 ( MUT‑CAV1)relative luciferase activity, see Figure 2c,d.The results indicated that LINC00662 inhibited the expression of miR‑106a‑5p through adsorption, while miR‑106a‑5p could target binding to LINC00662 and CAV1 mRNA and inhibit the expression of LINC00662 and CAV1.
3.3 Expression of LINC00662, miR‑106a‑5p, and CAV1 in sorafenib‑resistant cell lines
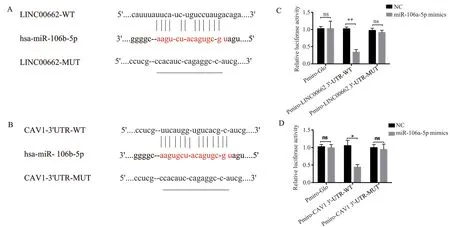
Fig 2 Dual luciferase reporter assay verifies miR‑106a‑5p targeting and regulation of LINC00662 and CAV1
To investigate the relationship between LINC00662, miR‑106a‑5p, CAV1 and sorafenib resistance, we analyzed the expression of LINC00662 in HepG2, HCCLM3, HepG2‑SR and HCCLM3‑SR cells, and the results showed that, compared with their control cells HepG2, HCCLM3 The results showed that the relative expression of LINC00662 in HepG2‑SR and HCCLM3‑SR cells was significantly higher compared to their control cells HepG2 and HCCLM3,and the differences were statistically significant (t=24.74, P<0.01 and t=51.70, P<0.01, respectively); the analysis of miR‑106a‑5p showed that the expression level of miR‑106a‑5p in HepG2‑SR and HCCLM3‑SR cells was significantly lower than their control cells HepG2‑SR and HCCLM3‑SR cells.The results of the analysis of miR‑106a‑5p showed that the expression levels of miR‑106a‑5p in HepG2‑SR and HCCLM3‑SR cells were significantly lower than those in their control cells HepG2 and HCCLM3, and the differences were statistically significant (t=13.59, P<0.001 and t=17.63, P<0.001, respectively); further analysis of the expression levels of CAV1 showed that the relative expression levels of CAV1 in HepG2‑SR and HCCLM3‑SR cells were significantly higher,and the differences were statistically significant (t=27.72, P<0.001 and t=27.58, P<0.001, respectively), and the protein level of CAV1 was significantly higher than that of its control cells, HepG2 and HCCLM3, with statistically significant differences (t=30.40,P<0.001 and t=16.89, P<0.01, respectively), the results are shown in Figure 3.The results indicated that There was a negative correlation between the expression of LINC00662 and miR‑106a‑5p in HepG2‑SR and HCCLM3‑SR cells and a positive correlation with the expression of CAV1, suggesting that the relationship between the expression of LINC00662, miR‑106a‑5p and CAV1 may be related to the drug resistance of sorafenib.
3.4 Inhibition of LINC00662 expression increases the sensitivity of HCC‑SR to sorafenib
To verify the effect of LINC00662, we used siRNA to study the effect of LINC00662 on the expression of miR‑106a‑5p and CAV1 as well as sorafenib resistance in hepatocellular carcinoma cells.The results showed that the expression of LINC00662 in HepG2‑SR and HCCLM3‑SR cells was significantly lower in the si‑LINC00662 group compared with the si‑NC group, with statistically significant differences (t=30.02, P<0.01 and t=19.94, P<0.05, respectively),while the expression of miR‑106a‑5p was significantly higher, with statistically significant differences.The differences were statistically significant (t=30.02, P<0.01 and t=19.94, P<0.05); analysis of CAV1 expression showed that CAV1 protein expression in HepG2‑SR and HCCLM3‑SR cells in the si‑LINC00662 group was significantly lower than that in their control group, and the differences were statistically significant (t=31.06, P< 0.01 and t=29.54, P<0.01), see Figure 4a,b,c.The IC50concentrations of sorafenib in HepG2‑SR and HCCLM3‑SR cells with knockdown LINC00662 were 30+0.25 μmol/L and 24+0.14 μmol/L, which were lower than those in their control groups, and the differences were statistically significant(t=3.06, P< 0.05 and t=2.14, P<0.05,), and the results are shown in Figure 4d,e.The results suggest that knockdown of LINC00662 can lead to reduced expression of CAV1 and can increase the sensitivity of hepatocellular carcinoma cells to sorafenib.

Fig 3 Expression of each gene in HepG2, HCCLM3 and corresponding drug‑resistant strains
3.5 Overexpression of miR‑106a‑5p increases the sensitivity of HCC‑SR to sorafenib
To confirm the role of miR‑106a‑5p, we investigated the effect of miR‑106a‑5p expression on sorafenib resistance in hepatocellular carcinoma cells.Compared with its effect on their control mimics‑NC, the level of CAV1 protein expression was reduced in HepG2‑SR and HCCLM3‑SR cells transfected with miR‑106a‑5p mimics,with statistically significant differences (t=28.40, P<0.01 and t=28.59, P<0.01), see Figure 5a,b,c, and the sensitivity to sorafenib sensitivity was significantly higher, reaching IC50 at 29+0.15 μmol/L and 19+0.04 μmol/L for sorafenib, with statistically significant differences (t=2.41, P<0.05 and t=2.39, P<0.05, respectively), and the results are shown in Figure 5d,e.The results indicated that miR‑106a‑5p could improve the sensitivity of HCC‑SR sensitivity to sorafenib.

Fig 4 siLINC00662 increases the sensitivity of HepG2‑SR and HCCLM3‑SR cells to sorafenib

Fig 5 Overexpression of miR‑106a‑5p increases the sensitivity of HepG2‑SR,HCCLM3‑SR to sorafenib
4.Discussion
LINC00662 has been found to be associated with tumourigenesis,progression and treatment resistance[12].Among them, the expression of LINC00662 was significantly increased in various cancers such as gastric, lung, breast and colorectal cancers and was significantly associated with poor prognosis in these tumours[12,13].Meanwhile,LINC00662 was found to form a ceRNA mechanism with the miR‑497‑5p/YAP1 axis to promote the proliferation and metastasis of gastric cancer cells, and more severely exacerbate their resistance to chemotherapeutic drugs[14].In the present study, LINC00662 was highly expressed in hepatocellular carcinoma sorafenib‑resistant cells (HCC‑SR), and interference with LINC00662 expression in HCC‑SR cells enhanced their sensitivity to sorafenib, suggesting that LINC00662 expression may be associated with sorafenib resistance in hepatocellular carcinoma cells.
Studies have shown that LncRNAs can inhibit miRNA action through the mechanism of competing endogenous RNAs (ceRNAs),and LINC00662 has oncogenic functions and plays an oncogenic role as ceRNA in the development of some tumors[15,16].To explore the relationship between LINC00662 expression and sorafenib resistance in hepatocellular carcinoma cells and its mechanism, we predicted the miRNA binding site of LINC00662 by bioinformatics and found that LINC00662 existed with miR‑106a‑5p binding site, and dual luciferase reporter gene assay and RT‑qPCR showed that LINC00662 was sensitive to miR‑106a‑5p, indicating that LINC00662 may affect the expression of its target mRNA through negative regulation of miR‑106a‑5p.
MiR‑106a‑5p is a member of the miR‑17 family.Recent studies have shown that MiR‑106a‑5p is aberrantly expressed in a variety of tumours, both as an oncogene[17,18] and as a tumour suppressor gene[19], and is closely associated with resistance to chemotherapy[20], but the relationship with sorafenib resistance in hepatocellular carcinoma is not yet clear.Our study found that miR‑106a‑5p expression was reduced in HCC‑SR and overexpression of miR‑106a‑5p increased HCC‑SR sensitivity to sorafenib and inhibited cell survival.The presence of binding sites for miR‑106a‑5p to CAV1 was predicted by bioinformatics sites, suggesting that miR‑106a‑5p may affect hepatocellular carcinoma cell sensitivity to sorafenib by targeting CAV1.Caveolin‑1 (CAV1) is a structural protein of cell membrane foci, a membrane protein associated with endocytosis, extracellular matrix organization, cholesterol distribution, cell signaling, and a regulator of liver function.cAV1 also has dual oncogenic and oncogenic effects[21,22].There is growing evidence that high CAV1 expression can promote drug resistance in some cancers and is associated with poorer response to chemotherapy[23].In our study, we found elevated CAV1 expression in HCC‑SR cells, suggesting that CAV1 expression may be associated with drug resistance in hepatocellular carcinoma.Further by dual luciferase reporter gene and Western Blot assays, we found that miR‑106a‑5p expression in HCC‑SR cells was negatively correlated with CAV1 expression and positively correlated with HCC‑SR sensitivity to sorafenib, indicating that miR‑106a‑5p could increase hepatocellular carcinoma cell sensitivity to sorafenib by targeting the inhibition of CAV1 expression sensitivity.
Based on the above results, we hypothesized that LINC00662 counteracted the effect of miR‑106a‑5p through ceRNA mechanism to increase the expression of CAV1, thus promoting the development of sorafenib resistance in hepatocellular carcinoma cells.Therefore,we further knocked down the expression of LINC00062 in HCC‑SR cells by siRNA technology to observe the effect of knocking down LINC00062 on the expression of miR‑106a‑5p and CAV1 in hepatocellular carcinoma cells and the sensitivity of hepatocellular carcinoma cells to sorafenib.The results showed that compared with control cells, the expression of miR‑106a‑5p was increased in the siLINC00062 group of cells, while the expression of CAV1 was decreased and the sensitivity to sorafenib was increased, suggesting that LINC00662 counteracted the effect of miR‑106a‑5p through the ceRNA mechanism and promoted the expression of CAV1, which in turn inhibited the sensitivity of HCC cells to sorafenib sensitivity of HCC cells.
5.Conclusion
In conclusion, this study found that LINC00662 and CAV1 expression increased and miR‑106a‑5p expression decreased in hepatocellular carcinoma sorafenib‑resistant cell lines (HCC‑SR),and that inhibition of LINC00662 enhanced the sensitivity of HCC‑SR to sorafenib by a mechanism related to the regulation of the miR‑106a‑5p /CAV1 axis, and this finding also provides new implications for the regulation mechanism of LncRNA involvement in drug resistance in HCC This finding also provides new implications for the involvement of LncRNAs in the regulation of drug resistance in HCC.
Conflict of interest statement for all authors
Wangwei Cai and Bozen Chen: responsible for experimental design, experimental supervision, paper writing and article review;Man Xiao and Dongjing Yan: provided technical guidance for the experiments; Tong Chen and Na Liang were responsible for experimental implementation, index testing and data analysis; all authors declare that there is no conflict of interest.
杂志排行
Journal of Hainan Medical College的其它文章
- Research progress of traditional Chinese medicine in the treatment of complications related to spinal cord injury
- Research progress on the safety of nail placement in adolescent idiopathic scoliosis surgery
- Study on the protective mechanism of Yizhiren regulating lipid metabolism in mice with diabetic nephropathy
- Differential expression analysis of coronary heart disease related genes in Hainan residents
- Experimental study of the anti-inflammatory activity of some compounds in Berchemia lineata(L.)DC
- Mechanism of Qiliqiangxin capsule on the regulation of IP3Rs/GRP75/VDAC1 gene in myocardial infarction rat heart
