The Relationship Between Ultraviolet B and DNA Methylation in Skin Cancers
2023-10-20YiLinWuYueYueZhangYongHeQunLyuLiMingLiMingJunJiang
Yi-Lin Wu, Yue-Yue Zhang, Yong He, Qun Lyu, Li-Ming Li,*, Ming-Jun Jiang,*
1 Jiangsu Key Laboratory of Molecular Biology for Skin Diseases and STIs, Hospital for Skin Diseases (Institute of Dermatology),Chinese Academy of Medical Sciences, Peking Union Medical College, Nanjing, Jiangsu 210042, China; 2 Outpatient Department,Affiliated Jinling Hospital, Medical School of Nanjing University, Nanjing, Jiangsu 210042, China; 3 Department of Dermatologic Surgery, Hospital for Skin Diseases (Institute of Dermatology), Chinese Academy of Medical Sciences, Peking Union Medical College, Nanjing, Jiangsu 210042, China.
Abstract
Keywords: DNA methylation, DNA methyltransferases, skin cancers, ten-eleven translocations, ultraviolet B
lntroduction
The UV spectrum is divided into UVA (320-400nm),UVB (280-320nm), and UVC (220-280 nm).Only UVA and UVB can pierce the atmosphere and reach the general populace.However, UVB barely reaches the basal layer1.Although UVB accounts for only 10% of solar UV radiation, it has been proved to be the key factor in photochemical damage to cellular DNA2In addition,accumulation of DNA damage and mutations without efficient treatment may lead to the initiation of skin carcinogenesis.
Therefore, overexposure of human skin to UVB radiation from the sun and the resulting DNA damage are considered the most significant environmental factors for the development of skin cancers, including basal cell carcinoma (BCC), squamous cell carcinoma (SCC),and malignant melanoma.In this review, we searched PubMed using the keywords “UVB” and “DNA methylation” to investigate and summarize the changes in DNA methylation of skin cancers caused by UVB and the mechanism by which UVB regulates DNA methylase.And the Searching strategies are (“DNA Methylation“[Mesh] AND “Ultraviolet Rays”[Mesh]) and (((Ray,Ultraviolet) OR (Ultraviolet Ray) OR (Ultra-Violet Rays) OR (Ray, Ultra-Violet) OR (Ultra Violet Rays) OR(Ultra-Violet Ray) OR (UV Light) OR (Light, UV) OR(Actinic Rays) OR (Actinic Ray) OR (Ray, Actinic)OR (Ultraviolet Light) OR (Light, Ultraviolet) OR(UV Radiation) OR (Radiation, UV) OR (Ultraviolet Radiation) OR (Radiation, Ultraviolet) OR (Ultraviolet Radiations) OR (Black Light, Ultraviolet) OR(Ultraviolet Black Light) OR (Ultraviolet Black Lights))AND ((DNA Methylations) OR (Methylation, DNA)OR (Methylations, DNA))).The data source from 2000 to 2021 was searched.This information may provide new ideas for subsequent research on UVB regulation of DNA methylation.
DNA methylation and demethylation
Epigenetic alterations generally represent the interactions between the environment and the genome.3DNA methylation influences the expression of genes; therefore, it regulates a wide range of biological processes, including cell proliferation, cell death, mutation, tumorigenesis, tumor promotion,and tumor progression.4Hypermethylation at promoter regions is a prominent epigenetic mechanism for decreasing the expression of tumor suppression genes, which play crucial roles in tumorigenesis.Because both hypermethylation and hypomethylation can cause changes in cellular gene expression, proper regulation of the DNA methylation status is essential for maintenance of tissue homeostasis.
DNA methylation patterns and DNA
methylation modifier enzymes
The processes of DNA methylation and demethylation are respectively influenced by DNA methyltransferases (DNMTs) and ten-eleven translocation (TET)family proteins, which are often located in cytosine-phosphate-guanine (a cytosine followed by a guanine nucleotide, CpG)-enriched regions called “CpG islands” in a promoter sequence.5Gene transcription has been shown to be inhibited when the promoter is methylated.Hence,DNA CpG methylation is regarded as a crucial hallmark of epigenetic modification in mammals.6
DNMTs
DNMTs, including DNMT1, DNMT3A, and DNMT3B,methylate DNA and maintain genomic methylation patterns7(Fig.1).DNMTs catalyze methylation of the 5-carbon of cytosine to form 5-methylcytosine (5-mC).DNMT1 is the enzyme considered responsible for the maintenance of mammalian DNA methylation during DNA replication by copying the methylation pattern from the parent strand to the daughter strand.However, recent research provides a different viewpoint that DNMT1 is also involved in the process of de novo methylation.8DNMT3a and DNMT3b,coded by different genes, are de novo methylases.Although DNMT2 is structurally related to the other DNMTs, it possesses poor DNA methylation activity.However, it catalyzes 5-mC in transfer RNA very efficiently.
TETs
DNA demethylation is initiated by TET family proteins, including TET1, TET2, and TET3, which belong to the α-ketoglutarate-dependent and iron(II)-dependent dioxygenase superfamily (Fig.2).TETs catalyze 5-mC to form 5-hydroxymethylcytosine,85-formylcytosine,and 5-car-boxylcytosine by three consecutive oxidation reactions.The 5-formylcytosine and 5-carboxylcytosine marks are then recognized by thymine DNA glycosylase,which activates the base excision repair pathway.In this process, the modified cytosine is replaced by an unmodified cytosine.9
UVB and DNA methylation in skin cancers
Relationship between UVB-induced DNA damage and DNA methylation
UVB is directly absorbed by nuclear DNA, leading to DNA damage.UVB induces GC to TA transitions, formation of cyclobutane pyrimidine dimers (CPDs), and formation of (6-4) photoproducts (6-4PPs).CPDs(rather than 6-4PPs or other lesions) are thought to be responsible for most UVB-induced mutations,10such as mutation of p53 in UVB-induced skin cancers.11CPDs are excised at slower rates than 6-4PPs, although the latter is also responsible for adverse effects of UV radiation including sunburns, skin photoaging, and cutaneous cancer.12Following UVB radiation, a network of DNA damage-response mechanisms triggers a signal transduction cascade to regulate various genome-protection pathways including DNA damage repair, cell cycle control,apoptosis, transcription, and chromatin remodeling.13Additionally, accumulated DNA damage results in the occurrence of skin cancers.However, CPDs preferentially form at pyrimidines containing 5-mC in mammalian cells that have been irradiated by UVB or sunlight.14This suggests that 5-mC plays an important role in UVB-induced DNA damage.

Figure 1.The process of DNMT1, DNMT3a, and DNMT3b in DNA methylation.DNMT1 maintains DNA methylation; while DNMT3a and DNMT3b de novo DNA methylation.5-c: 5 carbon of cytosine; 5mC: 5-methylcytosine; DNMT: DNA methyltransferases.
UV signature mutations are consequences of the interplay among CPD formation, cytosine deamination, and DNA repair.15DNA reparation is essential for preventing mutagenesis and skin cancer.Under normal physiological conditions, the nucleotide excision repair (NER)pathway is a DNA repair mechanism that functions to remove these bulk UVB-induced photoproducts, such as CPDs and 6-4PPs.16However, defects in the NER pathway may be associated with a higher risk of skin tumors.In patients with several rare human genetic disorders characterized by defects in the NER pathway, such as xeroderma pigmentosum-Cockayne syndrome complex,UV-sensitive syndrome, and the photosensitive form of trichothiodys-trophy,13cells are hypersensitive to UV light.Meanwhile, the incidence of all types of skin cancer, including melanoma, is increased by several orders of magnitude in patients with xeroderma pigmentosum,17suggesting that pyrimidine dimers contribute to melanoma and non-melanoma skin tumors.
Repair of pyrimidine dimer damage may remove methylated cytosines during the excision repair step, potentially contributing to altered DNA methylation.13Thus,the DNA methylation patterns at susceptible gene loci change.Both the NER pathway and base excision repair pathway play an important role in DNA demethylation by excising 5-carboxylcytosine to form cytosine.18
UVB-induced DNA methylation in skin cancers
DNA methylation is one of the hallmark epigenetic events most frequently studied in cancers.However, the DNA methylation in UVB-induced melanoma has received less attention than molecular signaling.Moreover, some researchers did not distinguish UVB-induced SCC from UVB-induced BCC, instead summarizing both as nonmelanoma skin cancers.Therefore, more research is needed in this area to determine whether any differences in DNA methylation exist among UVB-induced skin cancers in animal models.
UVB-induced inheritable DNA methylation changes can lead to an altered phenotype and provide a selective advantage to cells, perhaps when combined with UVB-induced mutations.The occurrence of such changes could thus be viewed as a tumor-driving event, such as when genes affecting growth control and/or DNA repair become involved.19A total of 6,003 genes exhibit a >2-fold change in CpG methylation in the classic skin carcinogenesis model.According to importance-performance analysis, the top canonical pathways are those related to cancer development, cyclic AMP (cAMP)-mediated signaling, G protein-coupled receptor signaling, and gene of phosphate and tension homology deleted on chromosome ten (PTEN) signaling.Compared with the normal epidermis, RNA splicing factor RNA binding fox-1 homolog 1 (RBFOX1) is the gene that shows the greatest methylation status change in UVBinduced tumors.RBFOX1, an RNA-binding protein, is highly expressed in the cytoplasm.Interestingly,RBFOX1is reported to be a susceptibility gene for hydrolyzed wheat allergy20and may play a role in neuronal development21;however, its role in skin cancer development is unclear.We have summarized the DNA methylation alterations in UVB-irradiated cells and mouse models in Table 1.
Interestingly, UVB irradiation does not directly induce detectable changes in DNA methylation in human keratinocytes.Most of the methylation differences observed in cancer are rather passenger events than driver events.22These events might be secondary to specific genetic events, or they could occur during the processes of enhanced cell proliferation.DNA methylations might also be induced by other processes, such as immune responses, inflammation, or formation of reactive oxygen species,23or other types of cells such as Langerhans cell may be involved in these processes.24
Intriguingly, the methylation profiles are completely opposite and the regulation of DNA methylation enzymes is reversed between skin cancers and systemic lupus erythematosus (SLE).As a result, potent therapies for skin cancer that work by downregulating DNA hypermethylation, such as honokiol, should be further studied to determine the risk of adverse effects such as lupus-like symptoms.Further research is needed to validate whether cell type-specific DNA methylation patterns exist.Several reports have shown that aberrant DNA methylation in circulating T cells is associated with an increased risk of post-transplant skin cancer.25This phenomenon may be explained by the fact that T cells play an important role in the tumor microenvironment during the process of tumorigenesis.Further studies are needed to determine whether UVB affects DNA methylation in circulating T cells in skin cancers as it does in SLE.
UVB and DNA methylation modifier enzymes
UVB affects expressions and activities of DNMTs and TETs

Figure 2.The roles of TETs in DNA demethylation.TETs catalyze 5mC to form 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC),and 5-carboxylcytosin (5-caC) by three consecutive oxidation reactions.Then, the 5-fC and 5-caC marks are recognized by thymine DNA glycosylase (TDG), which activates the base excision repair pathway (BER).5mC: 5-methylcytosine; TETs: ten-eleven translocations.
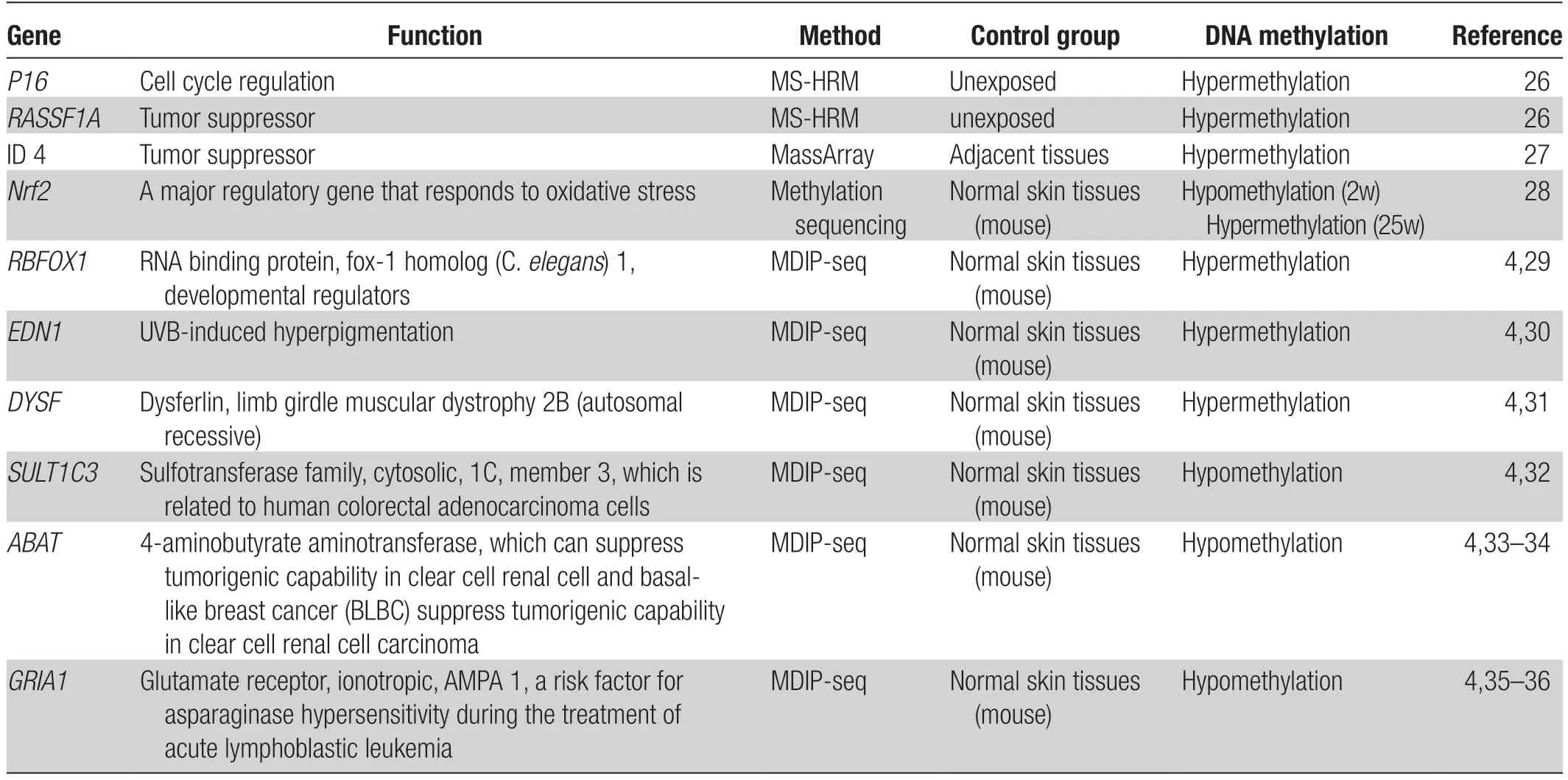
Table 1 DNA methylation alterations in UVB-irradiated cells and mouse models.
UVB irradiation can affect DNA methylation patterns through the expression and activities of DNMTs and TETs.In our previous study of UVB-induced cutaneous SCC, we found that DNMT1 is upregulated and TETs are down-regulated, resulting in increased methylation of inhibitor of DNA binding/differentiation 4, a downstream mediator of the Transforming growth factor-β (TGF-β)/bone morphogenic protein (BMP)/SMA (for small body size) and mothers against decapentaplegic (MAD) related protein (SMAD) signaling pathway and a regulator of the growth and differentiation of embryonic tissues.23Other studies have produced similar findings.37However, the expression of TET1, TET2,and TET3 mRNA and proteins was significantly upregulated in the UVB-exposed HaCaT cells.The difference in the expression of TETs may be due to accumulative doses of UVB or chronic exposure to UVB.38Yamadaet al.39showed that DNMT1 was downregulated after UVB irradiation.Different cell lines and mass UVB doses in the short term may have contributed to these inconsistent results.
Mechanism of UVB regulation of DNA methylation modifier enzymes
Few studies to date have focused on the mechanism of UVB regulation of DNA methylation modifier enzymes in skin cancers.van Doornet al.40found that UVB can increase the expression of DNMT1 and DNA methylation activity by inactivation of p53, which can directly repress transcription of DNMT1.
Inflammation may play a part in the regulation of DNA methylation modifier enzymes.Additionally, UVBinduced immunosuppression may contribute to the survival of these cells with aberrant DNA methylation, which results in the development of cutaneous malignancies.One study showed that UVB upregulated the activities and expression of DNMTs by increasing the levels of cyclooxygenase 2, prostaglandin E2, and prostaglandin E2 receptors.41Antiinflammatory agents and antioxidants, such as sulforaphane,42Chlamys farreri,22and honokiol,37can inhibit DNMT expression and upregulate the level of TET enzymes to alter DNA methylation.They can also reduce the incidence and number of tumors.However, the molecular mechanisms of how these agents are involved in regulating DNA methylation modifier enzymes are unclear.
In SLE, UVB induces the decreased activity of DNMT1 through aryl hydrocarbon receptor (AhR) activation-dependent silent mating-type information regulation 2 homolog 1 suppression.43AhR plays an important role in UVB-irradiated keratinocytes and critically contributes to skin photocarcinogenesis in mice.44However, whether AhR also plays a similar role in UVB-induced skin cancer remains to be further studied.
Conclusion
The studies discussed in this review show that UVB not only alters the genetic blueprint but also causes epigenetic modifications of skin.More specifically, UVB affects the expression of DNA methylation modifier enzymes.Our previous study indicated that UVB induces cutaneous SCC progression by de novo inhibitor of DNA binding/ differentiation 4 methylationviamethylation regulating enzymes.However, precisely how UVB affects the expression of DNA methylation modifier enzymes remains unknown.
The differences in DNA methylation among various types of UVB-induced skin cancers are not well understood.Some researchers did not distinguish UVB-induced SCC from UVB-induced BCC in mice models, and the scarcity of research on DNA methylation of UVB-induced melanoma makes investigating this topic more difficult.The dynamic changes in DNA methylation in UVBinduced skin cancers are still unclear.The research to date has only analyzed the early (2 and 15 weeks) and late(25 weeks) stages of UVB-induced non-melanoma.Under physiological conditions, however, the differences in DNA methylation during the occurrence and progression of UVB-induced skin cancers may be dynamic.Studying these two time points alone is not adequate.Further research of the dynamic changes in DNA methylation will be crucial for understanding the role of DNA methylation and gene functions in the occurrence and development of UVB-induced skin cancers.Our research group plans to address this topic as a next step.
Our review has the following limitations: (1) This article uses a search strategy that combines subject terms and entry terms to filter.There may be articles that use descriptions that are neither subject terms nor entry terms but are not included.(2) Only English articles that can be viewed in full text are included, and articles in other languages are not included.
Because epigenetic modifications are reversible, research on epigenetic modifications may reveal potential therapeutic markers or targets, leading to the development of novel strategies for preventing or treating UVB-related skin cancers.Such targeting could be achieved through further research about the mechanism of UVB regulation of DNA methylation modifier enzymes, which would provide more precise and efficient therapy to correct aberrant DNA methylation patterns and restore normal growth control in skin cancers.This may also help to maintain DNA methylation homeostasis in tissues.
However, currently available epigenetic treatment may be limited by global effects leading to adverse reactions that outweigh potential benefits.Further research may be needed to make these drugs more tissue-specific or only locally effective to maintain the proper methylation pattern and reduce systemic toxicity.
Source of funding
This work was supported by the Natural Science Foundation of Jiangsu Province (No.BK20191136), the Fundamental Research Funds for the Central Universities(No.3332019104), and the Open Project of Jiangsu Biobank of Clinical Resources (No.JSSWYB2020-05-003).
杂志排行
国际皮肤性病学杂志的其它文章
- Tuberculosis Verrucosa Cutis on the Buttocks:A Case Report
- Livedoid Vasculopathy Secondary to Protein C Deficiency: A Case Successfully Treated With Rivaroxaban
- Neutrophilic Panniculitis Associated With Myelodysplastic Syndrome/Myeloproliferative Neoplasm: A Case Report and Literature Review
- The Role of Endoplasmic Reticulum Stress in Melanoma
- Basal Cell Carcinoma Excision Guided by Dermoscopy: A Retrospective Study in Macau
- Teledermatology During the COVID-19 Pandemic in a Developing Country: Could This Be the Answer to Improving the Reach of Dermatology Care?
