Response of fungal communities to afforestation and its indication for forest restoration
2023-10-07KichunHungZhenliGuoWenZhoChnggeSongHoWngJunningLiReyilMuminYifeiSunBokiCui
Kichun Hung, Zhenli Guo, Wen Zho, Chngge Song, Ho Wng, Junning Li,Reyil Mumin, Yifei Sun, Boki Cui,*
a School of Ecology and Nature Conservation, Beijing Forestry University, Beijing, 100083, China
b Key Laboratory of Ministry of Education for Coastal and Wetland Ecosystems, Xiamen University, Xiamen, 361102, China
Keywords:
ABSTRACT Soil fungi in forest ecosystems have great potential to enhance host plant growth and systemic ecological functions and services.Reforestation at Saihanba Mechanized Forest Farm,the world's largest artificial plantation,has been integral to global forest ecosystem preservation since the 1950s.To better assess the ecological effects of soil microbiology after afforestation, fungal diversity and community structure (using Illumina sequencing) from forests dominated by Larix gmelinii var.principis-rupprechtii, Pinus sylvestris var.mongolica and Picea asperata, and from grassland were surveyed.In total, 4,540 operational taxonomic units (OTUs) were identified, with Mortierella and Solicoccozyma being the dominant genera of grassland soil and Inocybe, Cortinarius, Piloderma,Tomentella, Sebacina, Hygrophorus and Saitozyma dominating the plantation soil.Principle coordinate analysis(PCoA) and co-occurrence networks revealed differences in fungal structure after afforestation.Significantly,more symbiotroph guilds were dominated by ectomycorrhizal fungi in plantations under the prediction of FUNGuild.The community composition and diversity of soil fungi were significantly influenced by pH via redundancy analysis(RDA)and the Mantel test(p<0.01).This finding emphasizes that soil pH has a strong effect on the transition of fungal communities and functional taxa from grassland to plantation, providing a novel indicator for forest restoration.
1.Introduction
Since the 1950s, China has implemented a large-scale afforestation program,which has effectively alleviated the problem of desertification and significantly improved the quality of the ecological environment(Wang et al., 2012; Li et al., 2022).The Saihanba Mechanical Forest Farm,located on the southern edge of the Otindag Sandy Land,is one of the works in the afforestation program conducted in 1962.It has a special ecosystem process and provides ecosystem services(Zhang et al.,2018).Soil nutrient conditions improved significantly with the increase of artificial vegetation (Kou and Jiao, 2022).The reciprocal feedback between plant communities and soil microbial communities is closely related to the regulation of ecosystem processes (Wardle et al., 2004;Trivedi et al.,2020).Soil microbial communities are important mediators that ensure the ecological processes and functions of biogeochemical cycles and are highly sensitive to environmental changes (Bardgett and Van Der Putten, 2014; Osburn et al., 2019).Soil fungi have strong environmental tolerance and can form a symbiotic relationship with plant roots (Zechmeister-Boltenstern et al., 2015; Man et al., 2022; Ren et al., 2022; Sokol et al., 2022; Tu et al., 2022; Zhang et al., 2022a,2022b).The interactions between saprotrophs and AMF for litter decomposition may be an important mechanism for promoting plant fitness under low soil nutrient availability(Cao et al.,2022).By adjusting the trade-off of the community construction process, it affects the functions and services in the terrestrial ecosystem, including the cycle of elements (Clemmensen et al., 2013; Geisen, 2016; Jiao et al., 2022).Therefore,understanding soil fungal communities and their relationship to habitat and soil physicochemical properties will provide fundamental data for assessing the status of forest restoration.However, researches into the diversity, structure, and co-occurrence network of plantation ecosystems are still limited, preventing a more comprehensive understanding of fungi and their possible ecological functions.
The structure and diversity of soil fungi are influenced by the soil properties, and changes in the microbial community can also affect soil quality(Lombard et al.,2011;Nakayama et al.,2019).Microbial diversity is a potential indicator of above-ground vegetation degradation(Wu et al.,2021).However, the ecological relationship between soil microbial diversity and the environment has received less attention than that of the aboveground component.Studies have shown that tree species richness could increase fungal diversity and alter community structure(Liang et al.,2016;Khlifa et al.,2017;Wu et al.,2019).Nevertheless,little information is available on changes in fungal diversity following plantation restoration.Therefore,this study aimed to elucidate the effects of afforestation on soil fungal structure and diversity,and to assess the ecological benefits of forest restoration and ecosystem function of planted forests.Understanding soil properties after afforestation is essential for assessing soil nutrient status and promoting sustainable plantation development because soil nutrients play a critical role in the spatial distribution of above-ground plant communities.Soil microorganisms are more sensitive to changes in soil nutrients than plants and can better reflect changes in soil quality (Deng et al., 2019).The assessment of soil microbial diversity and community composition as ecological indicators of forest restoration has been a topic of interest in recent years.Despite some progress in this field, a gap still exists,and further research is needed to better understand the implications of soil fungal diversity for forest restoration.In this study, we aimed to assess the effects of habitat differences and soil environmental factors on the diversity and functional characteristics of soil fungi and to investigate their potential as a novel indicator of forest restoration.
To achieve our objectives,we collected soil samples at a depth of 20 cm from different plantations in the Saihanba region, and determined their chemical properties,including pH,soil organic carbon(SOC),total nitrogen(TN),available phosphorus(AP),and cation exchange capacity(CEC).We also characterized the structural composition, diversity and functional characteristics of soil fungi using high-throughput sequencing technology.Based on previous reports on differences in soil fungal diversity in plantation habitats(Wang et al.,2022a),we hypothesized that soil fungal diversity and community structure in different plantations may differ due to the presence of different plant types and soil properties.We also intend to clarify the status of soil fungal functional traits and co-occurrence networks in different habitats and their contribution to the different tree species planted.
2.Materials and methods
2.1.Study area
This study is based on field experiments conducted at Saihanba Mechanized Forest Farm (42°21′–42°31′N, 116°53′–117°31′E), Hebei Province, China (Fig.1).With an elevation range of 1,010 to 1,939 m above sea level, the area has a semi-arid and semi-humid temperate climate,characterized by an average yearly air temperature of -1.4°C,along with an annual precipitation of 450.1 mm(Wang et al.,2010).The total operating area of the forest farm is 93,333 ha,including 74,667 ha plantation area,which makes it the largest plantation forest farm in the world.Artificial forests, natural secondary forests and grasslands in the Saihanba area account for more than 90% of the total area (Wei et al.,2018).The typical afforestation tree species in the forest farm are Larix gmelinii var.principis-rupprechtii, Picea asperata and Pinus sylvestris var.mongolica.The soil in this area is composed of brown soil, meadow soil and swamp soil.
2.2.Soil sampling and property analysis
Soil samples were collected in July 2022.Four sites comprising one grassland and three typical plantations were selected, that is, grassland(GL),Larix gmelinii var.principis-rupprechtii(L.gmelinii,LF),Picea asperata(SF)and Pinus sylvestris var.mongolica(P.sylvestris,PF).Fifteen sampling plots(20 m×20 m)were established with 10 m buffer zones separating each site.After removing the surface litter, we randomly collected nine soil cores(20 cm in depth)from each sampling plot and then mixed them to form a composite sample.In total, 60 samples (four sites × fifteen samples) were collected.All samples were quickly filtered through a 2-mm sieve to separate stones and other debris.The sieved soil was packed into sealed bags and centrifuge tubes, and the encapsulated samples were stored in an insulated box filled with dry ice for rapid transport to the laboratory.Samples in centrifuge tubes were stored at-80°C for DNA extraction,and samples in sealed bags were air-dried at room temperature for chemical determination.The soil chemical properties were determined with reference to Bao Shidan's soil agrochemical analysis(Bao,2000).
2.3.Soil DNA extraction, gene amplification and sequencing
Total DNA was extracted from soil samples using the DNeasy Power Soil Pro Kit (QIAGEN, Germany), and stored at -20°C until further analysis.The quality and quantity of extracted DNA were assessed using the NanoDrop ONE spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and 1.2% agarose gel electrophoresis.To amplify the ITS1 regions, the common fungal primers ITS1F (5-GGAAGTAAAAGTCGTAACAAGG-3) and ITS2 (5-TCCTCCGCTTATTGATATGC-3)were used.The 20 μL system was used for fungal amplification,and the steps of purification and quantification were based on the experimental method of Zhao et al.(2023).All samples were pooled in equal amounts and subjected to pair-end 2 × 250 bp sequencing using the Illumina MiSeq platform by Shanghai Personal Biotechnology Co.,Ltd.(Shanghai,China).

Fig.1.Sampling located in Saihanba Mechanized Forest Farm, Hebei Province, China.Four sites including larch, pine, spruce forests, and grassland were surveyed.Fifteen sampling plots (20 m × 20 m) were established, with 10 m buffer zones separating each site (60 samples in total).
2.4.Bioinformatic processing of the sequences
The original sequence was obtained by Illumina MiSeq doubleterminal sequencing.Microbiome bioinformatics was performed with QIIME2(https://docs.qiime2.org/2021.2/tutorials/).DADA2 plugin was used to screen low quality and noise reduction and the vsearch plugin was used to obtain operational taxonomic units (OTUs) with 97% similarity.OTUs were aligned with mafft and used to construct a phylogeny with fasttree2, and then species were classified and annotated based on the UNITE reference database.The original sequencing data are available from the NCBI database under accession number PRJNA940304.
2.5.Statistical analysis
We normalized the library using scaling with ranked subsampling using the “SRS” function, and R packages (version 4.1.1) were used to visualize alpha diversity and beta diversity data.Beta diversity was estimated according to Bray-Curtis distances across samples in habitats.Principal coordinate analysis (PCoA) was performed in the vegan packages.The beta NTI was calculated using the NST package for assessing the stochastic or deterministic processes of community construction(Ning et al., 2020).SPSS version 27.0 software (SPSS Inc., Chicago, IL,USA) was used for the analysis of variance, with p < 0.05 considered statistically significant.Under the condition that the test data met the homogeneity of variance, one-way analysis of variance (ANOVA) and post hoc Tukey tests were conducted to compare the soil properties.The effects of different locations on fungal diversity indices were tested by two-way ANOVA with post hoc Tukey tests.The Pearson correlation coefficient was used to show the correlation between community diversity,dominant genera,functional groups and environmental factors.A nonparametric Kruskal-Wallis test was used to assess differences between habitats for community data that were not normally distributed.Venn diagrams at genus taxa between four habitats were drawn in the VennDiagram package.To identify the community differences between habitats, a linear discriminant analysis effect size (LEfSe) was used(Kruskal-Wallis value of p< 0.05,LDA score>3.0).
The FUNGuild tool was utilized to assign fungal OTUs to ecological functional groups(Nguyen et al.,2016).Redundancy analysis(RDA)and Mantel tests were used to explain the relationship between the environmental factors and microbial composition.The significance of differentiation of microbiota structure among groups was assessed by permutational multivariate analysis of variance (PERMANOVA).The co-occurrence network based on the top 100 genera abundance genus level of fungal communities was performed using the Spearman correlation coefficients (R > |0.6| and p < 0.05) constructed with the genescloud tools (https://www.genescloud.cn).Gephi (https://gephi.org/)was used to calculate the topology properties of networks(including the number of nodes and edges, average degree, degree centrality, modularity,clustering coefficient,eigenvector centrality,average path length,and closeness centrality).Higher degrees in the habitat were screened for keystone species.The sampling point map was drawn by ArcGIS 10.7 software (ESRI, USA).Charting was performed with Origin 2022b software(OriginLab Corporation,USA).
3.Results
3.1.Soil fungal community composition of the Saihanba area
The soil fungal ITS genes were analyzed by Illumina MiSeq sequencing.After sequence denoising, screening low quality, and the pumping equality process, a total of 3,953,167 high-quality fungal sequences were obtained from 60 samples.Standardized leveling was performed at a sampling depth of 38,367 (minimum frequency) by the SRS function, including 14 phyla, 44 classes, 107 orders, 242 families,538 genera and 4540 OTUs.
The fungal community composition varied significantly between habitats (Fig.2, Tables S1–S4), with significant differences observed in relative abundance at the phylum, class, order, and family levels (Kruskal-Wallis, p < 0.05).Basidiomycota and Ascomycota accounted for 62.89%–98.36%of the abundance at the phylum level.At the class level,GL had a significantly higher relative abundance of Mortierellomycetes and Eurotiomycetes compared to plantations (p < 0.05).The relative abundance of Agaricomycetes, Pezizomycetes, and Tremellomycetes differed significantly among habitats (p < 0.05).Agaricales had the greatest abundance in SF,PF,and GL,while Atheliales and Thelephorales were significantly higher in LF and SF than in PF and GL at the order level(p < 0.05).The dominant families also varied significantly between habitats(p<0.05).Mortierellaceae,Clavariaceae,and Piskurozymaceae became the main families in GL.LF had the highest relative abundances of Atheliaceae and Pyronemataceae,while Inocybaceae,Thelephoraceae,Hygrophoraceae, and Cortinariaceae were enriched in the SF.Sebacinaceae had the greatest abundance in PF.
The composition of soil fungal genera and distinct taxa in different habitats were further analyzed(Fig.3).A total of 538 genera of soil fungi were identified in this study,with significant differences in the dominant genera among different habitats (Fig.3a) (Kruskal-Wallis, p < 0.05).Planted forests and grassland shared 99 genera, the abundant top five were Inocybe,Cortinarius,Piloderma,Tomentella and Mortierella.There are 7 genera shared among the three plantations, including Otidea, Ascochyta,Gymnopus,Prosthemium,Tubaria,Trichopeziza and Coemansia.The results of abundance and variance showed that Mortierella and Solicoccozyma were enriched in GL; Piloderma, Saitozyma, Humaria, and Pseudotomentella were enriched in LF; Inocybe, Hygrophorus, Cortinarius,Tomentella, and Amphinema were enriched in SF; and Sebacina and Helvellosebacina were enriched in PF.
3.2.Fungal community structure in different habitats
Soil fungal community structure was analyzed using principal coordinate analysis (PCoA) and beta dispersion analysis at the OTU level based on Bray-Curtis distance(Fig.4).The PCoA results explained 22.9%of the total variation in fungal community structure.There was a significant separation of fungal communities between grassland and planted forest, and statistical tests showed significant differences in community structure between habitats (p < 0.001).The results of beta dispersity analysis showed that the fungal community structure within GL,LF and PF were mainly dominated by homogenizing dispersal (stochastic contribution:77.14%,86.67%,and 72.38%,respectively).In contrast,SF was controlled by homogeneous selection with a contribution of 70.48%.In addition,the beta NTI values(except GL-PF)differed between habitats to a significant level(p <0.001).
3.3.Fungal alpha diversity in different habitats
Shannon diversity,Chao1 richness and Pielou evenness indices were selected based on OTU levels to assess the alpha diversity of different habitats in the Saihanba area (Fig.5).As the number of sequences increased,the rarefaction curves of different habitats all tended to level off(Fig.5a).The richness of fungal communities was greatest in GL and least in SF.The results of one-way ANOVA showed that the diversity of GL was significantly higher than that of plantation forests(p<0.05),and there were no significant differences between LF and PF.Alpha diversity was significantly positively correlated with pH (p < 0.001) and negatively correlated with AP(p <0.05,Table 1).
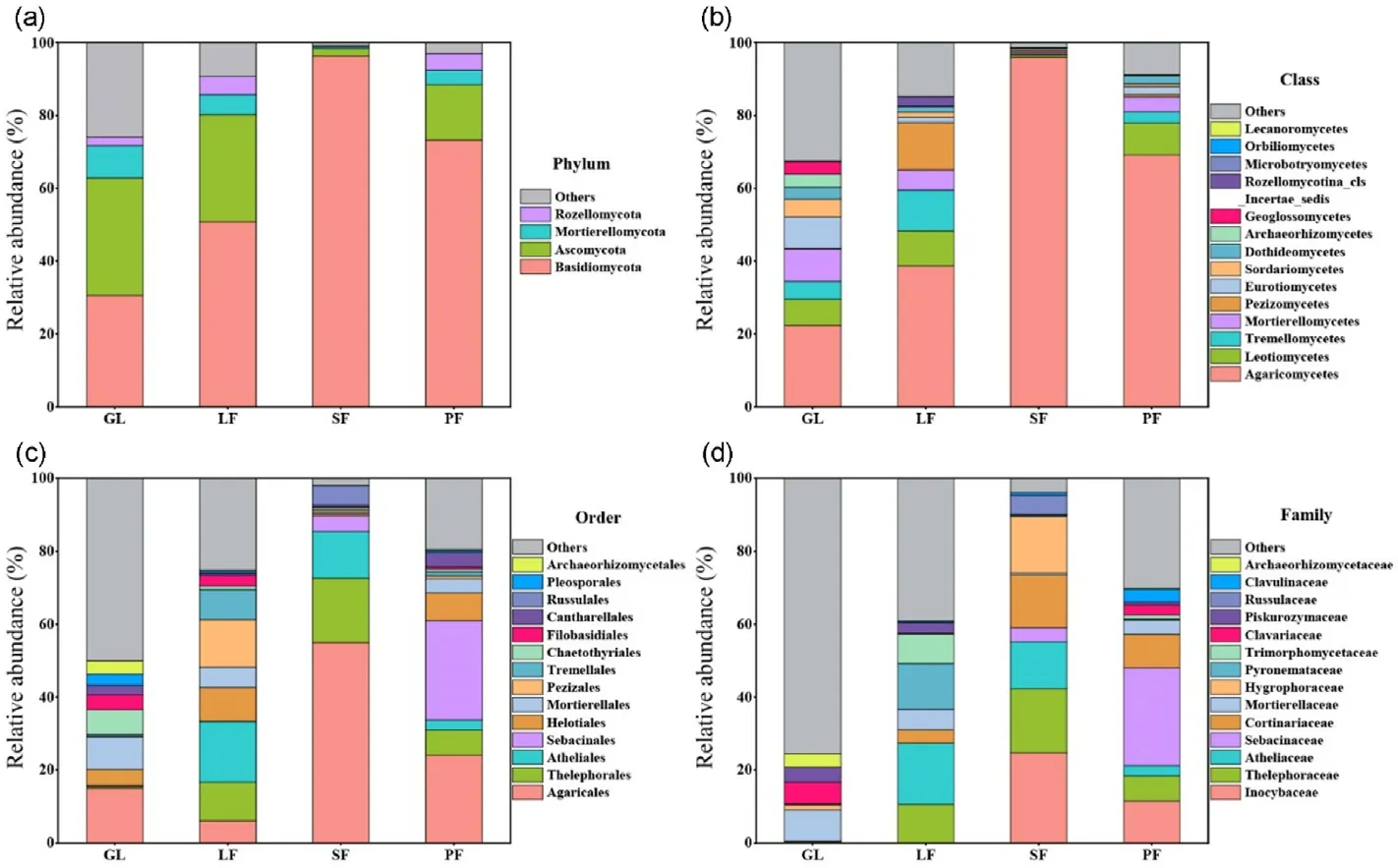
Fig.2.Relative abundance of soil fungi at phylum level (a), class level (b), order level (c) and family level (d) in different habitats.GL = grassland, LF = L.gmelinii forest, PF = P.sylvestris forest, SF = P.asperata forest.
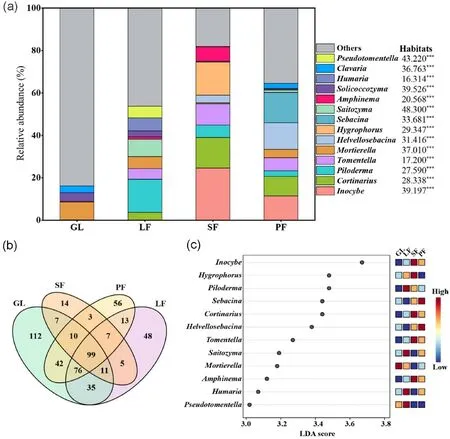
Fig.3.Relative abundance of fungal genus(a)in different habitats,Kruskal-Wallis tests showed the effect of habitats on the relative abundance of fungi(nsp>0.05,*p<0.05,**p<0.01,***p<0.001);(b)Venn plot of genus taxa between the habitats;(c)LEfSe analysis of genus between habitats(LDA significance threshold>3.0).
3.4.Fungal community functional groups of the saihanba area
Fungal OTUs were further assigned to ecological functional groups using the FUNGuild tool.A total of 41.44% (GL), 62.44% (LF), 97.91%(SF), and 77.35% (PF) of OTUs were identified as trophic modes by FUNGuild.GL mainly includes saprotroph (14.10%), saprotrophsymbiotroph (11.82%) and pathotroph-saprotroph-symbiotroph(10.12%).In contrast, 40.48% (LF), 87.94% (SF), and 62.97% (PF) of symbiotrophs were the majority trophic modes in plantation forests(Fig.6).The relative abundance of the functional groups was different among the habitats according to Kruskal-Wallis tests(Table S5).
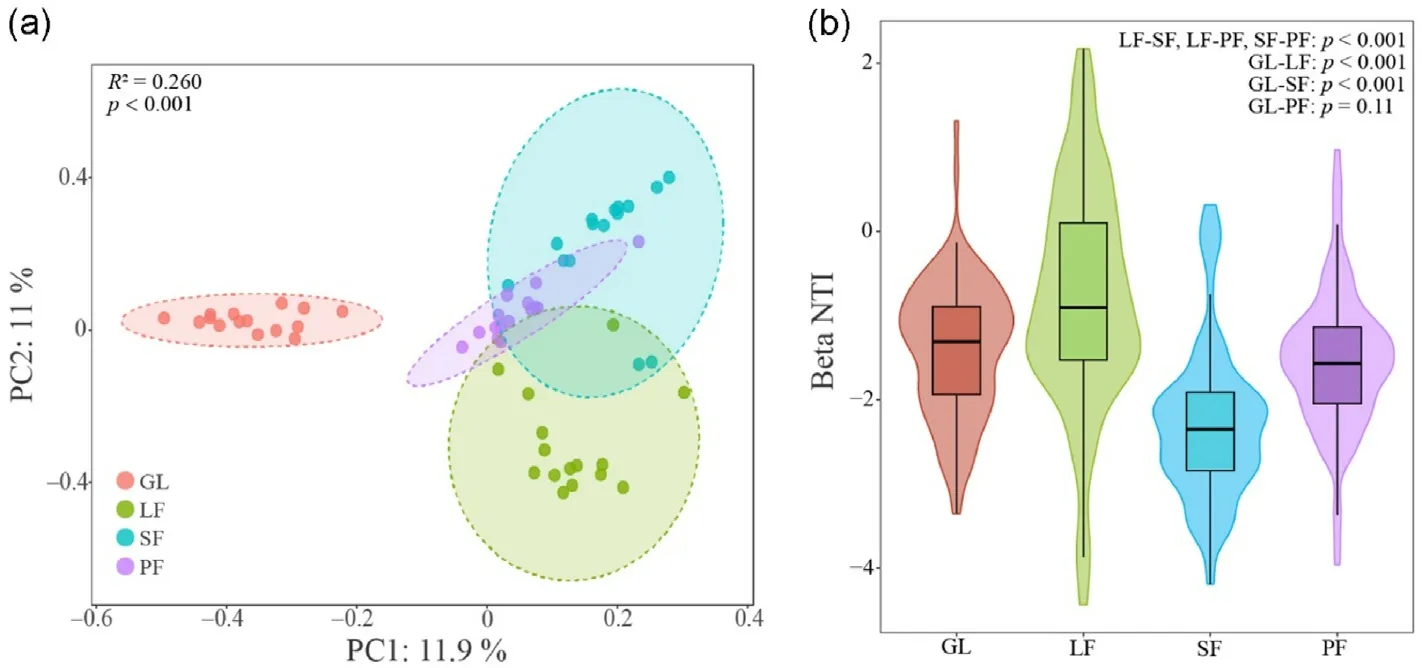
Fig.4.Beta diversity analyzed using PCoA (a) and beta dispersion analysis (b) based on Bray-Curtis distance.

Fig.5.Rarefaction curves of different habitats(a).Alpha diversity of fungal communities(b:Shannon diversity index,c:Chao1 richness,d:Pielou Evenness).Letters indicate significant differences in different habitats at p < 0.05 according to Tukey's HSD.
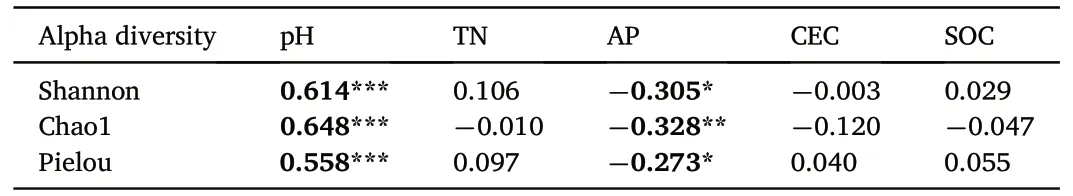
Table 1 Pearson correlation coefficient of alpha diversity to soil properties.
3.5.Correlations between environmental variables and fungal composition
The pH of GL and PF soil was weakly acidic and significantly higher than that of LF and SF.The AP of the LF and SF soils was significantly higher than that of the GL and PF soils.SOC had a maximum value in LF soil,and there was no significant difference between SF,PF and GL.The TN and CEC contents were the highest in LF and the lowest in PF, and there was no significant difference between SF and GL (Table 2).

Fig.6.Relative abundance of FUNGuild trophic modes between habitats.
RDA was used to shed light on the influence of variation in environmental variables on soil microbial community structure(genus level)according to the results of DCA.The community analysis results showed that RDA1 and RDA2 explained 10.520%and 6.996%of the community composition,respectively(Fig.7).The envfit of the Mantel test indicated that soil pH was the main environmental factor contributing to shifting the fungal community composition (R = 0.315, p = 0.001) (Table 3).Other soil factors affecting the fungal community include TN, AP, SOC and CEC.
3.6.Co-occurrence patterns of soil fungal communities
Co-occurrence networks were generated to explore the complexity of soil fungal connections between habitats, and topological properties were calculated (Fig.8, Table 4).The PF microbial network consisted highest edges (238) and average degree (5.231).The modularity of different habitats was greater than 0.4(0.563 for GL,0.579 for LF,0.531 for SF,and 0.533 for PF),indicating a modular structure.After Spearman correlations,different habitats were assigned to modules(7 of GL,10 of LF, 6 of SF, and 9 of PF).Moreover, the clustering coefficients of communities were 0.386(GL),0.325(LF),0.174(SF),and 0.311(PF),and the average path lengths were 3.741(GL),3.570(LF),4.372(SF),and 3.690(PF).
Keystone species were also altered in different habitats (Table S6),including Cadophora (GL), Solicoccozyma (GL), Humicola(LF), Trichocladium (LF and PF), Pseudosigmoidea (LF), Russula (SF), Tomentella (SF),Hygrophorus (SF), Coniochaeta (SF), Ilyonectria (PF), Alternaria (PF), and Venturia (PF).The species belonged to two phyla (Ascomycota and Basidiomycota) and four classes (Sordariomycetes, Saccharomycetes,Dothideomycetes, and Agaricomycetes).
4.Discussion
4.1.Effect of afforestation on the fungal composition
Simple fungal community composition of the plantation forest resulted in a high relative abundance of Ascomycota and Basidiomycota.The relative abundance among different habitats was significantly different, for example, the phylum Basidiomycota was significantly higher in SF than in other habitats (Fig.2a).This variation may be attributed to the spatial variability of the environment and the heterogeneity of the sampling sites (Zhang et al., 2020).Soil fungi played an important role in vegetation restoration.Ascomycota and Basidiomycota could participate in the element cycle of terrestrial ecosystems by degrading organic matter (Paungfoo-Lonhienne et al., 2015; Zhu et al.,2022).Enhancing adaptation to extreme environments through the formation of mycorrhizae, thus promoting the growth of sandy plants and the recovery of vegetation(Lu et al.,2016;Genre et al., 2020).
The Saihanba area has significant conservation value due to the unique fungal communities found in different habitats.Ectomycorrhizal species, such as Piloderma and Tomentella, play a crucial role in the nutrient acquisition of forest trees(Kohler et al.,2015).Their distribution in the SF, LF, and PF may be attributed to different soil properties,aboveground plant conditions, and genotype differences (Bonito et al.,2014; Yang et al., 2022).However, introducing exotic tree species mayhinder the formation of stable symbiotic relationships between ectomycorrhizal fungi and host plants in the region (Dickie et al., 2010; Alem et al., 2020; Tuo et al., 2022).Changes in microbial community composition are closely related to changes in ecosystem functions, and understanding this mechanism is important for revealing related ecosystem processes (Albright et al., 2020; Wahdan et al., 2021).Furthermore, species of the dominant genus Solicoccozyma in GL are usually obtained from soil, while the unique fungal communities in different plantations are mainly composed of mycorrhizal species of the genera Piloderma, Inocybe, Hygrophorus, Cortinarius, Sebacina, and

Table 2 Soil properties in different habitats (mean values ± standard error).
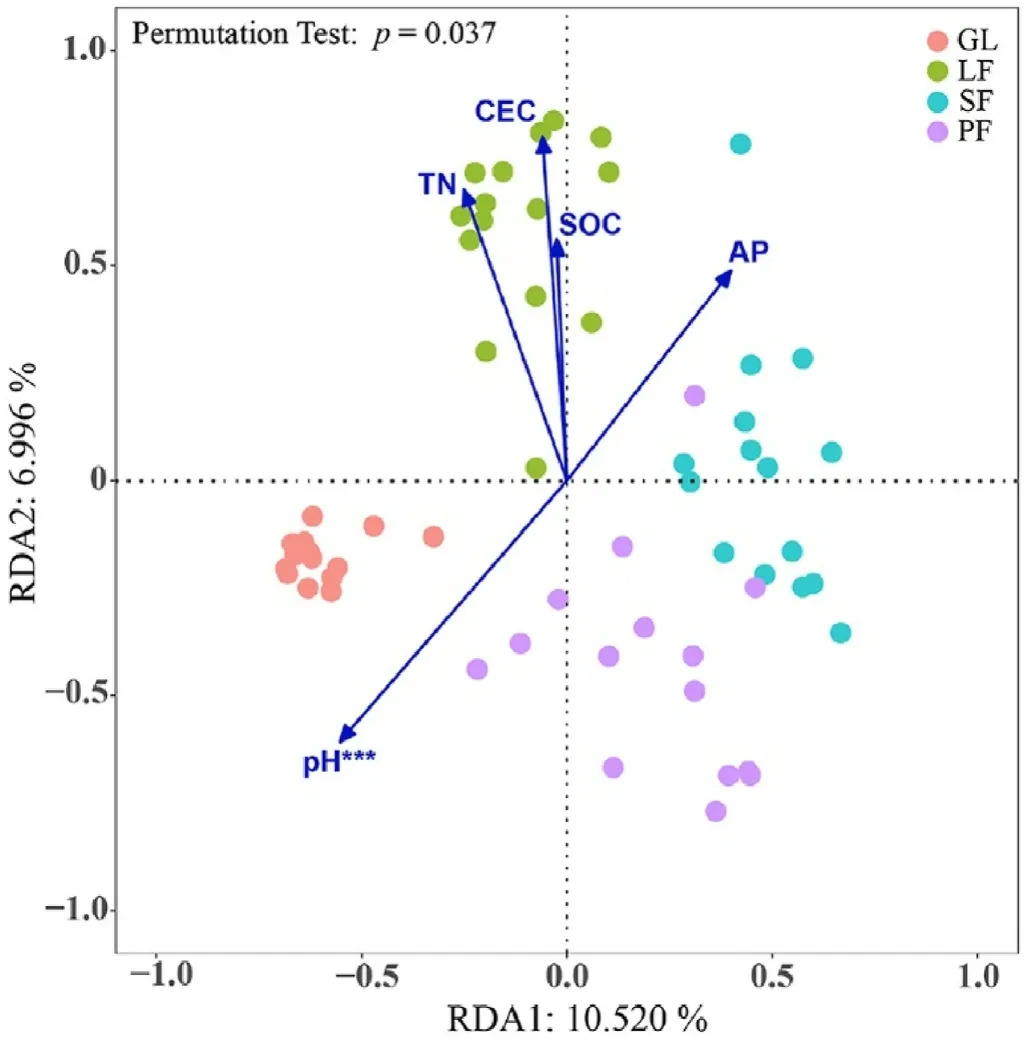
Fig.7.Redundancy analysis (RDA) of fungal communities at the genus level and environmental variables.Mantel tests were used to test the significance of correlations between fungal community structure and environmental variables.
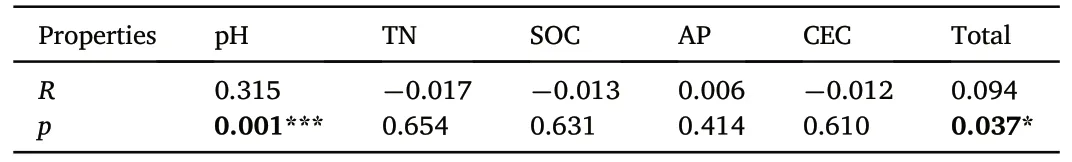
Table 3 Mantel Analysis of fungal community structure and soil properties.
Tomentella.Piloderma is a common symbiont of coniferous and broadleaf tree species, while Tomentella is frequently found in forests around the world(Kuhar et al.,2016;Lindahl et al.,2021;Lu and Yuan,2021).These findings highlight the importance of preserving the diverse fungal communities in the Saihanba area and understanding their ecological roles in forest ecosystems.By doing so, we can better inform sustainable forest management practices and promote the conservation of forest biodiversity.
Moreover,recent studies have shown that ectomycorrhizal fungi play a crucial role in maintaining the functional diversity of the regional forest ecosystem in the Saihanba area(Dang et al.,2018).Thus,distinguishing the ecological function of the soil fungal community, particularly ectomycorrhizal fungi, can be a valuable indicator of the ecological restoration of its trophic structure.To further elucidate the intrinsic connection mechanism between mycorrhizal species and the plantation ecosystem,additional empirical research can be conducted.
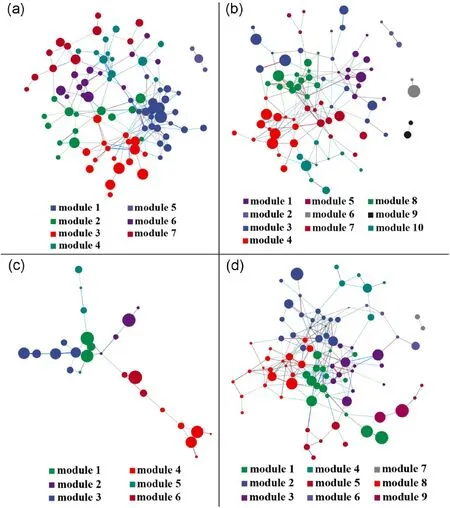
Fig.8.Co-occurrence networks of fungal communities at genus level of grassland(a),L.gmelinii forest(b),P.asperata forest(c),and P.sylvestris forest(d).The nodes are colored by modularity, and node size represents the degree of genus.The lines between the nodes are proportional to the Spearman correlation.
4.2.Effect of afforestation on the fungal community structure
PCoA analyses indicated significant changes in fungal community structure between grassland and typical plantation habitats in Saihanba(Fig.4a), and this difference may be caused by site-level differences in soil factors.Differences in soil habitat or the effect of spatial distance on the composition of fungal communities may lead to the differentiation of fungal niches (Feinstein and Blackwood, 2013; Cantrell et al., 2017).Changes in soil salinity,osmotic pressure and texture have an impact on the composition of microbial communities (Chowdhury et al., 2011;Mohamed and Martiny, 2011; Zhang et al., 2022).In this study, soil physical factors were not investigated.However, previous research has found that microorganisms adapt to changes in salinity and water to form special niches while ignoring the diffusion and random factors of species(Hussain et al.,2021;Chen et al.,2022).
The significantly different fungal community structures between habitats in Saihanba suggested that forest ecosystems may also have specific community assembly mechanisms and particular ecological functional taxa.Fungal communities in LF,PF and GL soils are dominated by the homogeneous dispersal of stochastic process and may be in the initial stages of community construction.The NTI values significantly less than zero indicate a possible overdispersal of the system (Stegen et al.,2012).Higher dispersal was also seen in the PCoA results.Homogeneous dispersal may be passive,and its significant effect on fungal community construction was also observed in previous studies (Chen et al., 2020;Wang et al.,2022b).In contrast,soil fungal communities in SF are mainly homogeneous selection processes, which are the result of physiological adaptations to the existing soil environment (Stegen et al., 2015; Zhou and Ning, 2017).Assessing fungal communities in different habitats is crucial to filter out the relative contribution of environmental factors to fungal composition in ecosystems (Tian et al., 2018).In summary, the soil community composition and structure of planted forest ecosystems are significantly different compared to grassland, posing more specific requirements for tree species selection and soil properties in terms of microbial ecology.
4.3.Fungal diversity is driven by environmental factors and affects system stability
The results showed that fungal alpha diversity in GL soil was significantly higher than that in plantation forests,and the lowest diversity was found under SF(Fig.5).Sun et al.(2018)found no significant differences in fungal communities between forest and grasses in the same region.However,the results of studies conducted in the Loess Plateau as well as the Tianshan region of China support the results of this study (Wang et al., 2019, 2021).The lower fungal diversity in plantation soils compared to grassland soils may be due to a certain degree of inhibition by the allelopathy secreted by the root system(Nagati et al.,2019).As a coniferous species, spruce apoplast contains a large number of organic components that are not easily decomposed, such as lignin, tannin and resin,which may further limit the growth and reproduction of spruce soil microorganisms (Gallet, 1994).Different fungal communities can affect the decomposition of ground litter(Voˇríšková and Baldrian,2013).This may lead to a lower organic matter content in forest soils than in grassland soils.Additionally,plantation forest growth requires nutrient uptake from the soil along with the release of secretions,which indirectly affect microbial structure and abundance by changing the soil environment(Marschner et al., 1997; Broeckling et al., 2008; Yin et al., 2016).Tree species had a significant effect on fungal community structure(Hartmann et al.,2013;Ma et al.,2014;Urbanová et al.,2015).The results showed that soil fungal diversity was significantly lower in the SF than in the LF and PF, indicating that microbial diversity may be influenced by tree species.
Alpha diversity was significantly correlated with pH and AP via Pearson correlation (Table 1).Previous studies have found a significant correlation between soil microbial community composition and soil nutrients (Tian et al., 2017; Yang et al., 2017).Soil nutrients were significantly higher in LF soil than in GL soil,but fungal diversity was lower in LF soil than in GL.LF soil has greater SOC and may not recover to predegradation or natural forest characteristic levels after afforestation in terms of organic carbon content (Degryze et al., 2004; Olszewska and Smal, 2008).However, broadleaf trees have a greater capacity for soil organic carbon accumulation, which may be related to the higher biomass returned by broadleaf roots and litter than conifers.The organic matter content of the soil usually reaches its maximum during the most active season of aboveground plant growth (Xu et al., 2018).The soil total organic carbon content of different forest types in the Saihanba area has obvious seasonal dynamics (Qian et al., 2016).However, whether more environmental factors affecting fungal communities have such seasonal effects needs to be verified by further experiments.
Redundancy analysis(RDA)and the Mantel test showed that pH was the most significant factor affecting soil fungal communities(Fig.7).This result aligned with previous research by Romanowicz et al.(2016) and Liu et al.(2018) that identified soil pH as the most important environmental factor influencing forest soil fungi.This indicates that the physicochemical characteristics of the soil itself significantly affect the fungal community diversity.Soils in the Saihanba area are weakly acidic,with pH values ranging from 5.90 to 6.68 in different habitats.Basidiomycota is one of the dominant groups of fungi;its richness can respond positively to changes in the soil pH value,and the richness of Ascomycota is more abundant in soils with higher pH (Tedersoo et al., 2014).Although soil nutrient status varies between habitats, it is not a driving factor influencing changes in microbial community diversity and structure.
Planted forests are rich in symbiotic trophic functional taxa,and the soil environment may be more stable.Symbiotic trophic diversity was more abundant in soils with lower pH and fungal diversity,which is quite different from previous studies (Lindahl et al., 2007; Sterkenburg et al.,2015).However, there is no doubt that the symbiotic relationship between fungi and their environment could enhance the stability of the ecosystem(Averill et al.,2014;Treseder et al.,2018;Aqeel et al.,2023).The relationship between biodiversity and ecosystem stability is one of the key theoretical issues in ecology and was proposed by MacArthur(1955)and Elton(1958)in the 1950s.Some studies have concluded that there is no clear positive correlation between fungal alpha diversity and the stability of soil systems or stand health (Deng, 2012; Guo et al.,2020).However, specific microbial taxa have a positive impact on community resilience or resistance (Van Ruijven and Berendse, 2010;Delgado-Baquerizo et al., 2017).Considering the relatively fixed scale range of microbial community diversity analysis,the conclusions drawn by describing dominant populations are largely consistent(Griffiths and Philippot, 2013).However, it has also been shown that the degree of influence of species diversity on the ecological function of the community needs to be integrated with environmental conditions that cannot be generalized (Li et al., 2019).In the results of this study, soil fungal diversity in planted forest ecosystems showed variability across sampling sites.However, it is unclear whether the variability in fungal diversity and restoration potential is local or global.
4.4.The shift in fungal networks and their indicative role for forest restoration
In this study, the complexity of the networks (with a high number of nodes,edges,and average degree)showed that PF>GL>LF>SF(Fig.8).PF have the most complex network and seem to be more adapted to the current environment than LF and SF.Complex co-occurrence networks had a stronger ability to resist environmental interference than simple networks (Santolini and Barabási, 2018; Gao et al., 2022).However, the modularity index of different habitats is greater than 0.4 and has a typical modular structure with resistance to environmental changes (Newman,2006;Messier et al.,2019).The co-occurrence network was simplest in SF,which may be related to the least number of unique fungal genera in the soil.In addition,the Coniochaeta genus has a high degree of connectivity in the network, and it is possible that some species of this genus produce secondary metabolites that alter soil fungal communities in SF.Previous study has revealed the influence of phytopathogenic fungi on microbial network complexity and community assembly(Gao et al.,2021).Although the complexity of the soil fungal co-occurrence network in SF was lower than that of other habitats, it cannot be denied that they have a special resistance stability to environmental disturbances.Additionally,soil might provide a heterogeneous environment for fungal communities through nutrient cycling by plant roots (Lareen et al., 2016; Islam et al., 2020;Bastida et al.,2021;Navarrete et al.,2023);thus,the microbial community environment might vary depending on the plant species (Milenge Kamalebo et al.,2019;Kajihara et al.,2022).We observed significant differences in soil chemistry across habitats.Thus,the co-occurrence network may be influenced by a combination of inter-root environmental conditions and soil chemistry.However,further studies are needed to reveal the effect of the loss of fungal alpha diversity after afforestation on microbial co-occurrence networks.
Keystone species identification through co-occurrence networks is an effective method for describing fungal community interactions and structural changes(Herren and McMahon,2018;Z.Li et al.,2022).The results identified significantly different key genera in the fungal communities of the four habitats investigated(Table S6).The key genera in GL are Solicoccozyma and Cadophora, where Solicoccozyma is commonly found in various environments, and Cadophora is a dark septate endophyte (DSE) that has a strong saprophytic ability to degrade complex carbohydrates (Knapp et al., 2018).In LF soil, the key genera were mainly involved in ecological functions such as organic matter decomposition and bioremediation,where Humicola plays an important role in plant matter decomposition(including lignin and cellulose)and nutrient cycling(Andlar et al.,2018).Trichocladium degrades lignocellulose while altering the soil fertility status and influences key groups of surface soil nitrogen cycling processes (Uke et al., 2021; Sun et al., 2022).Many species in Pseudosigmoidea are plant pathogens that cause leaf spot diseases on various agricultural crops, but they can also modify a plant's ability to accumulate heavy metals by uptake and are applied in the remediation of polluted environments (Diene et al.,2014).In SF understory soil, the key genera were Russula, Tomentella, and Hygrophorus.These are ectomycorrhizal fungi that form reciprocal relationships with tree roots, promoting nutrient cycling in the ecosystem (Kuhar et al.,2016;Looney et al.,2018;Lu and Yuan,2021).Although Coniochaeta is not an ectomycorrhizal fungus, some species in the genus can produce various secondary metabolites with extreme antifungal activity (Wang et al., 1995).The PF habitat was found to be dominated by key genera like Ilyonectria, Trichocladium, Alternaria and Venturia which are pathogenic fungi or common pathogens.Ilyonectria, for example, can cause root rot in trees, while Alternaria can cause diseases in many crops(Woudenberg et al.,2013;Androsiuk et al.,2022).Venturia is also known as the model species of common pathogens in plant pathology(González-Domínguez et al.,2017).
Interestingly, a large amount of colonization was observed in the litter under the forest,which was negatively correlated with most of the enzyme activities, leading to slow litter decomposition (Zhang et al.,2019).However,microbial populations of the same ecological niche are unlikely to coexist in the long term because of competition (Kennedy et al., 2002).On the other hand, the key genus of soil fungi in the SF habitat belongs to ectomycorrhizal fungi, as previous studies have suggested that mycorrhizal symbiosis may be a strategy for fungi to overcome various stresses in the soil(Pereira et al.,2014;Balami et al.,2020).Although the network structure of SF is relatively simple,they may have adapted to the current environment to reach the climax community.Therefore, it is important to consider the key genera of soil fungi while selecting tree species and in plantation management.The results also showed that the key genera of Pinus sylvestris var.mongolica forest contain pathogenic fungi, which may make trees susceptible to growth retardation, decline and even death.Overall, the identification of keystone species in fungal co-occurrence networks provides insights into the ecological functions and interactions of fungal communities in different habitats.By understanding the key genera in different habitats, we can better inform the management and conservation of forest ecosystems.
5.Conclusion
Afforestation plays an important role in combating desertification.However, the correlation and variability of the fungal microbiome in typical plantation forests in the Saihanba area are not well documented.The results showed that afforestation significantly changed the diversity and structure of the soil fungal community in the Saihanba area.This suggests that soil fungal communities can be used as an indicator of forest restoration.Based on redundancy analysis(RDA)and the Mantel test,we hypothesize that soil pH differences between habitats led to higher variation in fungal diversity, resulting in the separation of fungal community composition and structure and potential ecological functions.Future studies, including additional indoor experiments via macrogenomic and meta-transcriptomic analysis, would provide a deeper understanding of the functional soil microbial community in forest ecosystems.Finally, understanding the differences between the fungal microbial communities in typical plantation and grassland soils can provide a basis for forest management and conservation to maintain global forest productivity and sustainable development.
Funding
This research was supported by the National Natural Science Foundation of China(Nos.32270010,U2003211 and 31870008)and Beijing Forestry University Outstanding Young Talent Cultivation Project (No.2019JQ03016).
CRediT authorship contribution statement
Kaichuan Huang: Methodology, Conceptualization, Validation,Formal analysis, Investigation, Data curation, Writing – original draft,Writing–review&editing.Zhenli Guo:Methodology,Formal analysis,Writing – original draft, Writing – review & editing.Wen Zhao: Investigation, Formal analysis, Validation, Writing – review & editing.Changge Song: Resources, Investigation.Hao Wang: Resources, Investigation.Junning Li: Formal analysis, Validation, Writing – review &editing.Reyila Mumin: Resources, Formal analysis.Yifei Sun: Methodology, Writing – review & editing.Baokai Cui: Methodology,Conceptualization, Data curation, Writing – original draft, Writing – review&editing,Funding acquisition,Supervision.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.i.org/10.1016/j.fecs.2023.100125.
杂志排行
Forest Ecosystems的其它文章
- Climate and fire drivers of forest composition and openness in the Changbai Mountains since the Late Glacial
- Complexity responses of Rhododendron species to climate change in China reveal their urgent need for protection
- Differential seed removal,germination and seedling growth as determinants of species suitability for forest restoration by direct seeding – A case study from northern Thailand
- Words apart: Standardizing forestry terms and definitions across European biodiversity studies
- Regeneration of Nothofagus dombeyi (Mirb.) Ørst.in little to moderately disturbed southern beech forests in the Andes of Patagonia (Argentina)
- Spatial niche segregation between bird species in the Białowie˙za primeval forest (NE Poland)
