Core collection construction of tea plant germplasm in Anhui Province based on genetic diversity analysis using simple sequence repeat markers
2023-09-16TAOLinglingTlNGYujieCHENHongrongWENHuilinXlEHuiLUOLingyaoHUANGKelinZHUJunyanLlUShengruiWElChaoling
TAO Ling-ling,TlNG Yu-jie,CHEN Hong-rong,WEN Hui-lin,XlE Hui,LUO Ling-yao,HUANG Ke-lin,ZHU Jun-yan,LlU Sheng-rui,WEl Chao-ling
State Key Laboratory of Tea Plant Biology and Utilization/Key Laboratory of Tea Biology and Tea Processing of Ministry of Agriculture and Rural Affairs/Anhui Provincial Laboratory of Tea Plant Biology and Utilization,Anhui Agricultural University,Hefei 230036,P.R.China
Abstract
The tea plant [Camellia sinensis (L.) O.Kuntze] is an industrial crop in China.The Anhui Province has a long history of tea cultivation and has a large resource of tea germplasm with abundant genetic diversity.To reduce the cost of conservation and utilization of germplasm resources,a core collection needs to be constructed.To this end,573 representative tea accessions were collected from six major tea-producing areas in Anhui Province.Based on 60 pairs of simple sequence repeat (SSR) markers,phylogenetic relationships,population structure and principal coordinate analysis (PCoA) were conducted.Phylogenetic analysis indicated that the 573 tea individuals clustered into five groups were related to geographical location and were consistent with the results of the PCoA.Finally,we constructed a core collection consisting of 115 tea individuals,accounting for 20% of the whole collection.The 115 core collections were considered to have a 90.9% retention rate for the observed number of alleles (Na),and Shannon’s information index (I) of the core and whole collections were highly consistent.Of these,39 individuals were preserved in the Huangshan area,accounting for 33.9% of the core collection,while only 10 individuals were reserved in the Jinzhai County,accounting for 8.9% of the core set.PCoA of the accessions in the tea plant core collection exhibited a pattern nearly identical to that of the accessions in the entire collection,further supporting the broad representation of the core germplasm in Anhui Province.The results demonstrated that the core collection could represent the genetic diversity of the original collection.Our present work is valuable for the high-efficiency conservation and utilization of tea plant germplasms in Anhui Province.
Keywords: tea plant,core collection,genetic diversity,SSR markers
1.lntroduction
The tea plant [Camelliasinensis(L.) O.Kuntze] is a perennial woody plant with abundant secondary metabolites (Taietal.2015; Liang and Shi 2015).Tea plants are typical self-incompatible plants,processing high genetic diversity owing to long-term natural selection and cross-pollination for generations (Chen 2006).The tea plant growing areas are mainly distributed in the Huangshan and Dabie Mountain areas in Anhui Province,which are located on the Yangtze River Valley and Haihe Valley,respectively.These areas have the characteristics of both the north and south climatic zones in China and breeds rich and diverse tea plant germplasm resources (Suetal.2019),including “ZhuyeZhong”,“ShidaCha”,“QianFan”,and “JinJi” species.Many famous historical teas,such as “QiHong”,“Taiping Houkui”,“Lu’an Guapian”,and “Huoshan Huangya”,are made of the corresponding species.Furthermore,Anhui Province has a long history of tea plantation and retains some ancient tea plant germplasms growing in natural alpine communities.
Tea plants,as self-incompatible species,are difficult to protect and utilize,as well as costly to manage and maintain.A core collection needs to be established,which shows that a minimal number of germplasm resources represent the maximum level of genetic diversity and structure of the entire collection (Frankel and Brown 1984; Brown 1989).Studies on core germplasm have been conducted for many crops including apple (Kimetal.2019),apricot (Bourguibaetal.2017),rapeseed (Chenetal.2017),radish (Leeetal.2018),and chilli (Mongkolpornetal.2014).Core collection construction of tea plant germplasm has also been conducted in other provinces.Huangetal.(2022) collected 110 tea plant accessions from Hunan Province and constructed a core set that consists of 22 tea plant accessions.Niuetal.(2020) collected 415 tea plant accessions from Guizhou and developed a mini-core set,which consist of 37% of the whole collection.Taniguchietal.(2014) collected 788 tea plant accessions from 14 countries around the world and developed a core collection consisting of 192 tea plant accessions.Because tea plants are selfincompatible and have complex genetic diversity,more germplasms are needed to construct a core collection.The construction of core collection for tea plants can decrease genetic redundancy and help breeders reduce the cost of managing tea plants.Therefore,construction of a core set from different regions is urgently required to ensure retention of the maximum genetic diversity.At present,data sources for constructing core germplasms are mainly based on morphological and molecular markers (Herrmannetal.2017).Molecular markers based on DNA polymorphism such as restriction fragment length polymorphisms (RFLPs),random amplified polymorphisms (RAPDs),single nucleotide polymorphisms (SNPs),and simple sequence repeats (SSRs) (Wangetal.2016; Zhouetal.2020; Lietal.2023),are not affected by the period of plant growth and are more suitable for constructing core collection than morphological markers (Jiang 2013).SSR markers are characterized by a number of allelic variations,high information and polymorphism,and common dominant inheritance,and are the most widely used molecular marker to identify germplasm resources.Research by Tanetal.(2015) showed that 8 out of 30 SSR markers can be employed as a core marker set,which can clearly identify the fingerprints of 128 China cloned tea varieties.Liuetal.(2017) identified that 36 new highly polymorphic genomic SSR markers using whole genome-based SSR loci,and finally selected 5 SSR markers as core marker set for fingerprinting the 80 tea plants.Zhangetal.(2018) used SSR and three other molecular markers to analyze the genetic diversity of 50 tea plant in Qinba area.To accurately classify and retain germplasm resources from a molecular perspective,we selected SSR markers to construct the core germplasm of tea plants.Several provincial genetic diversity assessments and construction of core collections have recently been reported (Luetal.2021).However,an in-depth study on tea germplasm resources in Anhui Province is scarely.
In this study,60 SSR markers were used to analyze the genetic diversity and population structure of 573 tea accessions from Anhui Province,and the core collection was constructed.This core collection consisted of 115 tea individuals,including 31 ancient tea plants,80 landraces,and 4 cultivars,which will provide a reference standard and excellent germplasm resources for the protection,management,and utilization of tea plant germplasm resources in Anhui Province.
2.Materials and methods
2.1.Plant materials and DNA extraction
A total of 573 tea accessions were collected from Anhui Province,including ancient tea plants,landraces,and cultivars.Besides,14 tea accessions from Yunnan Province were used as the outgroup.Ancient tea plant here refers to the wild tea plants growing in natural forest land for over 100 years of cultivation (Jietal.2011).The samples were collected from local tea plantations with colony species called landraces,while national and provincial varieties are referred to as cultivars (Liuetal.2018; Nieetal.2021).To ensure that the collected tea plants were sufficiently geographically separated and to avoid collecting multiple samples from the same parent,fresh tea leaves were collected from 26 counties in six cities of Anhui Province (Table 1).Detailed information on these samples,including the sample number,accession name,and geographical location,is listed in Appendix A.
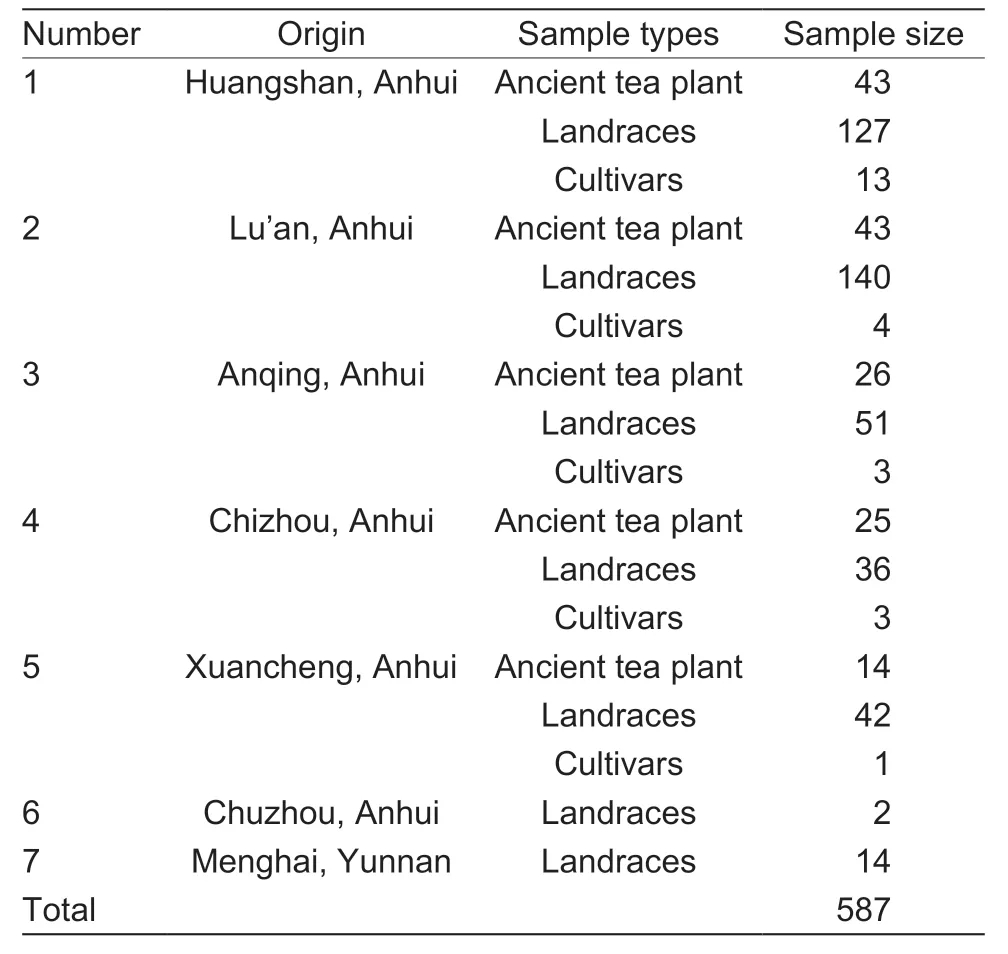
Table 1 Origins and sample sizes of tea plant germplasm for analysis
Fresh branches from different tea plants were inserted into wet flower mud,and their leaves were immediately preserved at –20°C within a refrigerator until further use.A modified cetyltrimethylammonium bromide method (Clarke 2009) was applied to extract genomic DNA from young leaves.The consistency and quality of the extracts were measured using a Nanodrop 2500 (Thermo Fisher Scientific,USA) and 1.0% agarose gel electrophoresis,respectively.The extracted DNA samples were stored at –20°C within a refrigerator.
2.2.Selection of SSR markers and PCR amplification
To ensure that the selected SSR markers were polymorphic,a total of 60 SSR markers were selected from previous reports or based on the genome of tea plants (Yaoetal.2012; Liuetal.2017; Zhuetal.2023).A total of 45 tea plants were used for preliminary screening,and all samples were used for final screening.Finally,60 highly polymorphic SSR markers were obtained to effectively distinguish tea plants in Anhui Province.The primer sequences and polymorphisms have been mentioned in Appendix B,and primers were synthesized by General Biological Co.,Ltd.(China).
DNA was diluted to 30–50 ng μL–1,and 10 μL PCR mixture contained 5 μL 2× Taq Plus Master Mix,3 μL double distilled H2O,1 μL genomic DNA,and 0.5 μL of forward and reverse primers each.PCR amplification procedure was as follows: pre-denaturation at 94°C for 3 min,32 cycles of 30 s at 94°C for denaturation; a reaction time of 30 s with the annealing temperature depending on primers,and extension for 35 s at 72°C,the extension time was determined according to the length of the amplification product.PCR products with tandem repeats were separated by automatic capillary electrophoresis (Fragment AnalyzerTM96,AATI,USA) as previously described (Liuetal.2019).PROSize Software v.2.0 was used to analyze the results of the Fragment AnalyzerTM96 system,and fragments with clear bands were used for counting.
2.3.Analysis of genetic diversity parameters
The results of capillary and artificial tape readings were used to access the clear and repeated bands in the range of target fragments.Based on the molecular mass,fragment lengths were converted into A,B,C,D and more every 4–6 bp.One band represented homozygosity,and two bands represented heterozygosity for each sample,indicating genotypes such as AA,AB,BC,BD,DE,and EE.The allele positions of different primers were different,and the amplified bands were sorted to form a matrix (Liuetal.2018).According to the matrix data,the polymorphism information content (PIC) and major allele frequency (MAF) were computed using PowerMarker v.3.25 (Liu and Muse 2005).Expected heterozygosity (He),observed heterozygosity (Ho),number of alleles (Na),and Shannon information index (I) were calculated using Popgen v.32 software.Nei’s genetic distance between tea individuals was also calculated using PowerMarker v3.25.
2.4.Phylogenetic tree and analysis of the population structure
Genetic distance was clustered using the unweighted pair group method with arithmetic mean (UPGMA) (Tamuraetal.2007).The ITOL website was used to enhance and edit the phylogenetic tree (Letunic and Bork 2019).Population structure was analyzed using the STRUCTURE Software,and the number of optimal populations was estimated.The population structure was based on the Bayesian model,and the maximum likelihood value was generated for the simulation results of everyK-value.A high maximum likelihood value indicated aK-value close to the real situation (Evannoetal.2005).The burn-in period and MCMC iterations were both set at 10 000.The grouping value (K-value) range of the subgroups was 2–10,and simulations for eachK-value were repeated thrice.Subsequently,data were uploaded to the Structure Harvester (http://taylor0.biology.ucla.edu/structureHarvester/) website to calculate the most suitableK-value based on the ΔKmethod.PCoA was performed using GenAlEx v.6.503 (Peakall and Smouse 2006).
2.5.Construction of core collection and evaluation of hereditary diversity
The R package Core Hunter can sample different and representative subsets from a large number of germplasm collections (Thachuketal.2009).Based on the genetic distance matrix,the numbers of core and reserved collections were calculated using the Core Hunter.Genetic diversity parameters of the whole collection and core collection were compared by one-way analysis of variance (ANOVA) using SPSS 25.
3.Results
3.1.Polymorphism of the molecular markers
A total of 487 alleles were detected using the 60 SSR markers (Appendix B),and the averageNavalue detected by each marker was 8.250.TheHoandHevalues were 0.424 and 0.680,respectively.The highest PIC of marker was 0.817 (TM255),while CsLAC36 marker has the lowest PIC value (0.316),with the average PIC being 0.642.Fifty-one out of the sixty selected markers had polymorphisms >0.5,representing highly polymorphic primers.In addition,the PIC values of nine markers were <0.5,representing moderately polymorphic markers and indicating that polymorphism of the selected markers is generally high.The averageIvalue of the population was 1.427.In summary,the collected germplasm resources had abundant genetic diversity.
3.2.Genetic diversity of different populations of tea plants
A total of 573 tea plants were collected from 26 counties of six major tea-producing areas in Anhui Province (Table 1),containing the representative local varieties of Anhui (Table 2).These sample included 151 ancient tea plants,397 landraces,and 25 cultivated tea plants (Appendix A).The genetic diversity of the three different tea populations was analyzed based on SSR markers (Table 3).The results showed that theIvalue of ancient tea plants,landraces,and cultivar groups were >1,indicating that three different populations had abundant genetic diversity.However,the PIC value andIvalue of ancient tea plants were higher than those of landraces and cultivar groups,which may be owing to the fact that ancient tea plants grow in natural communities and experience long-term natural selection and crosspollination,resulting in complex community structure and high genetic diversity (Yangetal.2022).
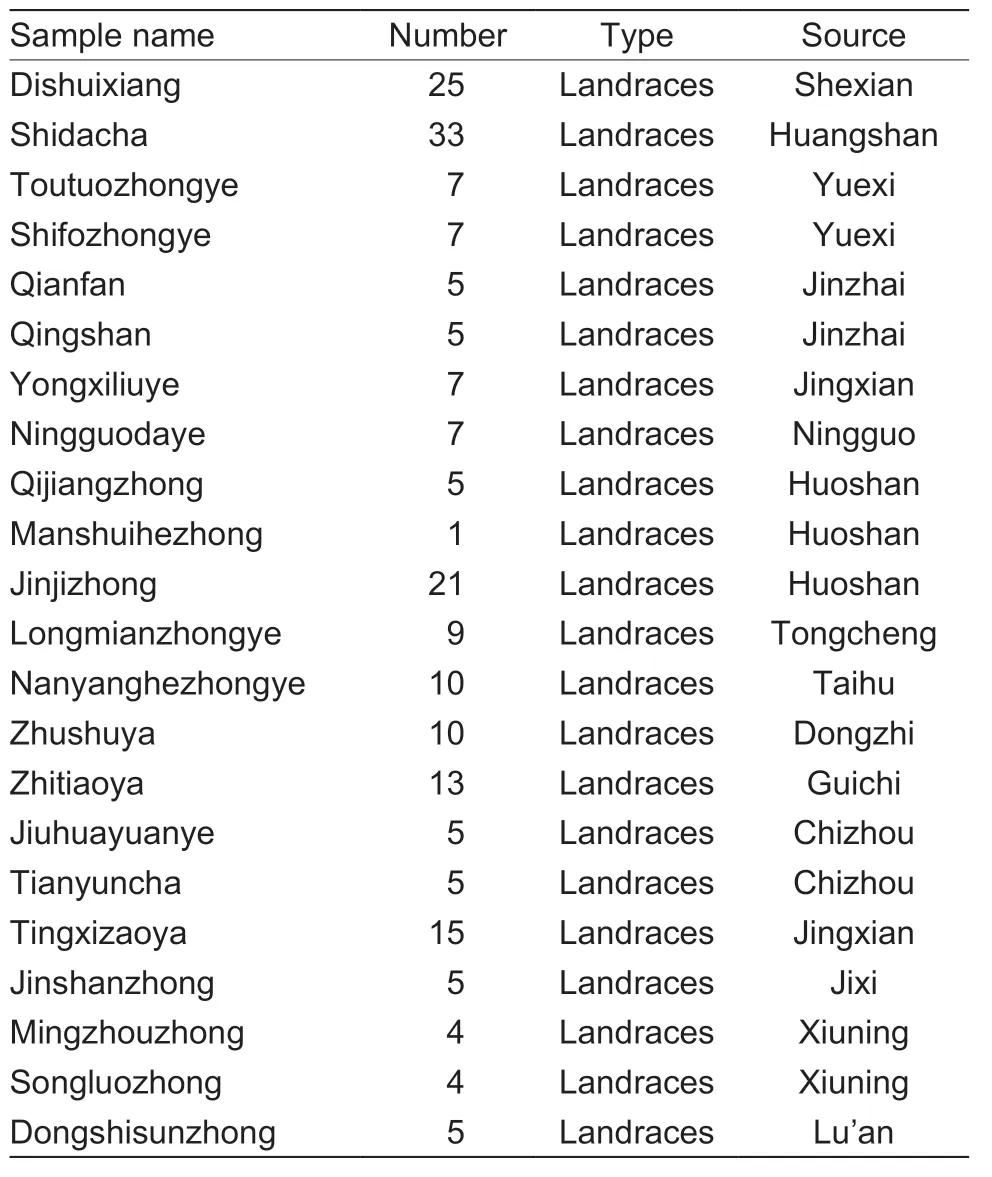
Table 2 Information of local representative germplasm resources in Anhui Province
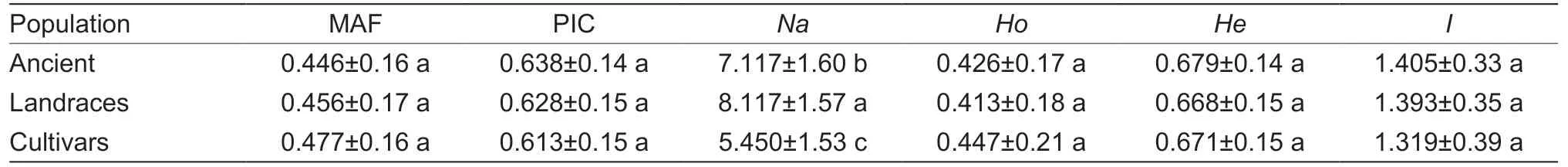
Table 3 Genetic diversity among different tea populations1)
3.3.Phylogenetic relationship of tea plants in Anhui Province
To probe the phylogenetic relationship among tea plants in Anhui Province,60 polymorphic SSR markers were used to amplify specific SSR loci of those tea plants.A phylogenetic tree was constructed using the UPGMA method based on Nei’s genetic distance,which explored the genetic relationship of 573 tea plants in Anhui Province,and 14 tea plants from Yunnan Province used as the outgroup (Fig.1).Group I included all tea plants in Yunnan and was divided into one branch separately.Group II was an admixed population,with ancient tea plants and landraces from Houkeng in Huangshan (9),ancient tea plants collected from Shucheng (7),and a few landraces from other cultivated regions.Group III was divided into two subgroups,and III-1 consisted of tea germplasm in Huangshan (28) and Chizhou (2).The small geographical distance between Huangshan and Chizhou supports that mutual introduction might have taken place in the early stage,because some tea plants are very similar in genetic diversity,leading to the aggregation of tea germplasms collected from these two regions.In subgroup III-2,80% of the tea plants were collected from Xianhualing,Qitoushan,and Bianfudong in Jinzhai County and the remaining 20% included 7 ancient tea plants from Zhushan Ancient Tea Estate in Yuexi County.Group IV included tea plants from Huangshan,Wangmantian Village in Shexian County of Huangshan,and Houkui strains collected from the Huangshan Tea Expo Garden.Huangshan has a long history of tea cultivation and plantation,causing the difference in genetic diversity between tea plants of Huangshan and other areas in Anhui,and the tea germplasms in Huangshan were separately grouped together.Tea plants from the tea area of southern Anhui,including Xuancheng (51),Chizhou (24),and Huangshan (45) counties formed Group V.As the geographical distance among these three places is small,tea plants in different regions must have been mutually introduced,leading to similar genetic diversity of tea plants collected from these areas and consequently phylogenetic clustering.Group VI tea germplasms were collected from the tea area of Dabie Mountain in western Anhui,including Lu’an and Anqing.In addition,15 ancient tea plants collected from Houkeng in Huangshan were also included,which indicated that tea plants in western Anhui were closely related to ancient tea plants in Huangshan,and tea germplasm in Lu’an and Anqing may have been introduced from Huangshan.
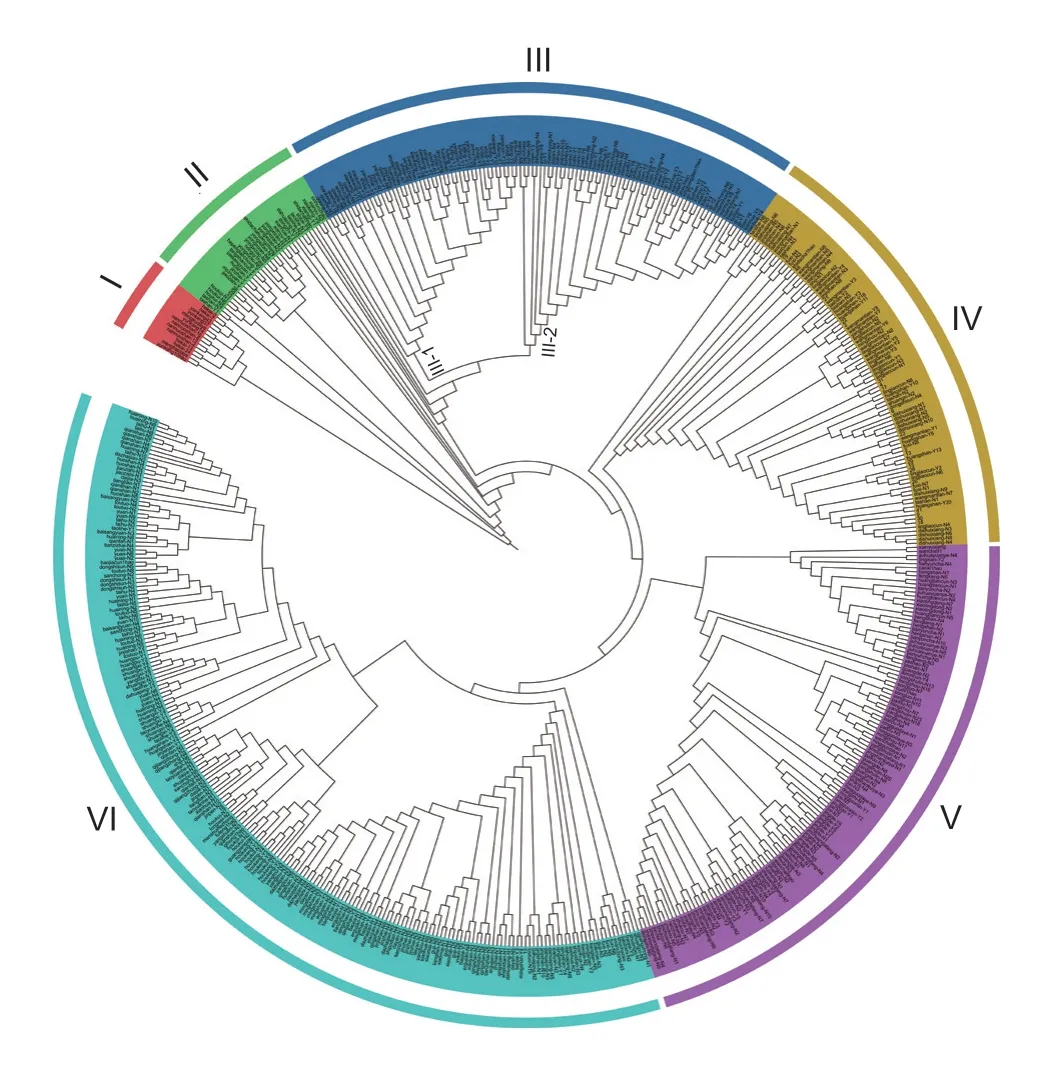
Fig.1 Phylogenetic relationship of 587 tea individuals with six different colors indicating six groups.The different colors represent different groups.Group I consists of 14 tea plants from Yunnan Province as outgroup.And 573 tea plants in Anhui Province are divided into several other groups.
These results suggest that tea germplasms from different geographical sources clustered in different groups,indicating that geographical isolation may have an impact on the genetic divergence of tea germplasms.However,no obvious distinction was noticed among ancient tea plant,landraces,and cultivars.If cultivated accessions were bred from landraces and the cultivated species were sexually propagated from ancient tea plants,they are expected to genetically cluster (Zhaoetal.2021).
3.4.Principal coordinate analysis and population structure of the tea germplasm
To validate the results of the phylogenetic analysis,587 tea plant individuals underwent PCoA with GenAlEx v.6.503.The results showed that the individuals were clearly clustered into six different groups (Fig.2),which was consistent with the results of the phylogenetic analysis.
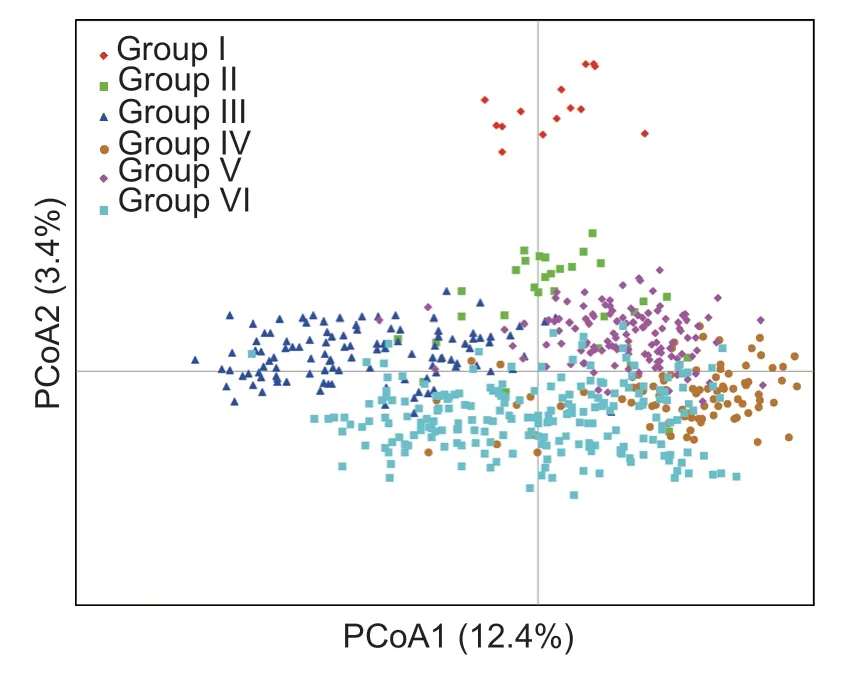
Fig.2 PCoA plot of 587 tea plant samples with six different colors indicating six groups obtained by analyzing the phylogenetic tree.Red represents the Yunnan population.Tea populations in Anhui Province are divided into five other colors.
Population structure analysis showed that ΔKvalue is the largest whenK=5,indicating that the fit is the best when the sample are divided into five groups (Fig.3).Group III comprised solely of the 14 tea accessions from Yunnan Province.The clustering results of the other four groups were consistent with those obtained by the UPGMA method,where tea germplasms from different geographical sources were clustered into different groups,particularly in the tea areas of the western and southern regions of Anhui.Tea individuals collected from Lu’an and Anqing cities were divided into group I and V,respectively.Group II contained tea plants principally from Xuancheng and Chizhou,while a few tea individuals were from Huangshan,corresponding to the clustering in the phylogenetic tree.Finally,most of the germplasms from Huizhou,Shexian,and Qimen of Huangshan were classified in group IV.The results indicated that genetic differentiation existed between tea plants from Huangshan and those from other regions,and geographical isolation caused inconsistent genetic diversity among tea plants of different regions.
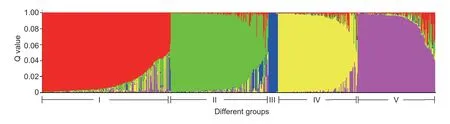
Fig.3 Analysis of population structure using Structure v.2.3.4.ΔK value is the largest when K=5.
3.5.Construction of tea plant core collection in Anhui Province
Using Core Hunter,the numbers of core collections and reserved varieties were calculated based on the genetic distance matrix among tea plants.One-way ANOVA was used to verify significant differences in the genetic diversity parameters between the constructed core and original subsets (Table 4).A total of 115 tea plant individuals were suggested to be preserved,accounting for 20% of the original collection,with no significant difference in genetic diversity parameters between the core and original collections.The genetic diversity of core and whole collection was evaluated using PCoA to confirm that the core collections were uniformly scattered across the plot (Fig.4).The germplasm from the major tea-producing areas of Anhui Province was retained,and detailed information on the core germplasm is provided in Appendix C.
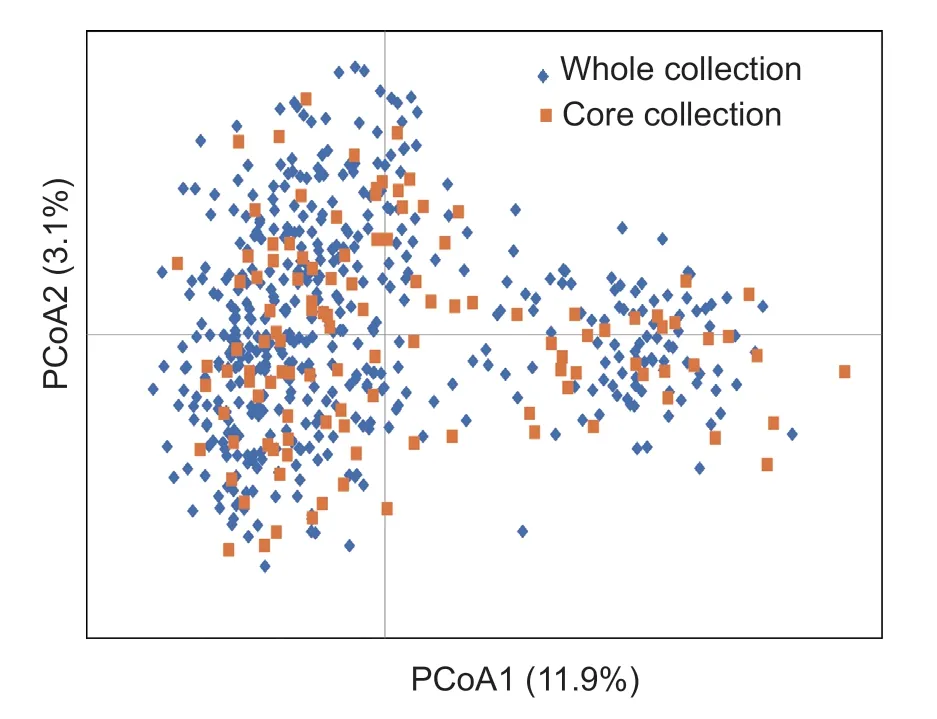
Fig.4 PCoA of the core collection (n=115) and whole collection (n=573) were based on genotypic data from SSR markers.The yellow represents the variety of core collection,and the blue represents the whole collection.

Table 4 Genetic diversity among different sets1)
A total of 31 ancient tea plants,80 landraces,and 4 cultivars were included in the core collection,and Appendix D showed some agronomic characters of ancient tea plants.A total of 39 individuals from the Huangshan region were retained,accounting for 33.9% of the core collection (Table 5),followed by Chizhou (15),Huoshan (15),Anqing (13),Shucheng (12),Xuancheng (11),and Jinzhai (10).As shown in Table 6,local tea plant resources from Anhui were retained in the core collection.The local germplasm consisted of all group species,and some plants showed similar genetic characteristics; therefore,one tea individual was sufficient to represent the accession.In addition,in the core collection of Huangshan “Shidacha” and Huoshan “Jinjizhong” reserved 9 tea individuals,respectively.The core collection retained genetic information of the original collection with the minimum number of individuals.The germplasms of 115 tea plants effectively represented the core germplasm of major tea-producing areas in Anhui Province.By establishing the core germplasm and reducing the genetic redundancy of tea germplasm in Anhui Province,these germplasms can be used for effectively breeding new varieties.
4.Discussion
Abundant and diverse germplasm resources lay an important foundation for germplasm innovation,genetic improvement,and variety breeding.The Anhui Province has a long history of tea plant cultivation,while these germplasm resources are limited to excavated and utilized.A large number of germplasm resources including 573 tea plant individuals were collected from Anhui Province,higher than that used for similar studies in other provinces,such as the genetic diversity of 126 tea plants analyzed in Guangxi Province (Guoetal.2021) and the population structure of 145 tea plants researched in Guizhou Province (Zhaoetal.2021).In this study,60 SSR markers were used to analyze the genetic diversity of 573 tea plants in Anhui Province,and the average PIC value was found to be 0.642,which is higher than those reported by previous studies for other markers,such as EST-SSR and SNP (Maetal.2012; Niuetal.2020; Huangetal.2022).It is possible that the collected tea plant genetic resources have complex background and rich genetic diversity.Another explanation is that the selected SSR markers are all reported markers with high polymorphism,and the higher resolution of the capillary DNA Fragment Analyzer™ 96,which greatly promotes the higher polymorphism of allele score.This indicates that tea plants in Anhui Province have high genetic diversity,which is worth further exploration in breeding.Analysis of the genetic diversity of different populations showed that ancient tea plants had a higher level of genetic diversity (I=1.405,PIC=0.638) than that of landraces (I=1.393,PIC=0.628) and cultivars (I=1.319,PIC=0.613),which is consistent with the results of a previous study (Luetal.2021).Long-term cross-pollination and natural selection result in high heterozygosity of ancient populations and rich genetic diversity,while artificial breeding reduces the genetic diversity among cultivated varieties (Jietal.2009).Compared with other tea plant-related studies,the tea germplasm resources in Anhui Province have relatively abundant genetic diversity (Qiaoetal.2010; Niuetal.2020; Huangetal.2022).
Population structure analysis showed that the highest clustering value wasK=5,and the phylogenetic tree demonstrated that 573 tea individuals from Anhui and 14 accessions from Yunnan were clearly divided into six groups,which was consistent with the results of PCoA.The 573 tea individuals were clustered in five other groups,and most of those from Anqing and Lu’an were assigned to one group.The varieties from Chizhou and Xuancheng were clustered into another group,while the rest of the varieties in Huangshan were clustered into a single group.Ancient tea plants,landraces,and cultivars from each region were all clustered in these groups,which indicates that landraces and cultivars might have been introduced from ancient local tea plants (Xiaetal.2020).In addition,the Yunnan population could be separated from Anhui populations,indicating that tea germplasms in Anhui Province has a relatively far genetic relationship with tea germplasm from Yunnan,which was consistent with the patterning owing to geographical distribution.The results showed that tea germplasm from different geographical sources gathered in different groups,and geographical isolation might lead to certain genetic differentiation among tea plants in different regions.The utilization of perennial self-incompatible plants by constructing a core collection is crucial for managing and conserving germplasm resources (Chen and Zhou 2005).At present,two national germplasm resource banks in Hangzhou and Menghai contain the majority of cultivars and varieties ofCamelliatea plants (Mao 2015).Therefore,constructing a core set of tea plants from Anhui Province is necessary to optimize the collection of tea plant germplasms.In this study,a core collection consisting of 115 tea individuals was constructed,accounting for 20% of the 573 original germplasms (Table 4).Compared with the entire collection,the retention rates of MAF,NaandHowere 92.9,90.9 and 91.3%,respectively,in the core collection,demonstrating that only insignificant differences were noticed in the genetic diversity indices between the core and whole sets.The core retention was similar to that of other woody plants,such as apricot and peach plants (Lietal.2007; Bourguibaetal.2017).Remarkably,the Huangshan region retained 39 accessions,accounting for 33.9% of the core collection (Table 5).Huangshan is a famous tea-producing area with abundant and diverse genetic resources of tea plants,such as “Shidacha” and “Dishuixiang” accessions,therefore it accounted for a relatively high proportion in the reserved collection (Jiangetal.2009; Nieetal.2021).
5.Conclusion
In this study,a comprehensive genetic diversity analysis was conducted on 573 tea plant germplasm including ancient tea plants,landraces and cultivars collected from Anhui Province,and a core collection of Anhui tea plants was constructed preliminarily.This study provides important information for breeders to protect and utilize tea plant genetic diversity,which will contribute to germplasm conservation,genetic research and breeding.More attention should be paid to core collections to improve the efficiency of germplasm exchange and utilization among tea plants.
Acknowledgements
This study was supported by the Project of Major Science and Technology of Anhui Province,China (202003a06020021),the National Key Research and Development Program of China (2021YFD1200200,2021YFD1200203),the National Natural Science Foundation of China (U20A2045),the Base of Introducing Talents for Tea Plant Biology and Quality Chemistry (D20026),and the Anhui Provincial Natural Science Foundation,China (2108085QC121).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.07.020
杂志排行
Journal of Integrative Agriculture的其它文章
- Combining nitrogen effects and metabolomics to reveal the response mechanisms to nitrogen stress and the potential for nitrogen reduction in maize
- Natural variations and geographical distributions of seed carotenoids and chlorophylls in 1 167 Chinese soybean accessions
- Carbon sequestration rate,nitrogen use efficiency and rice yield responses to long-term substitution of chemical fertilizer by organic manure in a rice–rice cropping system
- ldentification and epitope mapping of anti-p72 single-chain antibody against African swine fever virus based on phage display antibody library
- Dissecting the genetic basis of grain color and pre-harvest sprouting resistance in common wheat by association analysis
- The PcERF5 promotes anthocyanin biosynthesis in red-fleshed pear (Pyrus communis) through both activating and interacting with PcMYB transcription factors
