Dissecting the genetic basis of grain color and pre-harvest sprouting resistance in common wheat by association analysis
2023-09-16YANShengnanYUZhaoyuGAOWeiWANGXuyangCAOJiajiaLUJieMAChuanxiCHANGChengZHANGHaiping
YAN Sheng-nan,YU Zhao-yu,GAO Wei,WANG Xu-yang,CAO Jia-jia,LU Jie,MA Chuan-xi,CHANG Cheng,ZHANG Hai-ping
Key Laboratory of Wheat Biology and Genetic Improvement on Southern Yellow & Huai River Valley,Ministry of Agriculture and Rural Affairs/College of Agronomy,Anhui Agricultural University,Hefei 230036,P.R.China
Abstract
Pre-harvest sprouting (PHS) adversely affects wheat quality and yield,and grain color (GC) is associated with PHS resistance.However,the genetic relationship between GC and PHS resistance remains unclear.In this study,168 wheat varieties (lines) with significant differences in GC and PHS resistance were genotyped using an Illumina 90K iSelect SNP array.Genome-wide association study (GWAS) based on a mixed linear model showed that 67 marker-trait associations (MTAs) assigned to 29 loci,including 17 potentially novel loci,were significantly associated with GC,which explained 1.1–17.0% of the phenotypic variation.In addition,100 MTAs belonging to 54 loci,including 31 novel loci,were significantly associated with PHS resistance,which accounted for 1.1–14.7% of the phenotypic variation.Subsequently,two cleaved amplified polymorphic sequences (CAPS) markers,2B-448 on chromosome 2B and 5B-301 on chromosome 5B,were developed from the representative SNPs of the major common loci Qgc.ahau-2B.3/Qphs.ahau-2B.4 controlling GC/PHS resistance and PHS resistance locus Qphs.ahau-5B.4,respectively.Further validation in 171 Chinese mini-core collections confirmed significant correlations of the two CAPS markers with GC and PHS resistance phenotypes under different environments (P<0.05).Furthermore,the wheat public expression database,transcriptomic sequencing,and gene allelic variation analysis identified TraesCS5B02G545100,which encodes glutaredoxin,as a potential candidate gene linked to Qphs.ahau-5B.4.The new CAPS marker CAPS-356 was then developed based on the SNP (T/C) in the coding sequences (CDS) region of TraesCS5B02G545100.The high-density linkage map of the Jing 411/Hongmangchun 21 recombinant inbred lines (RILs) constructed based on specific locus amplified fragment sequencing markers showed that CAPS-356 collocated with a novel QTL for PHS resistance,supporting the role of TraesCS5B02G545100 as the potential candidate gene linked to Qphs.ahau-5B.4.These results provide valuable information for the map-based cloning of Qphs.ahau-5B.4 and breeding of PHS resistant white-grained varieties.
Keywords: common wheat,grain color,PHS resistance,GWAS,90K SNP,CAPS marker
1.lntroduction
Pre-harvest sprouting (PHS) in wheat (TriticumaestivumL.)is the phenomenon where the mature grains germinate directly on the spike before harvest.PHS severely reduces grain yield and the baking quality of dough by making it porous,sticky,and off-color due to increased activities of amylases,lipases,and proteases which can degrade starch,lipids,and proteins in the sprouting grains (Maresetal.2005; Andreolietal.2006; Kulwaletal.2012; Simseketal.2014; Zhouetal.2017; Alietal.2019).Seed dormancy is known to be one of the most influential factors affecting PHS resistance (Maresetal.1993).Warm and dry conditions during ripening will induce PHS by reducing the level of dormancy,and a moist and cool environment after grain ripening in the field is favorable for promoting germination (Reddyetal.1985).PHS has been reported in many countries around the world,such as China,India,the USA,Canada,Germany,Japan,and Australia.In China,the total area affected by PHS is 24.91 million hectares,which is 83% of the wheat planting area,especially in the northern spring wheat region,Yangtze River Valley,and northeastern spring wheat region which are characterized by frequent rainfall and high humidity during harvest (Xiaoetal.2002).In recent years,it has also become a serious problem in the Yellow and Huai Valley wheat regions due to climate changes (Alietal.2019).It is estimated that PHS damage is responsible for up to 1 USD billion in annual losses (Vetchetal.2019).Developing and growing wheat varieties with high levels of PHS resistance are necessary to mitigate the risk of PHS.
Grain color (GC) has long been associated with PHS,and red-grained wheat varieties are usually more tolerant to PHS than the white-grained varieties (Flintham 2000; Warneretal.2000; Himietal.2002).However,farmers and the flour processing industry in China prefer whitegrained varieties due to their high flour yield,more efficient flour extraction,a high ash content,a more favorable appearance and a less bitter taste in the final product (Langetal.2021).Therefore,understanding the correlation between GC and PHS resistance will provide valuable information for improving the PHS resistance of white-grained wheat varieties.
Various major and minor quantitative trait loci (QTLs)/genes on the 21 chromosomes jointly control PHS resistance in wheat.Of these,the major QTLs have been mapped on chromosomes 2A (Zhuetal.2016),2B (Munkvoldetal.2009),3A (Osaetal.2003),4A (Chenetal.2008),and 6B (Zhuetal.2019).In addition,six genes controlling PHS have been isolated by map- or homology-based cloning,includingTaVp-1encoding a B3-domain transcription factor (Yangetal.2007; Changetal.2009),TaMFT/TaPHS1encoding a phosphatidyl ethanolamine binding protein (Nakamuraetal.2011; Liuetal.2013),TaMKK3encoding a mitogen-activated protein kinase kinase 3 (Toradaetal.2016),TaQsd1encoding an alanine aminotransferase (Weietal.2019),andTaDOG1L1(Ashikawaetal.2010) andTaSdrwith unknown functions (Zhangetal.2014,2017).These studies associated the identified genes with diverse roles,indicating a complex genetic mechanism underlying PHS resistance.However,the functions of a few genes,includingTaDOG1L1andTaSdr,remain unclear.Therefore,exploring the candidate genes that control PHS resistance from different germplasms is necessary to fully comprehend the genetic mechanism underlying PHS resistance in wheat.
The pigmentation of red-grained wheat is presumed to be a result of the accumulation of the polyphenolic compound phlobaphene,which is derived from the precursor catechin and is an end-product of the flavonoid pathway (Miyamoto and Everson 1958; Gordon 1979).GC is primarily controlled by theR-1geneTamyb10,encoding an R2R3 MYB transcription factor at the end of the long arm of chromosomes 3A,3B,and 3D.Tamyb10,a transcription activator involved in flavonoid synthesis,regulates GC by modulating the expression levels of genes encoding chalcone synthase (CHS),chalcone isomerase (CHI),flavanone 3-hydroxylase (F3H),and dihydroflavonol 4-reductase (DFR) (Himi and Noda 2005; Himietal.2011).Recent studies have identified more QTLs (Alemuetal.2020) and marker trait associations (MTAs) regulating GC (Groosetal.2002; Kumaretal.2009; Linetal.2016; Zhouetal.2017; Alemuetal.2020; Rabieyanetal.2022),indicating that the genetic loci for GC include components other thanTamyb10,and the genetic mechanism underlying GC formation is complex.
Researchers have proposed two hypotheses for explaining the genetic relationships between GC and PHS resistance,including the “pleitropic” and “tightly linked” mechanisms.For example,Flinthametal.(2000) reported that the dominantR-1gene controlling GC enhanced seed dormancy and resulted in a high PHS resistance level in red-grained wheat.Groosetal.(2002) located a QTL associated with both GC and PHS resistance on the short arm of chromosome 5A in wheat.Kumaretal.(2009) also mapped a QTL associated with GC and PHS resistance at the end of the long arm of wheat chromosome 3B.Langetal.(2021) found that the geneMyb10-Dcontrolling GC confers PHS resistance by regulating the key geneNCED(9-cis-epoxycarotenoid dioxygenase) involved in the ABA biosynthesis pathway in wheat.These results indicated a direct association between GC and PHS resistance.However,various components of the genetic network regulating GC and PHS resistance remain unexplored in wheat.Therefore,it is necessary to delineate the genetic mechanism and identify the candidate genes regulating both traits from different germplasms to improve PHS resistance in whitegrained wheat.
In the present study,the objectives were (1) to analyze the phenotypic differences of GC and PHS in 168 wheat varieties (lines); (2) to detect QTLs associated with GC and PHS by association analysis; (3) to explore the association between GC and PHS; and (4) to identify the potential candidate gene for the major locusQphs.ahau-5B.4controlling PHS resistance alone,and develop a CAPS marker for marker-assisted selection in white-grained wheat breeding programs.The findings of this study will improve our understanding of the genetic network that simultaneously regulates GC and PHS resistance and provide valuable gene resources for enhancing the PHS resistance of white-grained wheat varieties.
2.Materials and methods
2.1.Plant materials
A set of 168 wheat varieties (168WV) with significant differences in GC and PHS resistance were used for genome wide association study (GWAS).These plants were grown at the Dayangdian Experimental Station of Anhui Agricultural University in Hefei (31°58´N,117°240´E) during the 2014–2016 wheat-growing seasons.
Meanwhile,171 Chinese mini-core collections (171CMCC) were used to validate the cleaved amplified polymorphic sequence (CAPS) markers developed in this study from the single nucleotide polymorphisms (SNPs) significantly associated with GC and PHS resistance.The seeds of the natural population (171CMCC) were sown at the Dayangdian Experimental Station during the 2014–2016 and 2020 wheat-growing seasons.
Another 174 recombinant inbred lines (JH-RILs) obtained from the cross between Jing 411 (J411; whitegrained and PHS susceptible) and Hongmangchun 21 (HMC21; red-grained and PHS resistant) were used to map the candidate gene and verify the associations of the developed gene-specific markers with PHS resistance.These RILs were planted at the Dayangdian Experimental Station during the 2014 and 2015 wheat-growing seasons.
The parents of the JH-RILs used for transcriptomes analysis,J411 and HMC21,were grown at the Dayangdian Experimental Station during the 2019–2020 wheat-growing seasons.The experimental fields were arranged in a completely randomized block design with two replicates,and plants were grown in each plot with two rows that were 2 m long and an inter-row spacing of 25 cm.
All plant materials were harvested at 45 days after heading to coincide with physiological maturity (Osaetal.2003; Appendix A).Physiological maturity was determined based on the loss of chlorophyll from the spike and at least 15 cm of the peduncle (Trethowan 1995).Thirty spikes were harvested from each line of 168WV,171CMCC,174RILs,J411 and HMC21,naturally air dried for 5 days,and hand threshed to avoid damage to the embryos.The hand threshed seeds of each genotype were stored in a freezer at –20°C to maintain dormancy until all genotypes were threshed (Liuetal.2008).After all the materials were threshed,some seeds of each genotype were selected and left at room temperature (23–25°C) for subsequent germination index (GI) determinations (Zhuetal.2014,2016).
2.2.Phenotypic evaluation
GC determinationThe GC of 168WV and 171CMCC were determined after harvesting by a colorimeter (Minolta CR-5 Konica Corporation,Shanghai,China),following the methods of Groosetal.(2002) and Zhuetal.(2019).This instrument evaluates GC (CIE 1931) (Wangetal.1999) using the L*a*b*color system,representing the quantitative relationship of the colors along three axes.In this system,the L value indicates lightness and is represented from 0 (black) to 100 (white); a measures green (–100) to red (100),and b measures blue (–100) to yellow (100).According to Groosetal.(2002),the a value determined by the colorimeter best reflects the grain color,because it represents “red”.Also,the gloss of the wheat grain should also be considered,so the a/L value was used as the reference value of grain color.
About 20 g of seeds per line were placed in a Petri dish (25 mm radius),and the GC value was represented as the average of three replicates.The GC values of 168WV were measured in 2015 and 2017 (15GC and 17GC,respectively) (Appendix B),and the GC values of 171CMCC were also measured in 2015 and 2017 (15GCCMCC and 17GC-CMCC,respectively).
PHS resistance evaluationAccording to the methods reported by Kottearachchietal.(2006) and Zhuetal.(2016),the seed germination index (GI) and field sprouting (FS) were used to evaluate the PHS resistance levels of the wheat varieties.The 30 seeds used for each GI assay were placed on filter paper moistened with 10 mL distilled water in Petri dishes (90 mm diameter),and incubated at 20°C under a photoperiod of 14 h day/10 h night and 80% relative humidity to simulate a day-night environment (Zhuetal.2014).The GI values were measured for portions of the seeds for each variety after 5,15 and 30 days at room temperature.The number of germinated seeds in each Petri dish was counted every day based on the appearance of a rupture on the seed coat,and the GI value was calculated as follows: GI (%)=(3×n1+2×n2+1×n3)/(3×N)×100,where N represents the total number of seeds used for the germination test; and n1,n2,and n3 represent the numbers of germinated seeds on days 1,2,and 3 (Walker-Simmons 1988).
The GI values of 168WV were determined at 5 and 15 days after harvest (DAH) during the 2015–2017 periods (15GI-5DAH,16GI-5DAH,17GI-5DAH,15GI-15DAH,16GI-15DAH,and 17GI-15DAH).In addition,the GI values of the 168WV were determined at 30 DAH in 2016 (16GI-30DAH).Meanwhile,the GI values of 171CMCC were determined at 5 and 15 DAH during 2015–2017 and 2021 (15GI5-CMCC,16GI5-CMCC,17GI5-CMCC,21GI5-CMCC,15GI15-CMCC,16GI15-CMCC,and 17GI15-CMCC) (Appendix C).In addition,the GI values were measured for 17CMCC at 30 DAH in 2016 (16GI30-CMCC) and for the 174 JH-RILs at 5 DAH in 2015 and 2016 (15GI-5DAH-JH and 16GI-5DAH-JH) (Appendix D).
In 2015 and 2016,rainfall occurred for more than one week during the harvest,resulting in severe PHS in the field.Ten prominent spikes were collected from each plot in the field and dried immediately in an oven (105°C for 2 h) to test FS,following the method of Zhuetal.(2014,2016).Sprouted grains were scored,in which the pericarp over the embryo was ruptured.The FS value was determined based on the number of germinated seeds/total number of seeds of 20 prominent spikes collected from two plots of each genotype.The FS values determined in 2015 and 2016 are represented here as 15FS and 16FS for 168WV and 15FS-CMCC,and as 16FS-CMCC for 171CMCC.
2.3.Statistical analysis of phenotypic data
Descriptive statistical analysis and the Mann-Whitney test (U-test) were performed for the GC,GI,and FS values of 168WV and 171CMCC using SPSS 26.0 Software (IBM,Armonk,NY,USA).The broad sense heritability (HB2) was determined using the ‘lme4’ package of the R4.1.2 Software (https://www.r-project.org/),and year was included as a random effect (Batesetal.2014; Wangetal.2017; Merk 2019).
2.4.Genomic DNA extraction and genotyping
Genomic DNA of wheat seeds was extracted from lines of 168WV,171CMCC,and the JH-RILs following the method reported by Kangetal.(1998).The genomic DNA samples were genotyped on an Illumina 90K iSelect SNP array containing 81 587 SNPs.Genome Studio Software (version 2011.1) was used to determine the genotype clusters for each SNP.Among the 81 587 SNP markers of the Wheat Illumina 90K iSelect SNP array,44 011 SNPs (53.9%) were polymorphic in 168 wheat varieties (lines).A total of 22 300 SNPs were chosen for GWAS based on a wheat consensus linkage map (Wangetal.2014) and physical map (IWGSC RefSeq 1.0).Of the 22 300 SNPs,3 085 SNPs were selected for the further analysis (withr2=0,P≤0.001 determined as the cutoff standard) using STRUCTURE version 2.3.4 Software (Pritchardetal.2000) with a Bayesian Markov chain Monte Carlo model clustering approach (Evannoetal.2005).The physical position of each SNP was obtained from the International Wheat Genome Sequencing Consortium data (IWGSCRefSeq1.0; https://urgi.versailles.inra.fr/blast_iwgsc/blast.php; IWGSC 2018 (Alauxetal.2018)).
2.5.Linkage disequilibrium,population structure,PCA analysis and association analysis
A genome-wide linkage disequilibrium (LD) analysis was performed across the A,B,and D subgenomes.The LD was calculated as the squared value of the correlations (r2) between pairs of all the 3 085 significant SNPs using the LD function of the TASSEL Software (Bradburyetal.2007).The LD decay curves of subgenomes A,B,and D were obtained by fitting the scatter plots ofr2and physical distances (Appendix E).Because the physical distance when the LDs of all subgenomes had decayed to half ofr2was estimated as 2 Mb,the MTAs within 2 Mb were defined as a single locus (Lietal.2022).Loci associated with the PHS or GC traits in all three years were considered as stable and major loci.
Following method of Mackay and Powell (2007),3 085 polymorphic SNP markers evenly distributed across the whole wheat genome were selected from the Illumina 90K iSelect SNP array to evaluate the population structure.The population structure was characterized using the mixed model in the STRUCTURE version 2.3.4 Software (Pritchardetal.2000; Evannoetal.2005); the number of subpopulations,K,was set from 1 to 11,and eachKvalue was run separately six times.Meanwhile,GraphPad Prism 9.0 (https://www.graphpad.com/scientific-software/prism/) combined with the TASSEL Software (Bradburyetal.2007) was used for the PCA analysis.Population structure was characterized with ADMIXTURE (Alexanderetal.2009) usingK=2.The genome-wide associations between the markers and traits were analyzed using the mixed linear model (MLM) (Muhammadetal.2020) of TASSEL Software,withQ(fixed effect) andK(kinship matrix,random effect) as the covariables; and a threshold ofP<0.001 was used to claim significant associations between SNPs and traits (Priceetal.2006; Chenetal.2016; Linetal.2016).
Circular Manhattan plots for the PHS and GC traits with the SNP-density plot were drawn using the rMVP package in R (version 4.1.2) (https://www.cran.r-project.org; Yinetal.2020).Rectangular Manhattan plots and Q-Q plots were also drawn for the PHS and GC traits (Appendix F).
2.6.Gene annotation of Qphs.ahau-5B.4 and Qgc.ahau-2B.3/Qphs.ahau-2B.4 as well as candidate gene screening of Qphs.ahau-5B.4
Gene annotation for 2 Mb on either side flanking the traitlinked markers ofQphs.ahau-5B.4andQgc.ahau-2B.3/Qphs.ahau-2B.4were based upon the IWGSC reference genome version 1.1.In addition,gene annotation within the interval ofQphs.ahau-5B.4was also based upon the durum wheat (AABB) reference genome (IWGSC 2018 (Maccaferrietal.2019); http://202.194.139.32/blast/blast.html).Annotated genes in theQphs.ahau-5B.4interval were screened using the hexaploid wheat expression database of ‘Chinese Spring development’ and ‘expression of embryo and endosperm in developing grain’ (IWGSC 2014; Weietal.2018; http://202.194.139.32/expression/wheat.html).Genes expressed in both developing grain and embryo were considered as potential candidate genes underlyingQphs.ahau-5B.4.
2.7.Transcriptome analysis
Seeds of the PHS susceptible variety J411 and the PHS resistant variety HMC21 were collected at 1,6,9,12,and 36 h after imbibition at 20°C (the water absorption state of most seeds reached the same level).Meanwhile,the seeds of the two JH-RIL pools (PHS-S and PHS-R; five extremely PHS susceptible and five extremely PHS resistant) and the two parents (J411 and HMC21) were collected at 4,6,and 10 h after imbibition at 20°C.The transcriptomes were analyzed using three biological replicates of the RNA samples extracted from the collected seeds described above.The RNA was isolated according to the manufacturer’s instructions for TRIzol reagent (Invitrogen,China).The quantity and quality were subsequently analyzed using an Agilent 2100 Bioanalyzer and an Agilent RNA 6000 Nano Kit (Agilent Technologies).Libraries were prepared according to the guide of the TruSeq RNA Sample Preparation v.2 (Illumina),and an average of 91.3 million paired-end reads (2×150 nucleotides) were sequenced on an Illumina HiSeq 2500 system for each library.Raw reads in ‘fastq’ format were first subjected to quality control using FastQC (v.0.11.9).Then the reads were mapped to the wheat reference genome (IWGSC RefSeq v.1.1) using HISAT2 v.2.0.5 (Perteaetal.2016) with default parameters.The read counts and DEGs were determined by DEseq2 (Liaoetal.2014; Loveetal.2014).The resulting read counts normalized by reads per kilobase million (RPKM) were used to measure the transcript abundances (Weietal.2018).
2.8.Cloning of the candidate gene TraesCS-5B02G545100
Two pairs of primers were designed using Primer Premier 5.0 (https://www.PremierBiosoft.com) to clone and sequence the coding sequence (CDS) ofTraesCS5B02G545100in J411 and HMC21 (Appendix G).
2.9.CAPS marker development
The enzyme digestion sites flanking the SNP sequences were identified using the DNAMAN Software (version 6.0; https://www.lynnon.com/dnaman.htmL).Specific primers for these enzyme digestion sites were designed using the Primer Premier 5.0 Software and synthesized by Shenggong Bioengineering Co.,Ltd.(https://www.sangon.com/cxOrder) (Appendix H).Subsequently,PCR was performed using 100 ng of DNA template in a 10-μL reaction mixture containing 5 μL Mix (Vazyme Biotech Co.,Ltd.),2.2 μL sterile water,and 0.4 μL of each upstream and downstream primer.The PCR was carried out using the following program: the first step pre-denaturation at 95°C for 5 min; the second step denaturation at 95°C for 15 s; annealing at 0.2°C for 15 s and at 62°C within 40 cycles; a 72°C extension 30 s (40 cycles); and a final 72°C extension for 8 min.The amplified PCR product (5 μL) was digested with 1 μL of 10×CutSmart buffer and 0.5 U of restriction endonuclease (NEB restriction endonuclease http://www.neb-china.com/; TaKaRa,https://www.takarabiomed.com.cn/) in a total volume of 10 μL at 37°C for 8 h.The digested products were detected by agarose gel (2.0%) electrophoresis.
2.10.QTL analysis
The high-density linkage map of JH-RILs constructed based on specific locus amplified fragment sequencing (SLAF-seq) was used for QTL analysis (Caoetal.2020).QTL IciMapping (version 4.1) was used to map the QTL (Lietal.2007; https://www.isbreeding.net),and the threshold value of LOD was set as 2.0 (Zhouetal.2011).
3.Results
3.1.Phenotypic variations in GC and PHS resistance
The ANOVA results for 168WV and 171CMCC are listed in Table 1.The 168WV-GI and 171CMCC-GI showed significant (P<0.001) differences among genotypes and environments (Wangetal.2021).The heritability of the GI trait was 0.89 in 168WV and 0.86 in 171CMCC,and the heritability of FS was 0.75 in 168WV and 0.70 in 171CMCC.For GC,the heritability of 168WV-GC was 0.79 and that of 171CMCC was 0.81 (Table 1).
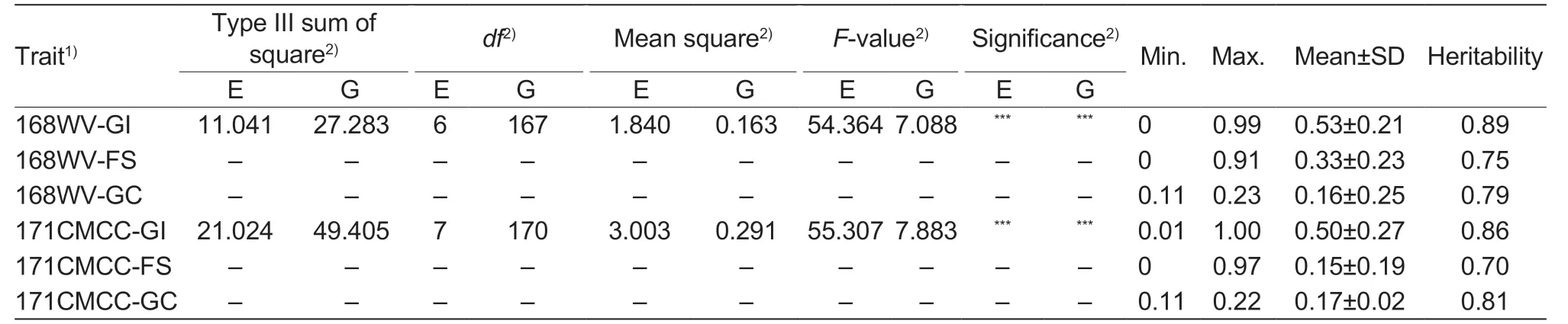
Table 1 Analysis of variance for germination index (GI) and heritability of GI,field sprouting (FS) and grain color (GC) in 168 wheat varieties (168WV) and 171 Chinese mini-core collections (171CMCC)
In the present study,the GC values varied less among the different years for 168WV,while they ranged from 0.13 to 0.23 (mean 0.18) for 17GC and from 0.11 to 0.17 (mean 0.14) for 15GC.However,the GI and FS values used to assess PHS resistance varied significantly (Appendices I and J).In 2015,2016,and 2017,the GI values ranged from 0.07 to 0.99 (average 0.52; 15GI-5DAH),0.04 to 0.90 (average 0.45; 16GI-5DAH),and 0 to 0.53 (average 0.36; 17GI-5DAH),respectively,at 5 DAH; and from 0.06 to 0.98 (mean 0.63; 15GI-15DAH),0.06 to 0.93 (mean 0.53; 16GI-15DAH),and 0.04 to 0.99 (mean 0.58; 17GI-15DAH),respectively,at 15 DAH.The population demonstrated a gradual increase in seed germination with the extension of post-ripening time.The average GI values at 5 and 15 DAH were 0.52 and 0.63 in 2015,0.45 and 0.53 in 2016,and 0.36 and 0.58 in 2017.Meanwhile,the GI ranged from 0.00 to 0.91 (mean 0.34) for 15FS and from 0.00 to 0.88 (mean 0.33) for 16FS.
Further analysis showed that for 171CMCC,the GC values showed limited variation under different environments.The GC values of 15GC-CMCC and 17GC-CMCC ranged from 0.11 to 0.20 (mean 0.20) and 0.15 to 0.22 (mean 0.22),respectively.In contrast,the phenotypic values of PHS resistance (GI and FS) varied widely.The GI values at 5 DAH ranged from 0.01 to 0.95 (average 0.2915; GI5-CMCC),0.01 to 0.98 (average 0.45; 16GI5-CMCC),0.05 to 0.99 (average 0.56; 17GI5-CMCC),and 0.07 to 0.98 (average 0.57; 21GI5-CMCC) in 2015,2016,2017,and 2021,respectively; and those at 15 DAH ranged from 0.04 to 1.00 (mean 0.55; 15GI15-CMCC),0.10 to 0.96 (mean 0.57; 16GI15-CMCC),and 0.14 to 1.00 (mean 0.7817; GI15-CMCC) in 2015,2016,and 2017,respectively.Meanwhile,the FS values ranged from 0.00 to 0.82 (mean 0.13) in 2015 and from 0.00 to 0.97 (mean 0.18) in 2016.This analysis showed an increase in seed germination with the extension of post-ripening time,similar to that in 168WV,indicating a gradual release of seed dormancy (Appendices I and J).
3.2.Correlation between GC and PHS resistance
Further analysis showed positive correlations for the GI values of the 168WVs among the different years (0.28–0.89).Significantly positive correlations were detected for GC between 2015 and 2017 (15GC and 17GC; 0.35).Meanwhile,negative correlations were detected between GC and GI values (0.12–0.32),suggesting that GC is positively correlated with PHS resistance,and the red-grained varieties have higher PHS resistance (Fig.1-A).

Fig.1 Pearson’s correlations between germination index (GI),field sprouting (FS),and grain color (GC) in 168 wheat varieties (168WV) and 171 Chinese mini-core collections (171CMCC).The notations 15GI-5DAH,16GI-5DAH,17GI-5DAH,15GI-15DAH,16GI-15DAH,and 17GI-15DAH represent the GI values of 168WV determined at 5 and 15 days after harvest (DAH) during the 2015–2017 seasons; 16GI-30DAH represents the GI value of the 168WV determined 30 DAH in 2016; 15FS and 16FS represent the FS values of 168WV determined in 2015 and 2016; 15GI5-CMCC,16GI5-CMCC,17GI5-CMCC,21GI5-CMCC,15GI15-CMCC,16GI15-CMCC,and 17GI15-CMCC represent the GI values of 171CMCC determined at 5 and 15 DAH during the 2015–2017 and 2021 seasons; 16GI30-CMCC represents the GI value of 17CMCC measured at 30 DAH in 2016; and 15FS-CMCC and 16FSCMCC represent the FS values of 171CMCC determined in 2015 and 2016.*,P≤0.05; **,P=0.01.
Meanwhile,the 171CMCC showed positive correlations for the GC values among the different environments (0.57).The GI values of the different years were also positively correlated (0.25–0.86).Meanwhile,significant negative correlations were detected between GC and GI (0.08–0.49),indicating a significant positive correlation between GC and PHS resistance for 171CMCC,which was consistent with 168WV (Fig.1-B).
3.3.Population structure,PCA analysis and association analysis
The ΔKvalue was plotted against the number ofKhypothetical subgroups.The highest ΔKwas observed forK=2 (the maximum membership probability; Fig.2-A).According to PCA analysis using TASSEL,the 168 WVs were also divided into two blocks (Fig.2-B).AtK=2,the curve reached its highest point,so 168WV was divided into two subgroups (Fig.2-C).
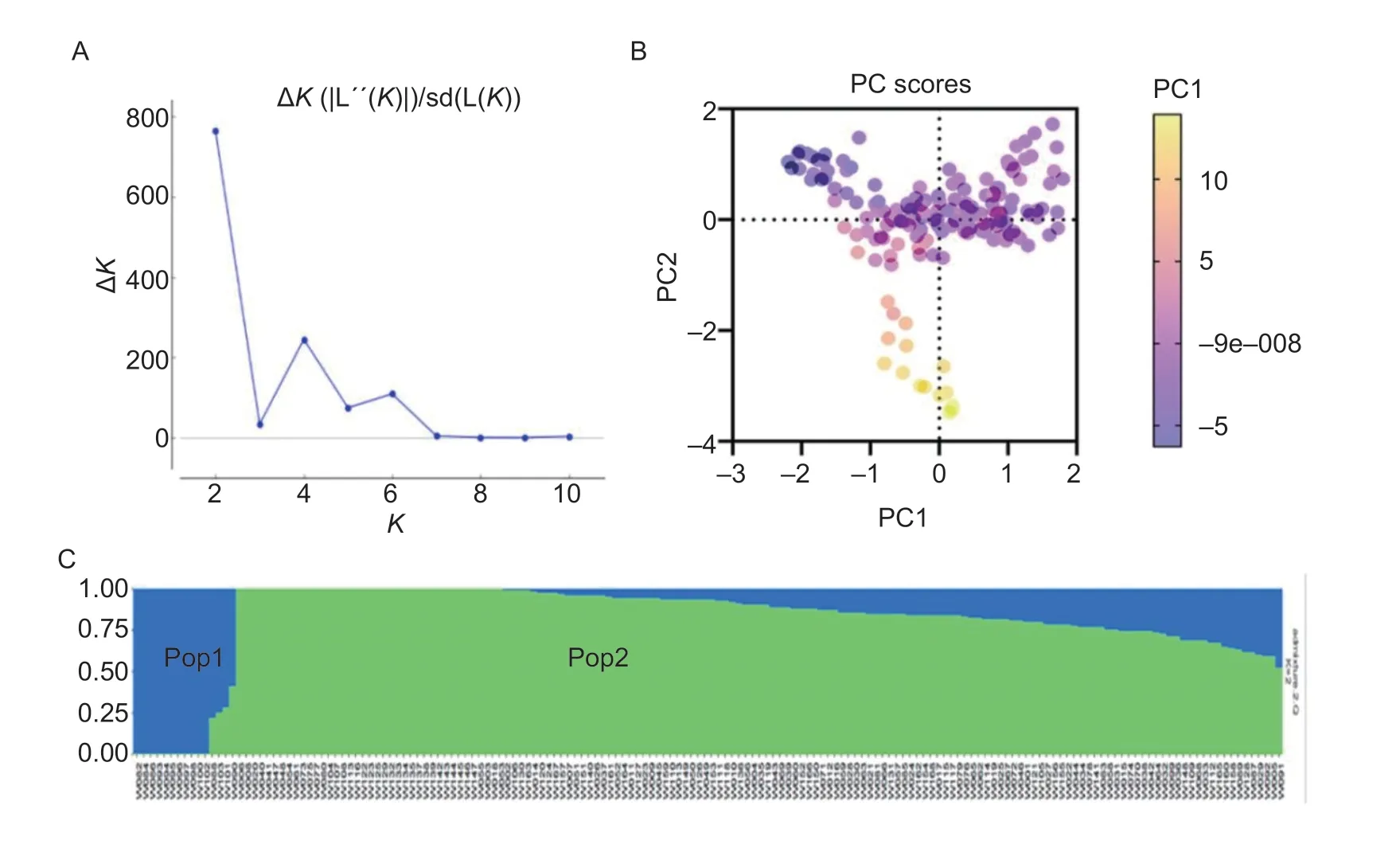
Fig.2 Population structure of 168 wheat varieties (lines) based on 3085 SNP markers.A,ΔK for STRUCTURE runs performed using six iterations for each K value (K=1–11).B,principal component analysis of 168 wheat varieties (lines).C,two major subpopulations from the 168 wheat varieties (lines).The x-axis represents the variety number,and the y-axis shows the sub-population membership probability; the colors of the bars (green and blue) indicate the two sub-populations identified through the STRUCTURE Program,and the area for the two different colors illustrates the proportion of each sub-population based on these SNP markers.
3.4.ldentification of loci associated with GC
A total of 168 wheat varieties were genotyped using an Illumina 90K iSelect SNP array.GWAS combined with the GC data from 2015 and 2017 significantly associated 67 MTAs with GC (in at least one year) based on MLM.These 67 MTAs were assigned to 29 loci and located on chromosomes 1A,1B,2A,2B,2D,3A,3B,5A,5B,5D,6A,7A,7B,and 7D,explaining 1.1–17.0% of the phenotypic variation in GC.Among them,five loci on chromosomes 2B,5A,and 7A were detected in both years (Table 2).The MTA BS00083078_51 forQphs.ahau-2B.4/Qgc.ahau-2B.3on chromosome 2B explained 9.9% of the phenotypic variation in GC in 2017 (Appendix K; Fig.3-A).
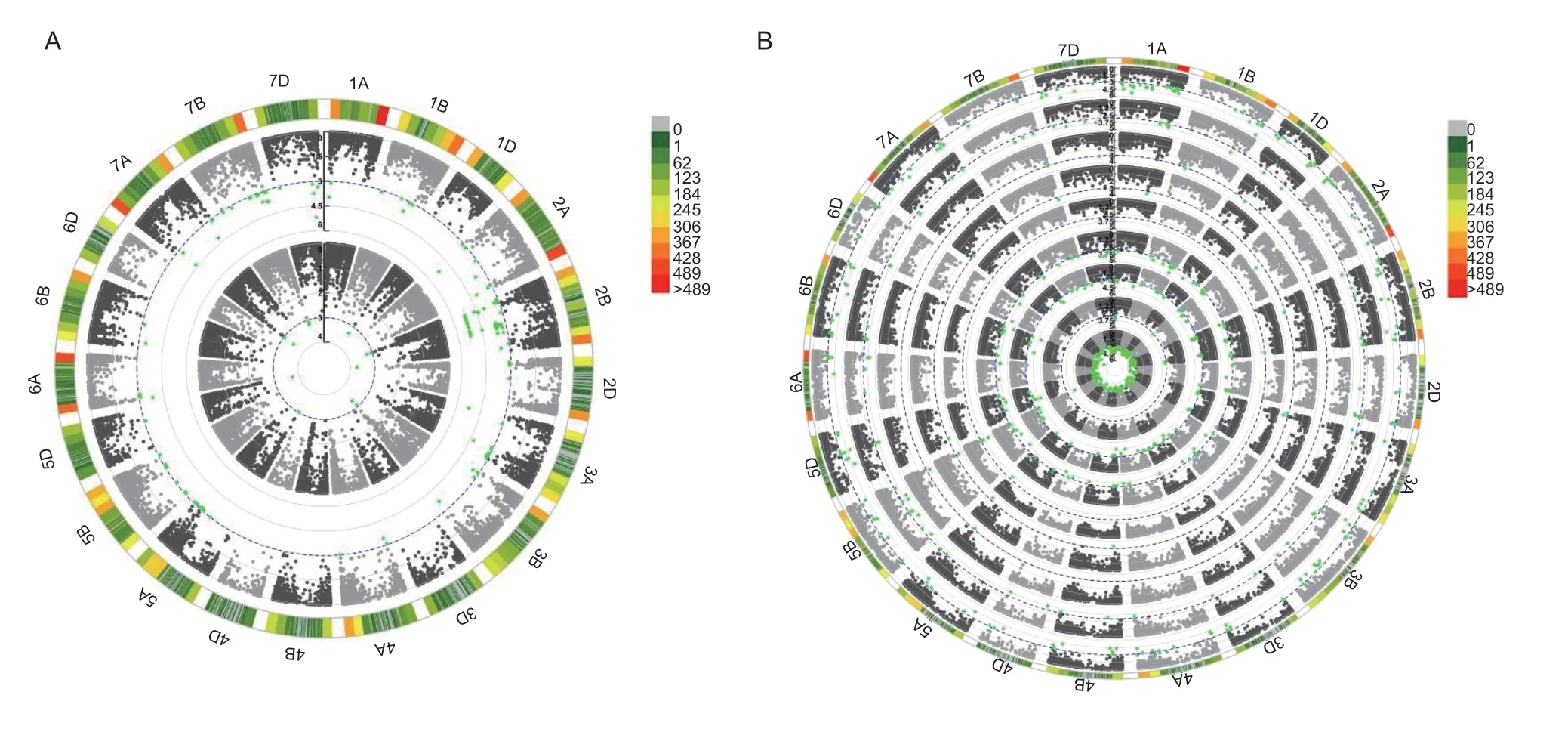
Fig.3 Circular Manhattan plots of 168 wheat varieties (lines) were drawn based on the mixed linear model (MLM) by the rMVP package in R (version 4.1.2) (https://www.cran.r-project.org).A,the circular Manhattan plot of 168 wheat varieties (lines),and the inner circle to the outer circle show 15GC and 17GC,respectively.B,the circular Manhattan plot of 168 wheat varieties (lines),and from the inner circle to the outer circle the bands are sequentially 15GI-5DAH,16GI-5DAH,17GI-5DAH,15GI-15DAH,16GI-15DAH,17GI-15DAH,16GI-30DAH,15FS and 16FS.The Manhattan plot indicates the -log10(observed P-value) for genomewide SNPs plotted against their respective positions on each chromosome.The threshold of 3.0 was adopted and is shown with a blue line.Marker density information is shown in the largest circle of Circular Manhattan plot.
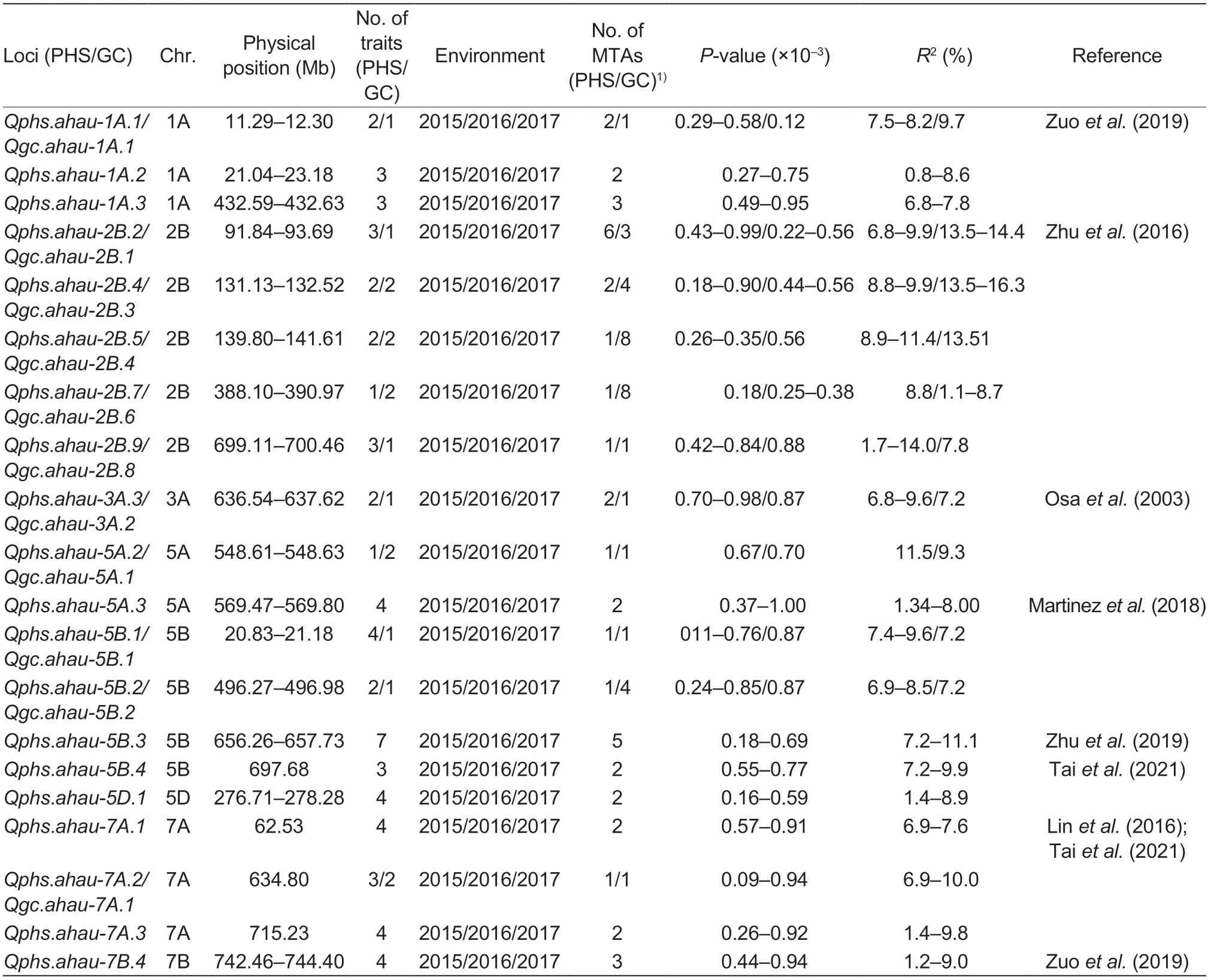
Table 2 Major loci for pre-harvest sprouting (PHS) and grain color (GC) identified by a genome-wide association study (GWAS) in 168 wheat varieties (lines)
3.5.ldentification of loci associated with PHS resistance
Meanwhile,the three-year PHS resistance data (GI and FS) and Illumina 90K iSelect SNP genotyping data of 168WV significantly associated 100 MTAs (54 loci) with PHS resistance by GWAS.These 100 MTAs were located on chromosomes 1A,1B,1D,2A,2B,2D,3A,3B,4A,5A,5B,5D,6A,7A,7B,and 7D,and explained 1.1–14.7% of the phenotypic variation in PHS resistance.Of these,10 loci on chromosomes 1A,2B,5A,5B,5D,7A,and 7B were stably identified in all three years (Table 2).In particular,the Tdurum_contig60189_263 forQphs.ahau-5B.4on chromosome 5B explained 9.9% of the phenotypic variation in PHS resistance in 2016 (Fig.3-B; Appendix L).
3.6.Common loci for GC and PHS resistance
In total,28 loci,including 96 MTAs,were associated with GC and PHS resistance; and these loci were distributed on chromosomes 1A,1B,2A,2B,2D,3A,3B,5A,5B,5D,6A,7B,and 7D.Of these,11 loci were stably associated with GC and PHS in different environments (Table 2).
3.7.Development and validation of the CAPS markers
The representative SNP (wsnp_Ra_c67199_65253620) on the major common locus for GC and PHS (Qgc.ahau-2B.3/Qphs.ahau-2B.4) and the representative SNP (Tdurum_contig60189_263) on the main locus for PHS resistance alone (Qphs.ahau-5B.4) were further converted into two CAPS markers,designated as2B-448and5B-301,respectively,and genotyped in 171CMCC (Appendix M).Two alleles were detected for the CAPS marker 2B-448,designated as2B-448-A(Hap1-2B) and2B-448-G(Hap2-2B).In addition,two allelic variations were detected for the CAPS marker 5B-301,designated as5B-301-C(Hap1-5B) and5B-301-T(Hap2-5B).Further,combining the GC and PHS resistance (GI and FS) and SNP genotyping data of 171CMCC,the Mann-Whitney U test showed significant differences in PHS resistance (17GI5-CMCC,17GI15-CMCC,and 21GI5-CMCC) and GC (17GC-CMCC) between the wheat varieties carrying2B-448-Gand2B-448-A(P<0.05).Similarly,significant differences were detected in PHS resistance (17GI5-CMCC,17GI15-CMCC,21GI5-CMCC,and 15FS-CMCC) between the wheat varieties carrying5B-301-Cand5B-301-T(P<0.05) (Table 3; Appendices N and O).
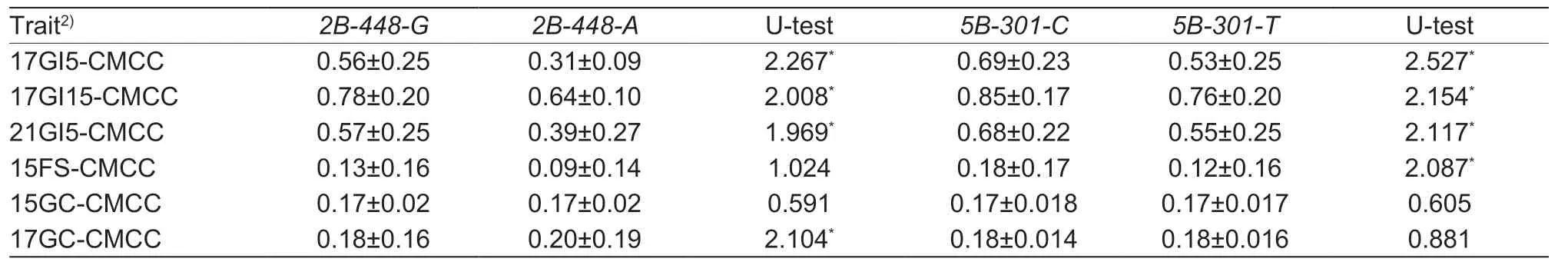
Table 3 Associations of the allelic variations of cleaved amplified polymorphic sequences (CAPS) markers 2B-448 and 5B-301 with the germination index (GI),field sprouting (FS),and grain color (GC) phenotypes in 171 Chinese mini-core collections (171CMCC)1)
3.8.Gene annotation of Qphs.ahau-5B.4 and Qgc.ahau-2B.3/Qphs.ahau-2B.4,as well as candidate gene screening of Qphs.ahau-5B.4
In the 2 Mb on either side flanking the trait-linked markers ofQphs.ahau-5B.4andQgc.ahau-2B.3/Qphs.ahau-2B.4,61 genes (Appendix P) and 53 genes (Appendix Q) were annotated in the Chinese Spring (AABBDD) reference genome,respectively.Considering thatQphs.ahau-5B.4was only for PHS and was independent of GC,which is favorable for breeding white-grained varieties with PHS resistance,we selected this locus for further analysis.To explore the candidate gene underlyingQphs.ahau-5B.4,we searched the durum wheat (AABB) reference genome,and found 54 genes within the interval ofQphs.ahau-5B.4.Of these,51 genes were also annotated within the interval ofQphs.ahau-5B.4in the Chinese Spring reference genome,and the remaining three genes were not in the Chinese Spring reference genome.Among these three genes,TRITD5Bv1G248590andTRITD5Bv1G247690encoding Aspartic proteinase nepenthesin and Mitochondrial import receptor subunit TOM22,respectively,were considered not to be the genes associated with seed dormancy and germination based on functional annotation information,while the third one isTRITD5Bv1G2481with unknown function (Appendix R).Therefore,we focused on the above 61 genes that were annotated in the Chinese Spring reference genome.Among them,37 genes were expressed in developing grain tissue according to the hexaploid wheat expression database,including 25 genes that were expressed in both developing grain and embryo (Appendix S).Notably,four genes (TraesCS5B02G543700,TraesCS5B02G544500,TraesCS5B02G545100,andTraesCS5B02G545200) showed relatively high transcript abundance (FPKM>1) in developing grain and embryo compared to the other genes,and thus were considered as the potential candidate genes underlyingQphs.ahau-5B.4(Appendix T).
3.9.Transcript abundance of the genes of Qphs.ahau-5B.4
The transcriptome sequencing results of J411 (PHS susceptible) and HMC21 (PHS resistant) showed the differential expressions ofTraesCS5B02G545100(encoding glutaredoxin) at 1,6,9,12 and 36 h after seed imbibition and ofTraesCS5B02G545200(encoding elongation factor TU-GTP) at 1,6 and 9 h after seed imbibition.In particular,TraesCS5B02G545100showed consistently higher transcript abundance in HMC21 than in J411 (Appendix U and V).Furthermore,the seeds of the two pools (5 PHS resistant and 5 PHS susceptible) from JH-RILs and the two parents (J411 and HMC21) at different imbibition stages (4,6,and 10 h) were used for transcriptome sequencing.The results indicated significantly different transcript abundances ofTraesCS5B02G545100andTraesCS5B02G545200at three imbibition stages between the two pools and two parents (Appendices W and X).In summary,the results based on the two transcriptomic analyses of this study indicatedTraesCS5B02G545100as a potential candidate gene linked toQphs.ahau-5B.4.In addition,TraesCS5B02G545200may also be a potential candidate gene in the target region,which can be further verified in the future.
3.10.Cloning and validation of the candidate gene
To further explore the association of the candidate geneTraesCS5B02G545100with PHS resistance,we cloned its CDS between the PHS susceptible J411 and the PHS resistant HMC21.In 498 bp of CDS sequence,nine SNP and two 6-bp InDel variations leading to eight amino acid differences were detected between J411 and HMC21.Among these amino acid changes,the changes of the 59th,67th and 117th amino acids occurred in the functional domain (glutaredoxin domain,76–139 aa) of TraesCS5B02G545100,which may affect the function of the encoded protein (Appendix Y).One SNP (T/C) within the CDS ofTraesCS5B02G545100was the same as the SNP (CAP7_c8713_356) from the Illumina 90K iSelect SNP array.The CAPS marker CAPS-356 was developed based on this SNP (T/C) and used to genotype the 174 lines of JH-RILs (Appendices Z and A1).Further,we analyzed the high-density linkage map of the JHRILs constructed based on specific locus amplified fragment sequencing (SLAF-seq) markers (Caoetal.2020) combined with the PHS resistance phenotypes under two environments (2015 and 2016).QTL analysis showed that the CAPS marker CAPS-356 was tightly linked with a QTL controlling PHS resistance (15GI-5DAH-JH and 16GI-5DAH-JH),which supports the idea ofTraesCS5B02G545100as a potential candidate gene underlying (Qphs.ahau-5B.4) (Appendices B1 and C1).
4.Discussion
4.1.Loci associated with GC and PHS resistance
Global warming and increased rainfall have led to an increase in the risk of PHS.Compared with chemical control,opting for varieties with high PHS resistance levels is economical,safe,and effective in minimizing the damage.PHS is more severe in white-grained wheat than in red-grained wheat.Therefore,in areas like China,where farmers prefer white grains due to high flour yield,it is necessary to develop and grow PHS-resistant white-grained varieties,which requires dissecting the genetic basis of GC and PHS resistance and the genetic relationship between these two traits.
Based on GWAS,the present study identified 29 loci for GC and 54 loci for PHS resistance,including 28 collocated loci (Appendix D1).Among the 29 loci that control GC,12 have been reported,such asQgc.ahau-1A.1(11.3–12.3 Mb) (Zuoetal.2019,9.6 Mb),Qgc.ahau-2B.1(91.8–93.7 Mb) (Zhuetal.2016,87.4–93.7 Mb),Qgc.ahau-2D.1(55.8–56.8 Mb) (Zhuetal.2016,55.8–56.8 Mb; Taietal.2021,59.9 Mb),Qgc.ahau-3A.2(636.5–637.6 Mb) (Osaetal.2003,636.0 Mb),Qgc.ahau-3A.4(693.2–693.5 Mb) (Zhuetal.2019,689.0–741.0 Mb),andQgc.ahau-6A.1(37.3–38.4 Mb) (Kumaretal.2009,38.4 Mb).We found thatQgc.ahau-3B.2 (756.7–756.8 Mb) is very close toTamyb10-3B(757.9 Mb),the key gene involved in grain color formation (Dongetal.2015),indicating the reliability of the present GWAS.Meanwhile,the remaining 17 GC loci identified on chromosomes 1B,2A,2B,3A,5A,5B,7A and 7B are novel,suggesting that other QTLs may also play key roles in regulating grain color.
In addition,the study identified 54 PHS resistance loci.Among these,23 have been reported,such asQphs.ahau-1D.1(18.2–20.0 Mb) (Albrechtetal.2015,19.0 Mb),Qphs.ahau-2A.2(12.7–14.9 Mb) (Zhuetal.2016; Zhangetal.2017,12.7–14.9 Mb; Martinezetal.2018,11.0 Mb),Qphs.ahau-2D.1(55.8–56.8 Mb) (Zhuetal.2019,55.8–56.8 Mb),Qphs.ahau-2D.2(641.1–643.8 Mb) andQphs.ahau-2D.3(644.1–645.1 Mb) (Zhuetal.2019; 637.0–644.0 Mb),Qphs.ahau-3A.1(11.5–13.9 Mb) (Liuetal.2008,11.0–15.0 Mb),Qphs.ahau-3A.4(655.6–655.7 Mb) (Osaetal.2003,657.0 Mb),andQphs.ahau-3A.5(693.2–693.5 Mb) (Zhuetal.2019,689.0–741.0 Mb),Qphs.ahau-3B.2(133.2–134.3 Mb) (Zhouetal.2017,126.0–663.0 Mb),Qphs.ahau-5D.2(434.5–434.5 Mb) (Taietal.2021,430.1 Mb),Qphs.ahau-5B.4(697.7 Mb) (Taietal.2021,699.6 Mb),andQphs.ahau-7A.1(62.5 Mb) (Linetal.2016,58.0–65.0 Mb).The other 31 PHS resistance loci on chromosomes 1A,1B,1D,2A,2B,3A,3B,4A,5A,5B,5D,7A,7B,and 7D were identified as novel and need to be further validated.
Considering thatR2values ofQgc.ahau-2B.3/Qphs.ahau-2B.4andQphs.ahau-5B.4were both high and stable,and the SNPs of the two major QTLs were also easier to convert to the CAPS markers,we developed the two corresponding CAPS markers based on the two SNPs linked to the above two loci.The validation of these CAPS markers with the traits in the 171CMCC revealed that the two alleles of the two markers significantly correlated with GC and PHS resistance,confirming the authenticity of the two loci.Compared with Taietal.(2021),determining whetherQphs.ahau-5B.4is a novel locus is difficult because of the proximal physical distance (2 Mb) between them (Appendix D1).
4.2.Candidate gene for the major locus Qphs.ahau-5B.4
To screen the potential candidate genes underlying the major locusQphs.ahau-5B.4,we searched the publically available wheat database to find their expression patterns in different tissues.We found that the four annotated genes,TraesCS5B02G545100(glutaredoxin),TraesCS5B02G545200(extension factor TU-GTP binding domain),TraesCS5B02G544000(protein kinase superfamily protein),andTraesCS5B02G544500(26S proteasome non-ATPase regulatory subunit),are highly expressed in wheat spikes and grains.Furthermore,according to the transcriptome data,we found thatTraesCS5B02G545100was stably and highly expressed in PHS resistant HMC21 and the PHS resistant pool at different seed imbibition stages; however,it showed obviously lower expression levels in PHS susceptible J411 and the PHS susceptible pool,suggesting thatTraesCS5B02G545100may be a potential candidate gene forQphs.ahau-5B.4.Therefore,we cloned and obtained the CDS sequence ofTraesCS5B02G545100from PHS susceptible J411 and PHS resistant HMC21.The CAPS marker CAPS-356,developed from a synonymous SNP (C/T) in the CDS sequence ofTraesCS5B02G545100,was tightly linked with a QTL for PHS resistance in the JH-RILs population.This finding supported the idea thatTraesCS5B02G545100encoding glutaredoxin is a potential candidate gene underlyingQphs.ahau-5B.4.
Glutaredoxins (GRX) are small heat stable proteins that are widely distributed in animals,plants,and microorganisms.Researchers have studied this protein in animals and humans,but less so in plants.A GRX usually contains a conserved CXXC/S active site,which can be assigned to one of three types (CPYC,CGFS,and CC) based on the amino acid sequence.In plants,glutaredoxin,the thioredoxin system,and the glutathione ascorbate cycle are involved in the maintenance of the redox state and the regulation of various cellular processes; and they also play essential roles in plant cell resistance to biological and abiotic oxidative stresses (Zhouetal.2018; Vermaetal.2020; Kakeshpouretal.2021; Kumaretal.2021).Notably,Xuetal.(2019) identified a dominant rice PHS mutantphs9-Dencoding a CC-type GRX.ThePHS9gene regulates rice PHS resistanceviareactive oxygen species and ABA signalling.Interestingly,the potential candidate geneTraesCS5B02G545100identified in the present study is a CGFS-type GRX.Studies have reported that CGFS-type GRXs play important roles in cytotoxin detoxification (Vermaetal.2020),cell growth and proliferation (Ehraryetal.2020),iron homeostasis regulation (Martinsetal.2020; Moseleretal.2021),Ca2+signal transduction (Maetal.2021),protein kinase C (PKC)-mediated stress response,and oxidative protection (Maetal.2020).However,the association of CGFS-type GRX with wheat PHS resistance is unclear.ForTraesCS5B02G545100,we performed a phylogenetic tree analysis based on the sequence information of the HMC21 type,which was consistent with the Chinese Spring sequence of the reference genome.This phylogenetic analysis showed a relatively distant genetic relationship between CGFStypeTraesCS5B02G545100and the rice CC-typePHS9(Appendix E1).Considering that these two genes belong to the GRX family,we speculate that they may have similar functions in mediating the PHS trait,but this needs to be verified by a transgenic wheat approach in future studies.
5.Conclusion
In summary,the present study combining GWAS,the wheat public expression database,transcriptomic sequencing,gene allelic variation analysis,CAPS marker development,and QTL mapping identified loci for GC and PHS resistance.This study further validated the associations of the candidate geneTraesCS5B02G545100encoding glutaredoxin onQphs.ahau-5B.4with PHS resistance alone using both biparental and natural populations.These results not only provide insight for understanding the complex genetic mechanism of the GC and PHS traits but they also provide useful gene resources and markers for breeding white-grained wheat varieties with high PHS resistance by the method of pyramiding PHS resistance genes.
Acknowledgements
This work was supported by grants from the University Synergy Innovation Program of Anhui Province,China (GXXT-2021-058),the National Natural Science Foundation of China (Joint Fund Projects,U20A2033),the Natural Science Foundation of Anhui Province,China (2108085MC98),the Jiangsu Collaborative Innovation Center for Modern Crop Production,China (JCIC-MCP),the key scientific and technological breakthroughs of Anhui Province (2021d06050003),and the joint key project of improved wheat variety of Anhui Province,China (21803003).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2023.04.017
杂志排行
Journal of Integrative Agriculture的其它文章
- Combining nitrogen effects and metabolomics to reveal the response mechanisms to nitrogen stress and the potential for nitrogen reduction in maize
- Natural variations and geographical distributions of seed carotenoids and chlorophylls in 1 167 Chinese soybean accessions
- Carbon sequestration rate,nitrogen use efficiency and rice yield responses to long-term substitution of chemical fertilizer by organic manure in a rice–rice cropping system
- ldentification and epitope mapping of anti-p72 single-chain antibody against African swine fever virus based on phage display antibody library
- The PcERF5 promotes anthocyanin biosynthesis in red-fleshed pear (Pyrus communis) through both activating and interacting with PcMYB transcription factors
- Core collection construction of tea plant germplasm in Anhui Province based on genetic diversity analysis using simple sequence repeat markers
