基于生物信息学方法双芯片联合分析阿尔茨海默病海马相关差异基因的研究
2023-07-14陈佳音钟斌贾芸菁李莎莎陈依烛李晨羽凌雁武
陈佳音 钟斌 贾芸菁 李莎莎 陈依烛 李晨羽 凌雁武


【摘要】 目的 利用生物信息學筛选阿尔茨海默病(Alzheimers disease,AD)海马组织的差异表达基因(DEGs),分析DEGs的生物学功能和信号通路,为AD的诊断、治疗靶点的筛选提供参考。方法 利用GEO(gene expression omnibus)数据库下载基因芯片GSE48350、GSE5281,使用数据库GEO2R工具在线分析,以P<0.05,|logFC>1|为条件筛选差异基因,筛选出与AD海马组织相关的DEGs,通过制作维恩图取交集获得两个芯片共同DEGs;使用R语言enrichGO函数对筛选出来的DEGs进行基因本体论(GO)功能富集分析;在Metascape网站做京都基因与基因组百科全书(KEGG)通路分析;使用STRING数据库和Cytoscape3.9.1软件进行蛋白-蛋白相互作用(PPI)网络分析。把筛选出来的共同DEGs作为关键基因,通过Cytoscape3.9.1软件中插件Mcode和CytoHubba分析后,按照节点度值大小排序,将前四个基因视为核心基因。最后,通过TargetScan与miRanda数据库评估核心基因相关的miRNA,并选用基因芯片GSE129053进行验证,构建核心基因的miRNA-mRNA调控网络。结果 两个基因芯片共有67个DEGs,其中上调基因36个,下调基因31个。GO分析结果显示,主要富集在生物过程中为突触囊泡介导的运输、神经元突触囊泡循环、网格蛋白介导的突触囊泡内吞机制等;在细胞定位中主要的过程有胞外囊泡、突触小泡等;分子功能主要过程有网格蛋白结合、葡萄糖的跨膜转运活性等。KEGG通路富集分析结果显示,DEGs富集在甲状腺激素信号通路(hsa04919);DEGs编码蛋白的PPI网络含节点10个;运用Cytoscape中CytoHubba插件和Mcode插件,把筛选出来的共同基因作为关键基因,前四个基因视为核心基因,分别为SNAP-25、SNAP-91、STMN2、GAP43,可以发现hsa-miR-153-3p关联的靶基因最多。结论 AD患者海马组织中关键的核心基因为SNAP-25、SNAP-91、STMN2、GAP43,hsa-miR-153-3p可能通过调控其靶基因SNAP-25、STMN2在AD患者海马组织中发挥重要作用,为AD的诊断和治疗提供了候选靶点。
【关键词】 阿尔茨海默病;生物信息学;GEO数据库;海马;铝致认知障碍
中图分类号:R749.1 文献标志码:A DOI:10.3969/j.issn.1003-1383.2023.06.003
A study on the analysis of differentially expressed genes in the hippocampus of Alzheimer's disease based on bioinformatics
CHEN Jiayin, ZHONG Bin, JIA Yunjing, LI Shasha, CHEN Yizhu, LI Chenyu, LING Yanwu▲
(School of Basic Medicine, Youjiang Medical University for Nationalities, Baise 533000, Guangxi, China)
【Abstract】 Objective To screen differentially expressed genes (DEGs) in the hippocampus of Alzheimer's disease (AD) by bioinformatics, analyze the biological functions and signal pathways of DEGs, and provide reference for the diagnosis and treatment target screening of Alzheimer's disease. Methods The gene chips GSE48350 and GSE5281 were downloaded from GEO (gene expression omnibus) database, database GEO2R tool was used to analyze them online. The differential genes were screened on the condition of P<0.05, |logFC>1|, DEGs related to AD hippocampal tissue were screened, and two chip common DEGs were obtained by making the Venn diagram to take intersection; R language enrichGO function was used to enrich the GO function of the selected DEGs; the pathway analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) was conducted on the Metascape website; STRING database and Cytoscape 3.9.1 software were used to analyze protein-protein interaction (PPI) network. The selected common DEGs were considered as key genes, and after analyzing them through the plugins Mcode and CytoHubba in Cytoscape 3.9.1 software, the common genes of the two were considered as key genes. Finally, the miRNA related to the core gene was evaluated through TargetScan and miRanda websites, and the gene chip GSE129053 was selected for verification to construct the miRNA-mRNA regulatory network of the core gene. Results There were 67 DEGs in the two gene chips, of which 36 were up-regulated genes and 31 were down regulated genes. The results of GO analysis showed that synaptic vesicle mediated transportation, neuronal synaptic vesicle circulation, and synaptic vesicle endocytosis mechanisms mediated by grid proteins mainly enriched on biological processes; the main processes in cell localization included extracellular vesicles, synaptic vesicles, etc; the main processes of molecular functions included the binding of clathrin and the transmembrane transport activity of glucose. The enrichment analysis of KEGG pathway showed that DEGs were enriched in the thyroid hormone signal pathway (hsa04919); the PPI network of DEGs encoding proteins contained 10 nodes; by using the CytoHubba and Mcode plugins in Cytoscape, the selected common genes were served as key genes, and the first four genes were considered as the core genes, namely, SNAP-25, SNAP-91, STMN2, and GAP43, respectively, and it was found that hsa-miR-153-3p was associated with the most target genes. Conclusion Key core genes in the hippocampus of AD patients are SNAP-25, SNAP-91, STMN2, and GAP43. hsa-miR-153-3p may play an important role in the hippocampus of AD patients by regulating its target genes SNAP-25 and STMN2, which provides candidate targets for the diagnosis and treatment of AD.
【Key words】 Alzheimer's disease(AD); bioinformatics; GEO database; hippocampus; aluminum induced cognitive impairment
阿爾茨海默病(Alzheimers disease,AD)是一种神经退行性疾病,症状发展从轻到重,主要表现为认知能力下降和大脑不可逆转的记忆丧失[1]。AD的发病机制有多种,比较公认的有β-淀粉蛋白异常沉积、tau蛋白的磷酸化、神经原纤维缠结(NFT)、氧化应激、胰岛素信号通路障碍、线粒体功能障碍等[2]。近年来,随着我国人口老龄化进程加剧,AD患者数量迅速增加,目前已超过700万人[3]。海马是中枢神经系统中参与学习和记忆贮存的重要组织,其改变与衰老、Aβ沉积、tau蛋白过度磷酸化、缺血等影响因素相关。本研究旨在通过生物信息学方法联合双芯片分析,探索AD患者海马组织中的关键基因靶点,为治疗AD提供新的思路。
1 材料与方法
1.1 基因芯片数据来源
在NCBI公共数据库GEO(https://www.ncbi.nlm.nih.gov/geo/)以“Alzheimer's Disease”为关键词搜索目标,下载GSE5281与GSE48350的基因芯片数据集,在这两个芯片中筛选60岁以上人群的海马样本,在GSE48350选取44个样本,其中19个AD样本和25个对照样本;在GSE5281选取23个样本,其中10个AD样本和13个对照样本,以上两个数据集平台都是GPL570。
1.2 差异表达基因(DEGs)的筛选
应用GEO数据库的分析工具GEO2R(www.ncbi.nlm.nih.gov/geo/ge2r)进行分析,GEO2R采用R语言软件GEOquery包读取数据,limma包用于筛选基因表达,去除没有相应基因符号的探针集或有多探针集的基因,以P<0.05,|logFC|>1作为条件筛选,两个芯片取交集获得共同DEGs。
1.3 DEGs的GO富集分析和KEGG通路分析
基因本体论(gene ontology,GO)从生物过程(biological process, BP)、细胞定位(cellular component, CC)和分子功能(molecular function, MF)三个方面提供基因的简单注释,使用R语言enrichGO函数对筛选出来的DEGs进行GO功能富集分析。京都基因与基因组百科全书(KEGG)是一个专门存储不同物种基因路径信息的数据库,将DEGs输入Metascape网站(http://metascape.org/)进行KEGG基因通路分析,Metascape是一个公开的数据库,有助于全面基因列表注释和分析。
1.4 构建蛋白-蛋白质互作网络
相互作用基因库检索工具STRING11.5(https://cn.string-db.org/)数据库能够寻找关键基因并构建编码蛋白质与预测蛋白质之间的相互作用,构建DEGs编码蛋白相互作用网络(protein-protein interaction,PPI)。筛选条件可信度为0.7。然后将PPI结果导入Cytoscape 3.9.1软件中进行可视化,运用Cytoscape中CytoHubba插件和Mcode插件,把筛选出来的共同基因作为关键基因,按照节点度值大小排序,前四个基因视为核心基因。
1.5 构建核心基因的miRNA-mRNA调控网络
用在线数据库TargetScan(www.TargetScan.org)与miRanda(www.microrna.org/microrna/home.do)寻找一些潜在的miRNAs,用于预测核心基因上游靶标。
2 结果
2.1 筛选的DEGs
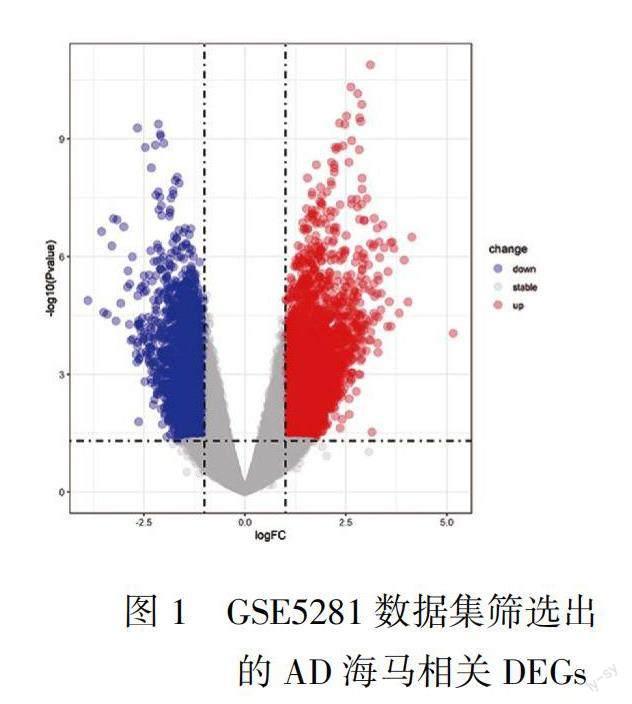
基因芯片GSE5281数据集共筛选出DEGs 5496个,其中上调基因3844个,下调基因1652个,见图1。基因芯片GSE48350筛选出DEGs共228个,上调基因144个,下调基因84个,见图2。这两个数据集有共同DEGs 90个,其维恩图见图3。排除GSE5281和GSE48350中表达趋势相反的基因后,最终我们得到了上调DEGs 36个,下调DEGs 31个,共67个。
2.2 DEGs的GO功能分析与KEGG通路分析
DEGs在R语言中的GO功能分析见图4,差异基因名称与GO富集生物功能的关系见图5。GO分析结果显示,DEGs主要集中于为网格蛋白介导的突触囊泡内吞机制、神经元突触囊泡循环等生物过程;富集在胞外囊泡、突触小泡等细胞过程;富集在网格蛋白结合、葡萄糖的跨膜转运活性、SNARE复合体、脂质转运体活性等分子功能。KEGG通路富集分析结果显示,DEGs富集在甲状腺激素信号通路(hsa04919),分析结果见图6。
2.3 DEGs调控蛋白网络与关键基因分析
以DEGs构建蛋白互作网络,互作关系的蛋白质共有10个,将蛋白互作网络可视化,如图7所示。我们把筛选出来的共同基因作为关键基因,分别为SNAP-25、SNAP-91、STMN2、GAP43、STON2、SYT13、AMPH、SNCB、MET、KITLG,其中前四个基因视为核心基因。
2.4 构建核心基因的miRNA-mRNA调控网
用在线数据库TargetScan、miRanda寻找一些潜在的miRNAs,用于预测核心基因上游靶标,并用基因芯片GSE129053来验证。GSE129053验证有3个AD与3个大鼠海马组织,平台GPL18058通过阵列进行非编码RNA分析,以P<0.05,|logFC|>1作为条件筛选,共有90个上调的miRNA,368个下调的miRNA,筛选出与核心miRNA重复的miRNA并构建核心基因的miRNA-mRNA调控网,可以发现hsa-miR-153-3p关联的靶基因最多。见表1。
3 讨论
AD是一种病因较多、病程较长、危险因素较多、发病机制较为复杂的神经退行性疾病,由于其发病机制研究尚未有定论,目前仍缺乏有效的治疗方法。
本研究通过生物信息学方法从GEO数据库中选取GSE5281与GSE48350双芯片联合分析AD患者海马组织关键的核心基因,用在线数据库TargetScan、miRanda寻找核心基因上游核心miRNA靶标,并选用基因芯片GSE129053对核心miRNA进行验证;构建关键核心基因的miRNA-mRNA调控网络;分析结果显示hsa-miR-153-3p可能通过调控靶基因SNAP-25、STMN2,在AD患者海马组织中是可行的生物标志物。
SNAP-25是DEGs海马组织中PPI网络连接节点数量最多的基因,与突触囊泡释放和再循环、神经轴突延伸、神经元修复和突触形成[4]等多种生理功能有关。许多参与突触囊泡运输和释放的基因随着AD患者年龄的增长发生了显著变化。SNAP-25直接介导突触小泡与突触前膜的融合调控突触囊泡转运和神经递质释放到突触中,因此对调节神经递质释放到突触间隙中是至关重要的[5]。有研究结果表明海马CA1区域中的SNAP-25参与了记忆巩固[6]。通过敲除SNAP-25并对海马突触内吞作用进行成像,发现SNAP-25参与网格蛋白依赖性内吞作用[7]。ZHANG等[8]研究表明SNAP-25和SNAP-25/Aβ42比率有望作为AD最早症状阶段的诊断和预后评估生物标志物。突触体相关蛋白91(SNAP-91)也称为AP180,HU等[9]通过基于疾病临床和病理状态的基因共表达网络分析,确定SNAP-91与AD发病机制的新关联。有研究表明,STMN2可以维持正常的轴突生长和再生[10]。GAP43是一种与神经发育和突触重塑密切相关的特异性蛋白,它在神经系统的海马体中大量表达,与长期学习和记忆有关[11]。有学者对死后AD患者的大脑研究报告发现海马体区域GAP43增加[12]。
随着我国广西铝矿开发规模的不断扩大,矿区高铝环境对周围居民的健康造成严重的威胁[13]。流行病学资料显示,长期接触铝可导致认知功能障碍(mild cognitive impairment,MCI),MCI是AD发病前阶段,早期发现并干预治疗有望延缓或阻止AD的发生[14]。有研究发现MCI患者体液或多个脑区已经发生了改变[15]。我们课题组前期通过对广西百色市平果市铝矿区12位60岁以上老人进行调查取样,其中6人为MCI患者,6人为对照组,利用高通量测序实验对上述样品进行深入分析,研究发现共有130个差异表达的miRNA,上调表达12个,下调表达118个,其中hsa-miR-153-3p表达下调,证明铝导致认知障碍发生发展与AD具有相似的发病机制,为进一步早期治疗MCI延缓成严重AD的发生提供新的治疗思路[16]。有研究表明miR-153-3p通过SRC依赖性MAPK信号通路,可改善认知功能障碍并减少脑损伤[17]。
综上所述,AD患者海马组织中关键的核心基因为SNAP-25、SNAP-91、STMN2、GAP43,同时hsa-miR-153-3p表達水平在AD海马组织中具有较高的诊断、预后评估价值,并可能通过调控靶基因SNAP-25、STMN2发挥作用。
参 考 文 献
[1] ISAEV N K, STELMASHOOK E V, GENRIKHS E E, et al. Alzheimer's disease:an exacerbation of senile phenoptosis[J].Biochemistry Moscow, 2015, 80(12):1578-1581.
[2]SHAH H, ALBANESE E, DUGGAN C, et al. Research priorities to reduce the global burden of dementia by 2025[J].Lancet Neurol, 2016, 15(12):1285-1294.
[3]JIA J P, WEI C B, CHEN S Q, et al. The cost of Alzheimer's disease in China and re-estimation of costs worldwide[J].Alzheimer's Dement, 2018, 14(4): 483-491.
[4]BARK I C, HAHN K M, RYABININ A E, et al. Differential expression of SNAP-25 protein isoforms during divergent vesicle fusion events of neural development[J].Proc Natl Acad Sci U S A, 1995, 92(5):1510-1514.
[5]JAHN R, SCHELLER R H.SNAREs:engines for membrane fusion[J].Nat Rev Mol Cell Biol, 2006, 7(9):631-643.
[6]HOU Q L, GAO X, LU Q, et al. SNAP-25 in hippocampal CA3 region is required for long-term memory formation[J].Biochem Biophys Res Commun, 2006, 347(4):955-962.
[7]ZHANG Z, WANG D S, SUN T, et al. The SNARE proteins SNAP25 and synaptobrevin are involved in endocytosis at hippocampal synapses[J].J Neurosci, 2013, 33(21):9169-9175.
[8]ZHANG H, THERRIAULT J, KANG M S, et al. Cerebrospinal fluid synaptosomal-associated protein 25 is a key player in synaptic degeneration in mild cognitive impairment and Alzheimers disease[J].Alzheimers Res Ther, 2018, 10(1):80.
[9]HU R T, YU Q, ZHOU S D, et al. Co-expression network analysis reveals novel genes underlying alzheimer's disease pathogenesis[J].Front Aging Neurosci, 2020, 12:605961.
[10]KLIM J R, WILLIAMS LA, LIMONE F, et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair[J].Nat Neurosci, 2019, 22(2):167-179.
[11]NEVE R L, FINCH E A, BIRD E D, et al. Growth-associated protein GAP-43 is expressed selectively in associative regions of the adult human brain[J].Proc Natl Acad Sci U S A, 1988, 85(10):3638-3642.
[12]REKART J L, QUINN B, MESULAM M M, et al. Subfield-specific increase in brain growth protein in postmortem hippocampus of Alzheimers patients[J].Neuroscience, 2004, 126(3):579-584.
[13]漆光紫,周敏,龐广福,等.职业性铝暴露与认知功能障碍的关系研究[J].右江民族医学院学报,2016,38(1):20-21.
[14]BRADBURN S, MURGATROYD C, RAY N.Neuroinflammation in mild cognitive impairment and Alzheimer's disease:a meta-analysis[J].Ageing Res Rev, 2019, 50:1-8.
[15]PAK V M, ONEN S H, BLIWISE D L, et al. Sleep disturbances in MCI and AD: neuroinflammation as a possible mediating pathway[J].Front Aging Neurosci, 2020, 12:69.
[16]李莎莎.广西铝暴露致认知障碍老年人群血清外泌体差异表达miRNAs的研究[D].百色:右江民族医学院,2022.
[17]LI Y M, PENG B, LI Y, et al. MiR-203a-3p/153-3p improves cognitive impairments induced by ischemia/reperfusion via blockade of SRC-mediated MAPK signaling pathway in ischemic stroke[J].Chem Biol Interact, 2022, 358:109900.
(收稿日期:2022-08-21 修回日期:2022-12-14)
(编辑:潘明志)
