MicroRNA transcriptome of skeletal muscle during yak development reveals that miR-652 regulates myoblasts differentiation and survival by targeting ISL1
2023-05-08ZHOUXuelanGUOXianLIANGChunnianCHUMinWUXiaoyunYANPing
ZHOU Xue-lan , GUO Xian , LIANG Chun-nian , CHU Min , WU Xiao-yun #, YAN Ping #
1 Key Laboratory of Yak Breeding Engineering of Gansu Province, Lanzhou Institute of Husbandry and Pharmaceutical Sciences, Chinese Academy of Agricultural Sciences, Lanzhou 730050, P.R.China
2 Key Laboratory of Animal Genetics and Breeding on Tibetan Plateau, Ministry of Agriculture and Rural Affairs, Lanzhou 730050, P.R.China
Abstract The growth and development of skeletal muscle also determine the meat production of yak, ultimately affecting the economic benefits.Hence, improving growth performance is a top priority in the yak industry.Skeletal muscle development is a complex process involving the regulation of several genes, including microRNAs (miRNAs).However, the transcription of miRNAs in yak skeletal muscle during prenatal to postnatal stages is unknown.We used small RNA sequencing (small RNA-Seq) to determine the global miRNAs of longissimus dorsi muscle from yak (the samples were collected from three fetuses and three adults).Totally 264 differently expressed miRNAs (|log2(fold change)|>1 and P-value≤0.05) were detected between the two groups.Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that differently expressed miRNAs-targeted genes participated in pathways associated with muscle development, such as MAPK, PI3K-Akt, and Hippo signaling pathways, etc.MiR-652, which was up-regulated in the fetal group, was transfected into C2C12 myoblasts to examine its role.miR-652 promoted (P≤0.05) proliferation and differentiation, but inhibited (P≤0.001) apoptosis at early period.Furthermore, miR-652 reduced (P≤0.001) the proportion of C2C12 myoblasts in the G1 phase while increasing (P≤0.01) the proportion of cells in the S and G2 phases.Dual-luciferase reporter assays indicated that ISL1 served as a target of miR-652.In general, these findings expand our understanding of yak skeletal muscle miRNAs, and suggested that miR-652 probably regulated myogenesis by regulating ISL1.
Keywords: skeletal muscle, small RNA sequencing, miR-652, C2C12 myoblast, ISL1
1.Introduction
Yak (Bosgrunniens), an ancient bovine species, is found throughout Himalayan Region of South-Central Asia and the Qinghai–Tibetan Plateau.There are nearly 14 million yaks in the world, with China accounting for 95% of them.Yak, as multipurpose livestock, provides meat, milk, wool, transportation, and fuel to indigenous people on the highaltitude Qinghai–Tibet Plateau.Yak growth rate and body weight are two important economic characteristics that are closely related to meat production.When compared to cattle (Bostaurus), yaks have a smaller body weight/size and growth rate (Wieneret al.2003).Thus, improving growth performance is a top priority in the yak industry.
Skeletal muscle is a major component of the body in mammals, which is related to body development.Vertebrate skeletal muscle development is a tightly coordinated and strict regulatory process that involves the multipotent precursor cells, myoblast proliferation, withdrawal from the cell cycle, differentiation, as well as the fusion of myoblasts to form multicellular myotubes (Bentzingeret al.2012).In the prenatal period, increasing fiber numbers is the main characteristic of skeletal muscle growth.After birth, muscle development mainly depends on muscle fiber hypertrophy (Picardet al.2002; Whiteet al.2010).The growth and development of skeletal muscle involve several genes.For instance, MYOD and MYF5 transcription factors are responsible for the myogenic specification (Graves and Yablonka-Reuveni 2000).MyoGandMRF4play a vital role in the terminal differentiation from myoblasts to myotubes (Hernandez-Hernandezet al.2017).Transcription factors are key regulators of gene expression.ISL1, a transcription factor, belongs to the LIM-homeodomain family and serves a major role in cell proliferation, differentiation, and survival during embryogenesis (Zhuanget al.2013).ISL1is also important for the different types of muscle development.Previous studies have reported thatIsl1acts as a pioneer factor in cardiomyocyte fate commitment,isl1+cardiac progenitor cells could differentiate into cardiomyocytes, smooth muscle cells, and endothelial cells (Caiet al.2003; Laugwitzet al.2005; Morettiet al.2006; Gaoet al.2019).Additionally,Isl1-derived satellite cells could regenerate the injured limb muscle of mice, andIsl1overexpression could repress head muscle differentiation in chick embryos (Harel et al.2009).Moreover, a lineage study revealed thatIsl1also marks a subset of limb progenitors and is required for hindlimb development (Yanget al.2006).
Epigenetic modifications and non-coding RNA also participate in the growth and development of vertebrate skeletal muscle.MiRNAs belong to a type of endogenous non-coding RNAs with sizes of 18–25 nucleotides.They can repress mRNA translation or inhibit their translation by interacting with the 3´ untranslated region (3´UTR) (Kim and Nam 2006).MiRNAs are highly conserved across different species and may involve diverse biochemical processes, including cell proliferation, differentiation, and apoptosis.Previous studies have verified that several miRNAs are involved in C2C12 cells proliferation, differentiation, and migration, such as miR-22 (Wanget al.2018), miR-92b-3p (Yeet al.2020), and miR-452 (Yanget al.2021).In recent years, several studies have explored the role of miRNA in muscle tissue development in cattle (Tewariet al.2021), sheep (Liuet al.2019), pig (Aliet al.2021), chicken (Chenet al.2020), and duck (Liet al.2020), etc., using small RNA-Seq.In yak, Jiet al.(2020) used high-throughput sequencing to analyze miRNA expression profiles in skeletal muscle at different postnatal stages of yak, and identified candidate miRNAs in skeletal muscle development.Huanget al.(2021) also examined miRNA expression profiles inlongissimus dorsi musclesbetween yak and cattle-yak and suggested that some differently expressed (DE) miRNAs might be involved in muscle development.The number of muscle fibers differs between prenatal and postnatal periods.However, few studies have examined the miRNA expression profiles in these two distinct stages of yak.
To identify miRNAs that may have a potential role during muscle development of yak, we characterized miRNA expression profiles in thelongissimusdorsimuscle of yak at prenatal and postnatal developmental stages using the small RNA-Seq method.The sequencing result showed that miR-652 was one of the up-regulated miRNAs in prenatallongissimusdorsimuscle.Therefore, it was hypothesized that miR-652 might play an important role in regulating yak skeletal muscle development.This study aimed to explore the function of miRNA in C2C12 myoblast proliferation, differentiation, apoptosis, and cell cycle to reveal the potential role of miR-652 in developing yak skeletal muscle.This study might improve our understanding of miRNA function and molecular mechanism during skeletal muscle development of yak.
2.Materials and methods
2.1.Animals and cell culture
The experimental animals used were a well-known elite native breed of Datong yak.Thelongissimusdorsimuscle tissue samples from six unrelated female yaks were divided into two groups included fetal group and adult group (three fetal samples, 90 days after fertilization; three adult samples, five years after birth) in the slaughterhouse from Datong Hui and Tu Autonomous County, Qinghai Province, China.Fetal age was estimated based on crown–rump length.Each sample was immediately frozen in liquid nitrogen freezing, then stored at –80°C before further processing.Procell Life Science & Technology Company (Wuhan, China) provided the C2C12 myoblasts.293T cell lines were obtained from Guangzhou Fitgene Biotech Co., Ltd.(Guangzhou, China).C2C12 myoblasts were kept within Dulbecco’s modified Eagle’s medium (DMEM) that contained 10% fetal bovine serum (FBS) at 37°C with 5% CO2.C2C12 myoblasts were transferred to DMEM and added with 2% horse serum (HS) for differentiation.C2C12 myoblasts were replenished with fresh medium every 48 h.293T cell lines were raised for the dual-luciferase activity assay.We cultivated 293T cells in DMEM that contained 10% FBS, and the medium was changed every 48 h.
2.2.RNA extraction and small RNA sequencing
Trizol reagent was used to extract total RNA (Invitrogen, CA, USA).NanoDrop 2000 spectrophotometer was employed for evaluating the purity and concentration of total RNA (Nanodrop Technologies, Wilmington, USA).The OD260/OD280ratio for each sample was about 1.8–2.0.The degradation and contamination of RNA were examined on 1% agarose gels.RNA integrity was observed using Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA).RNA Integrity Number (RIN) was ≥8.0.Then, the proprietary adapters were directly connected to 5´ and 3´ of total RNA and synthesized the complementary DNA (cDNA) by reverse transcription PCR (RT-PCR).Next, the cDNA fragments of about 18–40 bp were separated by 15% polyacrylamide gel electrophoresis for cDNA libraries, followed by library sequencing using the Illumina HiSeq™ 2500 platform and SE50 single-end module (Illumina Inc., San Diego, CA, USA).
2.3.Data processing
Sequencing adapters and poly-A tails were removed by the Cutadapt Software (Martin 2011), and the lowquality reads from raw reads were deeply filtered using the Fastx_toolkit (Hannon 2016).Afterward, the remaining clean reads were mapped to the yak reference genome (BosGru_v2.0) using Burrows-Wheeler Aligner (BWA) (Li and Durbin 2010).The mapped reads were annotated using BLAST Software (V2.2.23).The known miRNAs were explored by the alignment between the unique sequences and bovine mature and precursor miRNA sequences in the miRBase 21.0 Database (https://www.mirbase.org).Besides, the known rRNA, tRNA, small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA) could be detected by alignment with Rfam12.1 (https://rfam.xfam.org) Database.The known piRNA was identified by alignment with the piRNABank Database (https://pirnabank.ibab.ac.in).Furthermore, all unannotated clean reads were adopted for predicting yak underlying novel miRNAs with the application of Mireap Software (V.0.2) (Liet al.2012) (http://sourceforge.net/projects/mireap/).
2.4.ldentification and verification of DE miRNAs
This study investigated the differential expression of miRNAs in prenatal and postnatal yak skeletal muscles to better understand the regulatory effect of miRNAs within prenatal and postnatal skeletal muscle.MiRNA expression levels were measured based on the clean reads, that were fully aligned to mature miRNA sequences.Furthermore, the relative expression levels of miRNAs were standardized to reads per million (RPM) clean tags.Afterward, the DE miRNAs and their correspondingP-value were calculated using the edgeR Package (Robinsonet al.2010).To evaluate the significant difference of miRNAs expression, the |log2(fold change)|>1 andP-value≤0.05 were determined as the threshold value (Jiet al.2020).To verify the reliability of the sequencing results, we performed RT-PCR and realtime qualitative PCR (qPCR) analysis on 18 randomly selected miRNAs.The first-strand cDNA was obtained using miScript II RT Kit (QIAGEN, Germany).The RTqPCR was performed using miScript SYBR®Green PCR Kit (QIAGEN) on a LightCycler 96 RT-PCR System (Roche Diagnostics, Basel, Switzerland).The relative expression level of miRNAs was analyzed with the 2–ΔΔCtmethod and normalized usinglet-7a(Timonedaet al.2012).The sequences of primers are shown in Appendix A.
2.5.Target prediction of DE miRNAs and enrichment analysis
To comprehend the role of DE miRNAs during the development of skeletal muscle, the target genes prediction of DE miRNAs was performed by an online website (https://www.omicstudio.cn/analysis/targetGene) based on the Miranda and TargetScan algorithms (TargetScan score>50, and Miranda energy<–10).We obtained the overlap target genes of the two algorithms.Moreover, the DE miRNAs-targeted genes were utilized to conduct Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
2.6.Expression analysis of miR-652 and qPCR
The expression level of miR-652 in C2C12 myoblasts was detected at 0, 1, 3, 5, and 7 days after induced to differentiation.The total RNA of C2C12 myoblasts was processed into single-strand cDNA using a Reverse Transcription Kit (Vazyme Biotech Co.Ltd., China).The qPCR was carried out in triplicate with SYBR Green Master Mix (VAZYME) using ABI QuantStudioTM6 Flex System.The expression levels of microRNA and mRNAs were normalized with those ofU6and glyceraldehyde-3-phosphate D ehydrogenase (GAPDH), respectively.The 2–ΔΔCtalgorithm was used to determine the relative expression level of each gene (Livak and Schmittgen 2001).The sequences of primers are shown in Appendix B.
2.7.Cell transfection
To investigate the role of miR-652 in C2C12 myoblast proliferation, differentiation, apoptosis, and cell cycle.The miR-652 mimic (overexpression), mimic negative control (NC), miR-652 inhibitor (inhibition), as well as inhibitor NC were all synthesized at Guangzhou Ribo Bio Company (Guangzhou, China).Next, the RNA oligonucleotides were transfected into C2C12 myoblast under different confluence (30% for proliferation and 80% for differentiation) using LipofectamineTM 2000 (Invitrogen, USA), following the manufacturer’s instruction.
2.8.Western blot
C2C12 myoblasts were homogenized in ice-cold RIPA lysis buffer (Beyotime Institute of Biotechnology, China), including protease inhibitor (PMSF, Beyotime Institute of Biotechnology, China).The lysates were centrifuged, and the protein concentration of the supernatant was measured using the Enhanced BCA Protein Assay Kit (Beyotime Institute of Biotechnology, China).Subsequently, 5× protein loading buffer was added to the lysates prior to full denaturation in boiled water.In entirety, 40 μg of protein was electrophoresed on a 12% SDS-polyacrylamide gel, which was transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Darmstadt, Germany).This membrane was blocked in 5% defatted milk, followed by overnight incubation using primary antibodies under 4°C.The following day, the PVDF membranes were rinsed using Tris-buffered saline with Tween (TBST), incubated with a second antibody at 37°C for 2 h, and rinsed five times using TBST, as well as exposed and quantified based on BandScan 4.0.For Western blot, anti-MyoG (1:250; ab1835; Abcam), anti-MyHC (1:500; MAB4470; R&D Systems), and anti-GAPDH (1:1 000, Hangzhou Goodhere Biotech, China) antibodies were used.
2.9.Cell proliferation assay
C2C12 myoblasts were kept into 48-well plates, which were transfected with miR-652 mimics, mimics NC, miR-652 inhibitor, and inhibitor NC.A Cell Counting Kit 8 (CCK8, Beyotime, Shanghai, China) was adopted to measure the cell proliferation after transfection for 36 h in accordance with the manufacturer’s protocol.After each well was treated with 10 μL CCK8 reagent and incubated at 37°C for 2 h, the absorbance of each sample at OD450was identified using Flexstation®3 Microplate Reader (Molecular Devices, USA).An EdU assay was also performed to evaluate cell proliferation.After transfection for 36 h, C2C12 myoblasts were incubated with 10 μmol L–1EdU (Beyotime, Shanghai, China) for 2 h at 37°C.Subsequently, they were washed, fixed, and permeabilized.Click reaction solution was supplemented and incubated at room temperature for 30 min in the dark.At the same time, flow cytometry was used to determine the percentage of proliferative cells.
2.10.Cell apoptosis assay
We evaluated apoptosis of C2C12 myoblasts using Annexin V-FITC/PI Apoptosis Detection Kit (Vazyme, Nanjing, China).After transfection for 36 h, the C2C12 myoblast was stained with 5 μL AnnexinV-FITC mixed with propidium iodide (PI) for 15 min under ambient temperature.Afterward, flow cytometry was adopted for analyzing the cell apoptosis of C2C12 myoblasts.
2.11.Cell cycle analysis
C2C12 myoblasts were cultured in the 48-well plates.After 36 h of transfection, the cells were digested with trypsin, washed with PBS, fixed in precooled 70% ethanol, and preserved overnight under 4°C.Next, after resuspension with precooled PBS, cells were raised with 20 mL RNaseA solution for 30 min at 37°C.Finally, cells were raised with 400 mL PI staining solution for 30 min at 4°C in the dark.Flow cytometry was performed to explore the cell cycle status.
2.12.Dual-luciferase reporter assays
The wild (ISL1-WT) and mutant (ISL1-Mut) binding sites in the 3´UTR ofISL1were synthesized from GeneCopoeia (Guangzhou, China), and inserted into the siCHECK™ Vectors (Promega, Madison, USA), respectively.We cultured 293T cells in 48-well plates until they reached 80–90% confluence.Co-transfection of the dualluciferase vector with wild or mutant binding sites ofISL1into 293T cells, respectively, with miR-652 mimics, mimics NC using Lipofectamine 2000 (Invitrogen, CA, USA).The 293T cell lines were collected at 36 h after transfection.Luciferase activity values were determined using the Dual-Luciferase Reporter Assay System (Promega, Madison, USA) assayed according to the manufacturer’s instructions.
2.13.Statistical analysis
In this study, each set of experiments included three independent replicates, and each replicate was an experimental unit.Data in the expression of miR-652 at 0, 1, 3, 5, and 7 days after induced C2C12 myoblasts to differentiation was analyzed using one-way ANOVA.Differences among means were tested by the least significant difference (LSD) test.In addition, the orthogonal polynomial contrast method was used to test the linear and quadratic changes of miR-652 expressions with the time (day).An independent samplest-test was done to examine the statistical differences in relative mRNA and protein levels, cell growth rate, percentage of Edu-positive cell, percentage of apoptotic cell, percentage of cell population, and relative luciferase activity between treatment groups and corresponding negative control groups.All statistical analysis was performed by SPSS v.20.0 (IBM Corp., Armonk, NY) Software.P-value≤0.05 indicated statistical significance.
3.Results
3.1.Overview of the study and data
Six libraries were sequenced from thelongissimusdorsimuscle of yaks at prenatal and postnatal stages (n=3 in each stage).The raw data were submitted to the NCBI Sequence Read Archive (accession number PRJNA687328).After filtering, 13 049 146 to 15 025 033 clean reads in six libraries were acquired.We aligned altogether 97.49–99.65% of clean reads into (Bos Gru v2.0) yak reference genome (Appendix C).Among the aligned reads, 78.90 to 95.10% of then were mapped to bovine miRNA mature and precursor in miRBase 21.0 by BLAST search (Appendix D).More than half of sRNAs were distributed in the intergenic region (Appendix E).In total, 396 and 286 known bovine miRNAs were identified from prenatal and postnatal groups, separately.Additionally, we discovered 274 and 152 candidate novel miRNAs in prenatal and postnatal groups, respectively.
3.2.Enrichment analysis of DE miRNAs
After normalization, DE miRNAs were defined as those that were more than 2-fold different and had aP-value≤0.05.There were 264 DE miRNAs identified, with 235 miRNAs being up-regulated in prenatal muscle tissue compared to postnatal muscle tissue, and 29 miRNAs being down-regulated.Interestingly, 109 miRNAs were only found in the fetus stage, while 14 miRNAs were only found in the postnatal stage.These DE miRNAs contain vital miRNAs that involve in the muscle development process, like miR-1, miR-22-3p, miR-127, miR-133a, miR-206, and etc.In addition, the RT-qPCR results confirmed the expression pattern of 18 DE miRNAs were the same as sequencing results when adult stage compared to fetal stage (Fig.1-A).The result also verified the reliability of sequencing results.Overall, 5 183 DE miRNAs-targeted genes were predicted.GO and KEGG enrichment analyses revealed target genes were significantly (P≤0.001) enriched in the energy homeostasis, negative regulation of inflammatory response, Golgi apparatus, protein kinase binding GO terms (Fig.1-B), and MAPK, Rap1, and PI3K-Akt signaling pathway, etc.(Fig.1-C).
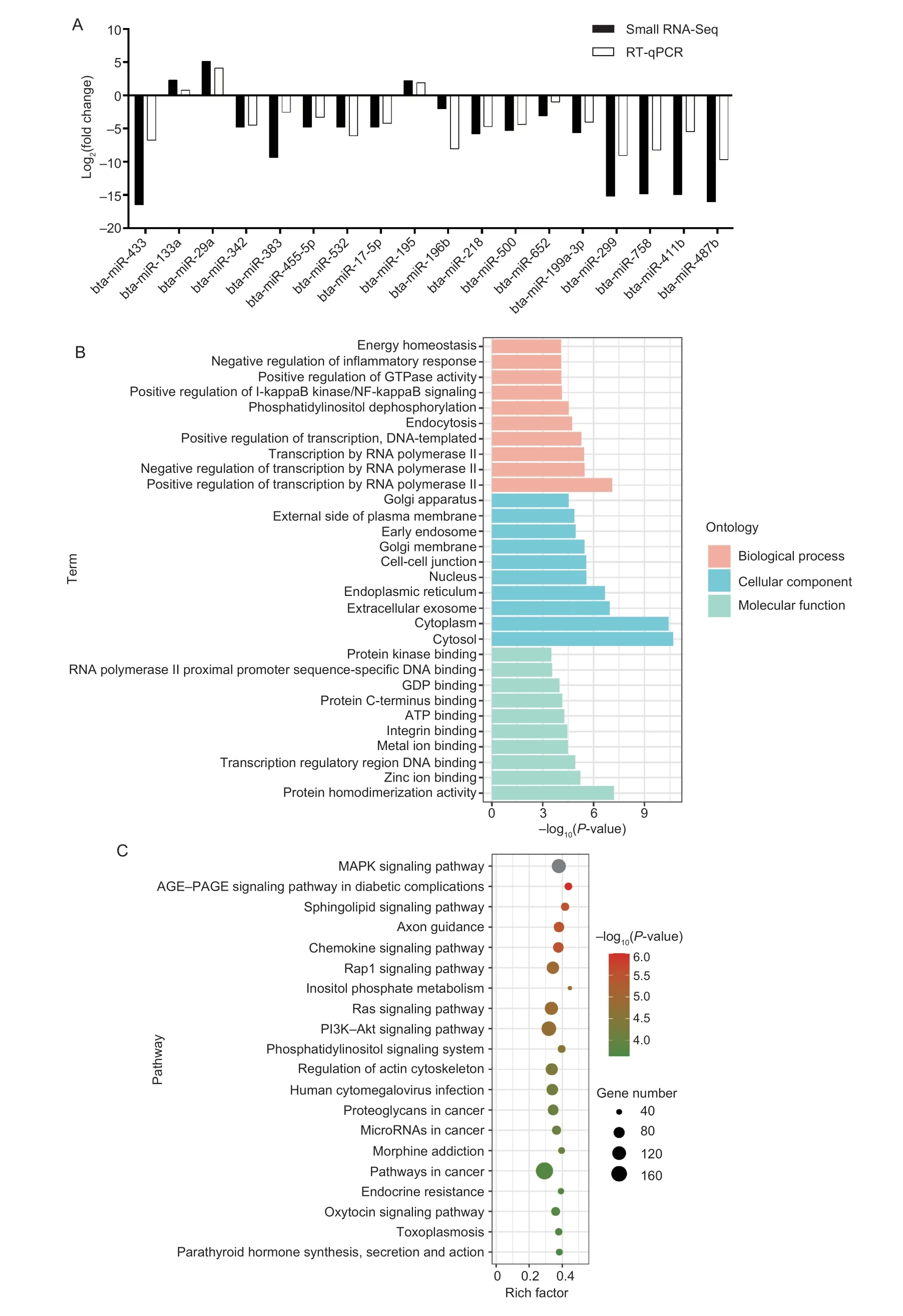
Fig.1 Verification and enrichment analysis of differently expressed (DE) miRNAs.A, validation of DE miRNAs by RT-qPCR, log2(fold change)>0 indicates the miRNA up-regulated in adult group (bta, cow).Log2(fold change)<0 is the opposite.B, top 30 GO terms of DE miRNAs-targeted genes.C, top 20 KEGG pathways of DE miRNAs-targeted genes.
3.3.Expression pattern analysis of miR-652 during C2C12 myoblasts differentiation
MiR-652, one of the up-regulated miRNAs in prenatal skeletal muscle tissue of yak, and the mature sequence is conserved in mammals (Fig.2-A), thereby suggesting its important roles in regulating muscle development.We used C2C12 myoblast as a research model for investigating the role of miR-652 during skeletal muscle development.After induced C2C12 myoblasts to differentiation, we found the expression level of miR-652 increased (P≤0.001) during C2C12 myoblast differentiation.The linear trend analysis demonstrate that the expression of miR-652 showed typical linear increase (P(linear)<0.001) trend (Fig.2-B).
3.4.MiR-652 promotes C2C12 myoblasts differentiation
The RT-qPCR results showed that miR-652 expression increased when C2C12 myoblast were stimulated to differentiate.The findings suggested that miR-652 plays a role in myoblast differentiation.MiR-652 mimics, miR-652 inhibitor, mimics NC, and inhibitor NC were separately transfected into C2C12 myoblasts using the differentiation medium to assess the impact of miR-652 on myoblast differentiation.In order to validate these results, mRNA and protein expression levels of myogenic transcription factors, such as MyHC and MyoG were evaluated.The mRNA level of marker genes (MyoGandMyHC) for differentiation were up-regulated (P≤0.05) under overexpression of miR-652 at 3, 5, and 7 days.However, these genes presented an opposite expression trend under the knockdown of miR-652 (Fig.2-C and D).Western blot assays also implied that miR-652 enhanced (P≤0.05) MyHC and MYOG expressions at the protein level (Fig.2-E–G).These results demonstrated that miR-652 efficiently stimulates myoblast differentiation and myotubes formation.
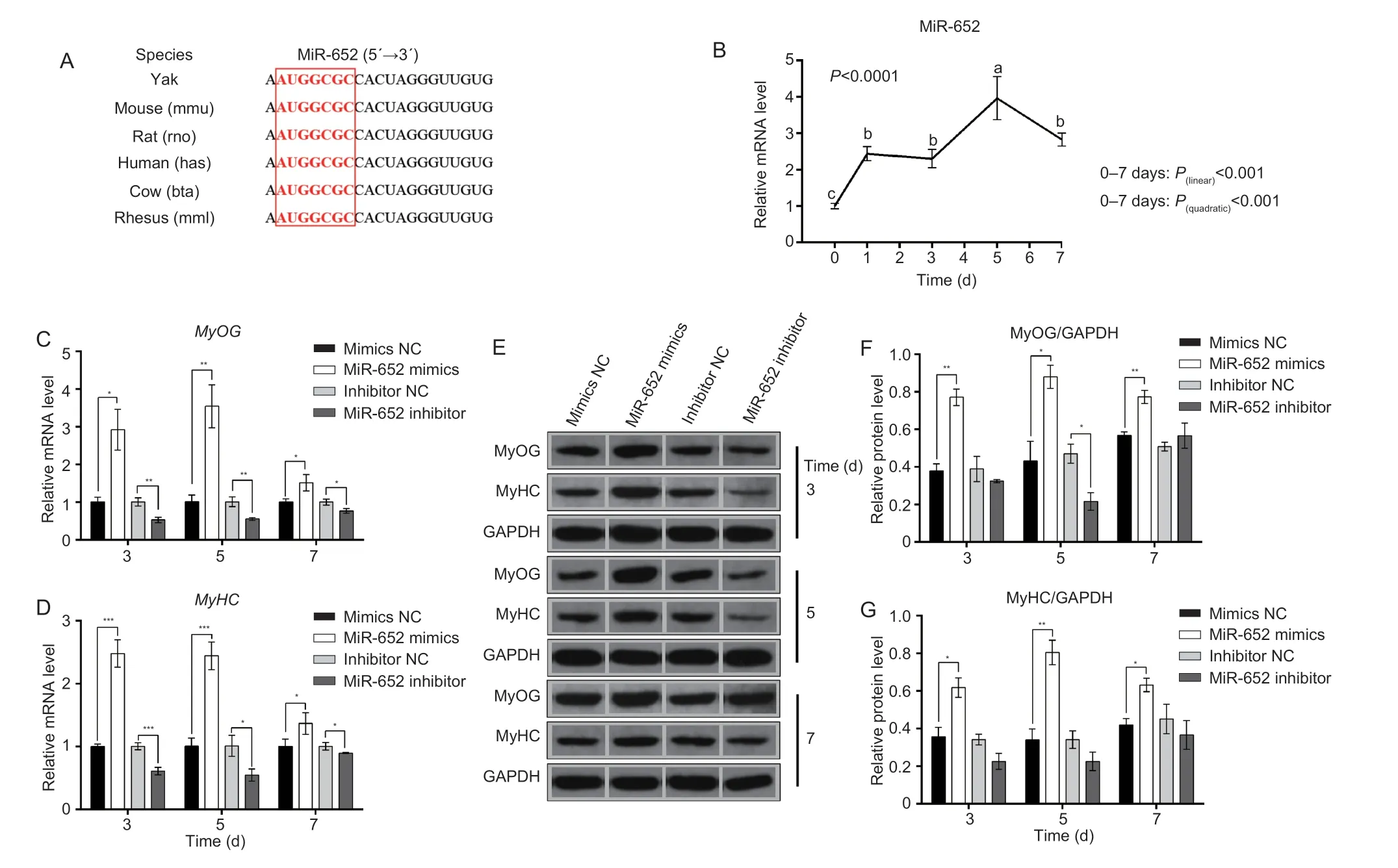
Fig.2 The role of miR-652 during C2C12 myoblast differentiation.A, miR-652 is conserved in mammals.B, the relative expression level of miR-652 during C2C12 myoblast differentiation was determined at 0, 1, 3, 5, and 7 days.C and D, the mRNA expression of MyOG and MyHC were detected at 3, 5, and 7 days by RT-qPCR after transfection with mimics NC, miR-652 mimics, inhibitor NC, and miR-652 inhibitor.E–G, the protein expression of MyHC and MyOG was measured at 3, 5, and 7 days by Western blot after transfection with mimics NC, miR-652 mimics, inhibitor NC, and miR-652 inhibitor into C2C12 myoblasts.The results are expressed as mean±SE (n=3).a, b, and c, P≤0.001; *, P≤0.05; **, P≤0.01; ***, P≤0.001.
3.5.MiR-652 promotes C2C12 myoblasts proliferation
To identify the role of miR-652 in myoblast proliferation, C2C12 myoblasts were correspondingly transfected with miR-652 mimics, mimics NC, miR-652 inhibitor, and inhibitor NC using LipofectamineTM2000.After treating the transfected cell with CCK8 reagent, miR-652 enhanced (P≤0.001) the cell growth rate, as shown in (Fig.3-A).CCK8 detection confirmed that miR-652 stimulated myoblast proliferation.The result was also confirmed in EdU staining assays, revealing miR-652 mimics increased (P≤0.001) the ratio of EdU-positive cells (Fig.3-B).
3.6.MiR-652 inhibits C2C12 myoblasts apoptosis
The FITC-AnnexinV/PI staining by flow cytometry assays demonstrated an enhanced (P≤0.001) number of apoptosis cells in the miR-652 mimics group as compared with the mimics NC group at an early stage (Fig.3-C and D).After inhibition of miR-652, the number of apoptotic cells reduced (P≤0.001) when compared with the inhibitor NC group both at early and later stages (Fig.3-E and F).These results elucidated the role of miR-652 in inhibiting muscle cell apoptosis.

Fig.3 The role of miR-652 during C2C12 myoblast proliferation and apoptotic.After transfection with mimics NC, miR-652 mimics, inhibitor NC, and miR-652 inhibitor into C2C12 myoblasts.A, C2C12 myoblasts growth ration detected at 450 nm with CCK8 reagent.B, the proportion of EdU-positive cells.C and D, percentage of apoptotic cells in early and later periods detected by annexin V-FITC/PI-staining flow cytometry after treated cells with miR-652 mimics or mimics NC separately.E and F, percentage of apoptotic cells in early and later periods detected by annexin V-FITC/PI-staining flow cytometry after treated cells with miR-652 inhibitor or inhibitor NC separately.The results are expressed as mean±SE (n=3).*, P≤0.05; **, P≤0.01; ***, P≤0.001.
3.7.MiR-652 regulates C2C12 myoblasts cell cycle
The PI staining by flow cytometry assays showed the miR-652 mimics induced C2C12 myoblasts cell arrest (P≤0.001) in the G1 phase and enhanced (P≤0.01) the percentage of S and G2 phase cells compared with the mimics NC group (Fig.4-A and B).Additionally, miR-652 inhibitor enhanced (P≤0.001) G1 phase cell proportion and lowered (P≤0.05) S and G2 phase cell proportions in comparison with the inhibitor NC group (Fig.4-C and D).Based on the results, miR-652 could modulate the cell cycle in muscle cells.
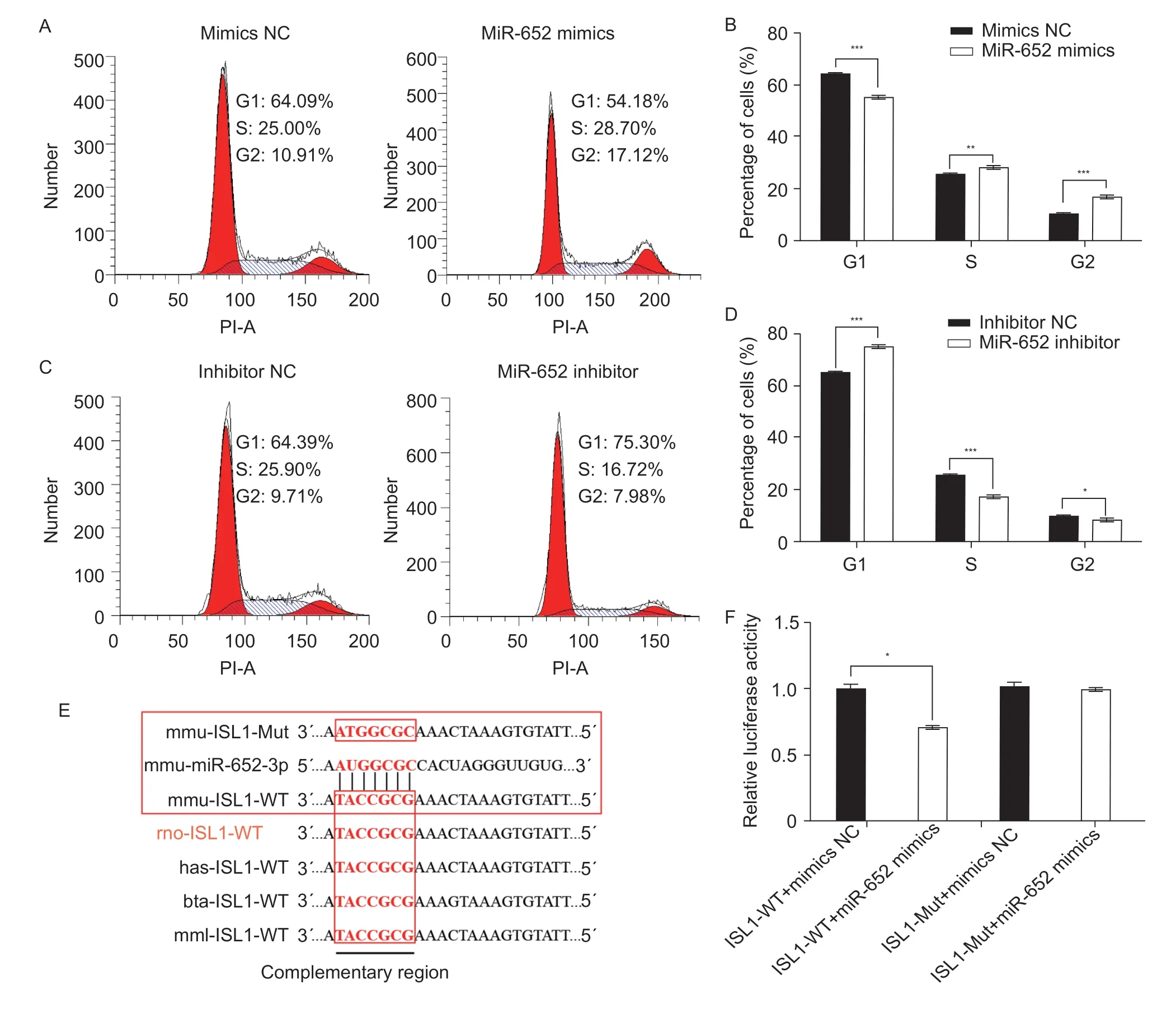
Fig.4 The role of miR-652 during C2C12 myoblast cell cycle.A and B, percentages of cells at different cell cycle stages detected by PI flow cytometry after transfection with mimics NC and miR-652 mimics.C and D, percentages of cells at different cell cycle stages detected by PI flow cytometry after transfection with inhibitor NC and miR-652 inhibitor.E, the putative miR-652 target site of the ISL1 3´UTR conserved in mouse (mmu), rat (rno), human (has), cow (bta), and macaque (mml).F, relative luciferase activity in 293T cells co-transfected with miR-652 mimics or control and the wild-type and mutated ISL1 3´UTR luciferase reporter.The results are expressed as mean±SE (n=3).*, P≤0.05; **, P≤0.01; ***, P≤0.001.
3.8.ISL1 is a target gene of miR-652
To explain the regulatory mechanism of miR-652 underlying C2C12 myoblast proliferation and differentiation, miR-652’s target genes were predicted using TargetScan Software.Bioinformatics analysis showed that the miR-652 was predicted to target at the 3´UTR of theISL1gene.The binding site was highly conserved among different species, and we found cow and mouse have the same miR-652 complementary region in the 3´UTR ofISL1(Fig.4-E).To ascertain whether miR-652 directly targetedISL1, a dual-luciferase reporter assay was performed.The luciferase activity was reduced (P≤0.05) from co-transfected with normal seed regions (ISL1-WT) and miR-652b-3p mimics.Whereas the luciferase activity was not affected by miR-652 mimics when co-transfected with mutant seed regions (ISL1-Mut) (Fig.4-F).These findings illustrated thatISL1is a direct target of miR-652.
4.Discussion
In this study, we compared the miRNA profiles between prenatal and postnatallongissimusdorsimuscle of yak by using a small RNA-seq approach.The result showed that multiple miRNAs expression was changed between prenatal and postnatal.It suggested that miRNA might play a vital role in skeletal muscle development of yak.Furthermore, we found miR-652 is significantly expressed in fetal group, functional investigation of miR-652 verified our hypothesis that miR-652 could influence the myogenesis of C2C12 cells.Target gene prediction and dual-luciferase reporter assay results indicate thatISL1is a target gene of miR-652.Previous studies also indicated thatIsl1is related to different types of muscle development (Caiet al.2003; Laugwitzet al.2005; Morettiet al.2006; Harelet al.2009; Gaoet al.2019).All evidence indicated that miR-652 might targetISL1to regulate muscle development and myogenesis.It suggested that miR-652 might be utilized as a target for molecular breeding to promote yak meat yield performance.
Numerous studies have revealed that miRNA is involved in the regulation of muscle development and myogenesis (Leeet al.2017, 2020; Zhanget al.2018; Wuet al.2019).Small RNA-Seq method could effectively identify key miRNAs in different biological processes.Houet al.(2012) analyzed the miRNAs expression of skeletal muscle from fetal to adult pigs and suggested that ssc-miR-378 was a new candidate miRNA for myogenesis and might targetBMP2andMAPK1.In this study, the small RNA-Seq results showed that miRNA accounts for a significant percentage of small RNA counts on the genome, which is similar to previous studies (Weiet al.2018; Wanget al.2021a).We detected 264 DE miRNAs, 235 miRNAs were up-regulated in prenatal muscle tissue compared with postnatal muscle tissue, as well as 29 miRNAs were down-regulated.Similar to a previous study in pigs, the number of DE miRNAs in embryonic was more than postnatal in the current study (Siengdeeet al.2015).The results indicated that the active expression of miRNA might exert a significant influence on prenatal muscle development.Functional enrichment analysis showed that DE miRNAs-targeted genes were significantly enriched in pathways related to skeletal muscle development and myogenesis, such as PI3K–Akt signaling pathway (Fuet al.2019), and MAPK signaling pathway (Fanet al.2018).
We found miR-652 was up-regulated in the prenatal group.Previous studies showed miR-652-3p plays important roles in several biological processes.Yanget al.(2016) verified miR-652-3p stimulated non-small cell lung cancer cells by targetingLgl1gene.Zhuet al.(2019) found that miR-652-3p could support bladder cancer cell proliferation, migration, and invasionviadirectly regulatingKCNN3.Jianget al.(2018) proved that the overexpression of miR-652-3p could promote lymphoblastic leukemia cell apoptosis.Shiet al.(2019) validated that miRNA-652-3p could regulate the ephrin receptor B4 level by targeting homeobox A9 to promote the HTR-8/SVneo cell line.Additionally, miR-652-3p/VEGFA axis influences the osteogenic differentiation of dental pulp stem cells (Wanget al.2021b).The finding supports the importance of miR-652 in cell proliferation, differentiation, apoptosis, and migration.Whether miR-652 has an effect on myogenesis, however, has yet to be determined.In general, mouse C2C12 myoblasts were used as a cell model for muscle development research, and sequence analysis revealed that miR-652 is conserved in both yak and mouse.Cell proliferation, differentiation, and apoptosis are critical processes in the development of skeletal muscle.The cell cycle is essential for the maintenance of normal myogenesis.In the present study, C2C12 myoblasts were treated with overexpressed miR-652, knockdowned miR-652, and their negative control.The gene and protein expression of muscle differentiation marker showed that miR-652 induced the differentiation of C2C12 myoblast; CCK8 and Edu assays verified miR-652 also promoted cell proliferation.Annexin V-FITC/PI staining flow cytometry experiment showed that miR-652 inhibited apoptosis.Besides, the PI staining flow cytometry experiment demonstrated that miR-652 could regulate the cell cycle in C2C12 myoblasts.Based on the above findings, miR-652 had a vital effect on modulating muscle development.
To understand how miR-652 modulates C2C12 myoblast development, we predicted and verified miR-652-targeted gene.ISL1is one of miR-652-targeted genes as predicted by TargetScan.Based on the dualluciferase reporter assay result, miR-652 directly targetedISL1.SFRP2is a Wnt antagonist that participates in myogenesis (Levinet al.2001).A previous study showed thatSFRP2is highly expressed in prenatal muscle and indicated thatSFRP2influenced porcine embryonic skeletal muscle development and was regulated by miR1/206 (Maet al.2016).The evidence showed that miRNA could regulate muscle development through target myogenesis related gene.Previous studies also indicatedISL1is related to muscle development.Hence, we speculated that miR-652 influenced the differentiation, proliferation, apoptosis and cell cycle of C2C12 myoblast may through regulate the expression ofISL1.
5.Conclusion
A genome-wide analysis of miRNAs was performed in prenatal and postnatal yaklongissimusdorsimuscle tissues in the current study.DE miRNAs are linked to muscle development based on GO and KEGG enrichment analysis of DE miRNA target genes.This information sheds light on the role of miRNAs in the development of yak skeletal muscles.We also investigated the role of miR-652 in the development of C2C12 myoblasts and suggested that miR-652 might targetISL1to regulate the proliferation, differentiation, apoptosis, and the cell cycle of C2C12 myoblasts.
Acknowledgements
This study was supported by the Agricultural Science and Technology Innovation Program, CAAS (25-LZIHPS-01), the China Agriculture Research System of MOF and MARA (CARS-37) and the National Natural Science Foundation of China (32102500).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethical approval
All of the experimental protocols and procedures were approved by the Animal Administration and Ethics Committee of Lanzhou Institute of Husbandry and Pharmaceutical Sciences of Chinese Academy of Agricultural Sciences (Permit No.SYXK-2014-0002).
Appendicesassociated with this paper are available on https://doi.org/10.1016/j.jia.2022.08.116
杂志排行
Journal of Integrative Agriculture的其它文章
- Herbicidal activity and biochemical characteristics of the botanical drupacine against Amaranthus retroflexus L.
- Developing a duplex ARMS-qPCR method to differentiate genotype l and ll African swine fever viruses based on their B646L genes
- The effects of maltodextrin/starch in soy protein isolate–wheat gluten on the thermal stability of high-moisture extrudates
- Elucidation of the structure, antioxidant, and interfacial properties of flaxseed proteins tailored by microwave treatment
- Effects of planting patterns plastic film mulching on soil temperature, moisture, functional bacteria and yield of winter wheat in the Loess Plateau of China
- lnversion tillage with straw incorporation affects the patterns of soil microbial co-occurrence and multi-nutrient cycling in a Hapli-Udic Cambisol
