Research progress on the drug action and resistance mechanism in Mycobacterium tuberculosis
2023-03-06GESaiSONGXinyiJIANGHuiyueLIZhaoyangZHUZhuangyanSUNManluan
GE Sai, SONG Xin-yi, JIANG Hui-yue, LI Zhao-yang, ZHU Zhuang-yan✉, SUN Manluan✉
Keywords:
ABSTRACT Tuberculosis (TB) is a chronic infectious disease caused by Mycobacterium Tuberculosis (MTB).It is the second largest single cause of death besides novel coronavirus pneumonia.Along with the abuse of antibiotics and extensive use of anti-tuberculosis drugs, multidrug-resistant(MDR) TB, drug-resistant (XDR) TB and totally drug-resistant (TDR) TB became obstacles to the tuberculosis eradication worldwide.According to the World Health Organization (WHO)statistics, China is not only a high burden tuberculosis country in the world, but also a country with a serious epidemic of MDR.Traditional drugs fail to meet the needs of tuberculosis control.Therefore, it is urgent to find new targets of anti-tuberculosis drugs and develop new anti-tuberculosis drugs.Hence, this paper systematically summarizes the mechanism of traditional and newly developed anti-tuberculosis drugs, in which stressing the research progress of drug resistance mechanisms.This work provides us with new insights of new antituberculosis drug developments, and may contribute to a reduction in the harm that tuberculosis brings to society.
Tuberculosis (TB) is a chronic infectious disease caused by Mycobacterium tuberculosis (MTB).The World Health Organization(WHO) pointed out in the ‘2021 Global Tuberculosis Report’that 71% of tuberculosis patients worldwide received rifampicin resistance test in 2020, of which 6.3% were victims of rifampicinresistant tuberculosis (RR-TB) or multidrug-resistant tuberculosis(MDR-TB).Meanwhile, up to 1.2% of patients were extensively drug-resistant tuberculosis (XDR) or pre-XDR.In 2020, the number of TB patients in China increased up by 842,000, and China is one of the countries with a high burden of pandrug-resistant/ rifampicinresistant TB[1].The emergence of drug-resistant tuberculosis bacteria has aggravated the difficulty of tuberculosis treatments.A study of nearly 10,000 MDR-TB patients in 25 countries around the world pointed out that the overall treatment failure rate of MDRTB patients worldwide is as high as 8%, and the mortality rate is as high as 14%, meanwhile, he overall failure rate is as high as 19%[2].Patients with drug-resistant tuberculosis often suffers from significantly prolonged hospital stay and poor prognosis.Compared with drug-sensitive tuberculosis patients, drug-resistant tuberculosis patients have low cure rates, recurrent attacks, and significantly increased mortality[3].The high cost of drug-resistant tuberculosis treatments, the side effects of second-line drug use, and the serious physical damage to patients have also made the preventions and treatments of tuberculosis urgent.Hence, the developments of antituberculosis drugs has also been a hot topic of global research.Due to the long treatment cycle of tuberculosis, patients often have problems such as poor medication compliance, leading to recurrent tuberculosis, and tuberculosis retreatments once again increases the risk of tuberculosis resistance.Studies indicated that poor treatment compliance, tuberculosis retreatment is one of the most important risk factors for MDR-TB, which is more than 11 times higher than the initial treatment of patients with MDR-TB risk[4].With the rapid developments of molecular biology technology and clinical diagnosis technology, more and more drug-resistant Mycobacterium tuberculosis has been reported, and its drug action mechanism and drug resistance mechanism have been gradually explored.This work summarizes the common first-line and second-line anti-tuberculosis drugs and the latest research and developments drugs from the aspects of drug action mechanism and drug resistance mechanism,to provide new ideas of exploring the molecular mechanism of drug resistance of Mycobacterium tuberculosis.
1.Anti-tuberculosis drugs action mechanism
The current clinical use of anti-TB drugs can be traced back to the 1940s (Figure 1).Many anti-TB drugs have been used for decades, which is one of the important causes of Mycobacterium tuberculosis resistance.According to the drug effect and adverse reactions, the commonly used drugs for the treatment of tuberculosis are divided as: first-line drugs and second-line drugs.Among them, the commonly used first-line drugs include isoniazid(INH), rifampicin (RIF), ethambutol (EMB), streptomycin (STR)and pyrazinamide (PZA).In order to prevent and strengthen the therapeutic effects of drug-resistant tuberculosis, first-line and second-line anti-tuberculosis drugs are used for combined chemotherapy.Common second-line drugs are kanamycin (Km),amikacin (Am), capreomycin (Cm), linezolid (LZD), ethionamide(ETH), prothionamide (PTH), fluoroquinolones (FQs), para-amino salicylic acid (PAS), D-Cycloserine (DCS), clofazimine (CFZ), etc.Human research and development of new anti-tuberculosis drugs has never stopped.Since 2012, new anti-tuberculosis chemical drugs,including Bedaquiline (BDQ), Delamanid (DLM, OPC-67683),Pretomanid ( PA-824 ), have been launched, and more than 20 other drugs entered the clinical trial stage[5].
According to the mechanism of drug action, common antituberculosis drugs are divided into five categories (Fig.2): (1)Inhibit cell wall synthesis (ethambutol, isoniazid, etc.); (2) affect cell membrane permeability (streptomycin, etc.); (3) Inhibit proteins synthesis (aminoglycosides, peptides, linezolid, etc.);(4) Inhibit nucleic acid synthesis (fluoroquinolones, rifamycins,etc.); (5) block folic acid metabolism (amino salicylic acid, etc.).Mycobacterium tuberculosis has evolved a corresponding resistance mechanism to resist drug ‘threat’ based on current anti-drug mechanism.In general, Mycobacterium tuberculosis resistance mechanisms can be divided as intrinsic resistance and acquired resistance ; innate resistance, known as natural resistance.For example, Mycobacterium tuberculosis is naturally insensitive to low concentrations of trimethoprim (TMP) due to its different folate metabolic pathways[6].Acquired resistance generally refers to the change of genes in bacteria caused by mutations, which leads to the change of biochemical mechanism in vivo, resulting in drug resistance.At present, the research on the molecular mechanism of drug resistance of Mycobacterium tuberculosis mainly focuses on the drug target and the related mutation genes, such as: (1) gene mutation leads to inactivation or passivation of drug metabolizing enzymes (pyrazinamide, etc.); (2) the change of target reduced the affinity with anti-tuberculosis drugs (ethambutol, etc.); (3) change the permeability of cell wall and cell membrane, resulting in drug penetration disorders (Delamani, fluoroquinolones, etc.); (4)change the active efflux mechanism, resulting in Mycobacterium tuberculosis directly discharged the drug through the efflux pump;(5) the formation of bacterial biofilm, blocking drugs into the body[7].
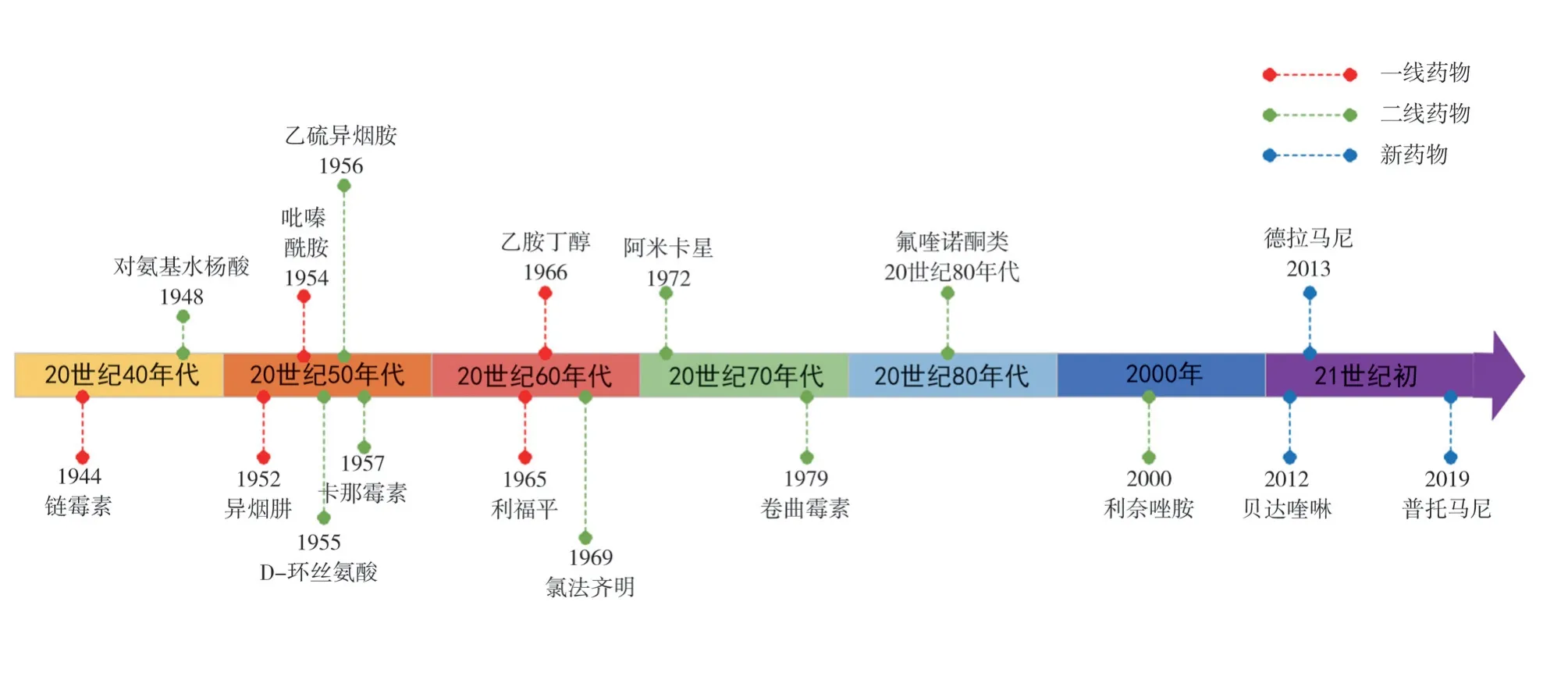
Fig1 Timeline of the application of anti-tuberculosis drugs

Fig 2 Anti-tuberculosis drugs action mechanism [8]
2.First-line drug
2.1 Isoniazid (INH)
Isoniazid (INH) has been used clinically since 1952 as a classic drug for the treatment of tuberculosis.Because of its cheapness,effectiveness, less side effects, and bactericidal effects on Mycobacterium tuberculosis in both reproductive and stationary phases, it is one of the most common anti-tuberculosis drugs in clinical[9].When INH enters MTB cell, it is first reduced by the catalase KatG and binds to NAD+to form an isonicotinic-NAD complex, which interacts with the NADH-specific enoyl acyl apolipoprotein and reductase InhA to inhibit the synthesis of branched mycolic acid in the MTB cell wall.It destroys cell wall integrity, accelerates cell rupture and eventually leads to MTB death during vigorous or resting metabolism[10].In addition, INH was identified to prevent tuberculosis infection by reducing oxidative stress ROS-induced immune cell death[11].
Studies on INH resistance have also focused on gene mutations,which related to InhA and KatG targets.For example, most commonly mutation happens in katG gene is the 315th residue mutates from Ser to Thr.This mutation could relieve the inhibitory effect of INH on mycolic acid synthesis, making MTB resist to INH[12].Similarly, the analysis of clinical INH-resistant strains showed that there were a large number of inhA gene promoter mutations in drug-resistant strains, the most common type of gene mutation was C15T[13].It was speculated that inhA gene mutation may also cause MTB resistance to INH[13].In addition, after INH enters cell, MTB may produce free radicals and affect its own metabolism, thereby reducing the sensitivity to INH.Moreover,InhA protein Thr266 can be phosphorylated to decrease its activity,which may change cell wall permeability and affect its resistance to INH[14].
The acetylation of iron regulatory protein HupB affect the expression of corresponding genes in MTB, resulting in drug resistance to INH[15].In recent years, it’s reported that mutations in the embB gene associated with ethambutol resistance may also cause mutations in MTB, but the specific mechanism needs further study[9].In addition,PknG, SahH, KasB and other proteins may also be involved in INH resistance[16].
2.2 Rifampicin (RIF)
Rifampicin (RIF), a semisynthetic antibiotic derived from rifamycins, inhibits both active and slow MTB metabolism.RIF mainly acts on the RNA polymerase β subunit of MTB, blocks the extension of mRNA, inhibits protein synthesis, affects transcription and kills MTB[17].Long-term clinical use of RIF injection is easy to cause patients to develop a certain degree of resistance to RIF.However, in practice, it is found that resistance to RIF alone is rare.In general, strains resistant to RIF are also resistant to other drugs (especially in combination with INH) ; thus, general RIF resistance is one of the hallmarks of MDR[1].RIF acts primarily on the RNA polymerase β subunit, so RIF-induced resistance is usually associated with mutations in the gene encoding rpoB[17].Mutation of rpoB gene leads to conformational change of RNA polymerase subunit protein and RIF resistance.At present, many studies suggest that the mutations associated with RIF resistance are mainly concentrated in the rifampicin resistance determining regions( RRDR ) of rpoB gene[18].According to WHO statistics, in 2020,the percentage of RIF-resistant TB cases confirmed by bacteriology in China was as high as 83 % ; in cases of relapse after treatment,the percentage of RIF-resistant TB cases confirmed by bacteriology is as high as 97%[1].Therefore, it is often recommended that RIF is combined with other drugs to prevent drug resistance and achieve better therapeutic effects.
2.3 Ethambutol (EMB)
Ethambutol (EMB) was discovered in 1961 and used in the treatment of tuberculosis in 1966.EMB mainly acts on active MTB and has no obvious bactericidal effect on MTB strains in dormant period.The mechanism of action of EMB is related to the synthesis of MTB cell wall.As a structural analogue of arabinose, it blocks the synthesis of arabinogalactan by inhibiting the arabinosyltransferase in the synthesis of MTB cell wall and reduces the permeability of cell wall[18].Studies have shown that the mechanism of EMB resistance is mainly related to the mutations ofembC,embAandembBgenes.The mutation of embB gene was the most common in EMBresistant clinical isolates.In addition, mutations in the embC andembC-embaAgene regions also led to EMB resistance[19].Similarly,protein post-translational modification can also indirectly regulate the transcription process of embABC operon, thereby regulating the change of EMB resistance[19].Clinically, EMB is mainly used for the treatment of patients with contraindications to aminosalicylic acid drugs, and is often used in combination with INH and RIF.
2.4 Pyrazinamide (PZA)
Structural analogues of quinolinic acid (QA), an important metabolite of pyrazinamide (PZA) in Mycobacterium tuberculosis NAD metabolism[20].PZA kills dormant and semi-dormant MTB in host macrophages.It is generally believed that PZA enters the bacteria through the membrane transport system, and is converted into its active form pyrazinoic acid (PO ) under the action of pyrazinamidase (PZase) encoded by the pncA gene.POA enters the extracellular acidic environment through the efflux mechanism to form protonated pyrazinic acid, which causes cell damage by affecting the membrane potential, thereby killing MTB[20].In addition, PZA can also affect the normal metabolism of MTB by affecting dehydrogenase dehydrogenation, resulting in antibacterial effect[21].In 2011, Shi et al.found that the in vivo active form of PZA, POA, can directly target the S1 protein in the ribosomal 30S subunit, affecting the protein translation process, thereby enhancing PZA activity[21].At present, the mechanism of PZA resistance is inconclusive, and the research is mainly focused on pncA gene mutation[25].In addition, rspA, panD, fadD2 and genes related to FAS-I metabolism may also be related to PZA resistance[22].
2.5 Streptomycin (STR)
Streptomycin (STR) is the first aminoglycoside first-line injection drug for tuberculosis treatment, which has bactericidal effect on MTB in logarithmic growth phase.STR can directly bind to 16S rRNA (encoded by rrs gene) and S12 ribosomal protein (encoded by rspL gene) in MTB ribosomal 30S subunit, hindering the binding of tRNA to ribosomes and inhibiting bacterial protein synthesis, leading to bacterial death.Therefore, mutation of rspL and rrs genes is one of the main causes of STR resistance in MTB[23].In the detection of 137 clinically isolated MTB strains, 86.13 % of STR-resistant strains carried rpsL or rrs gene mutations, of which rpsL gene mutations accounted for the majority (67.15 %).In addition, low levels of STR resistance have been linked to mutations in thegidBgene encoding 16S rRNA methyltransferase[24].
3.Second-line drugs
3.1 Fluoroquinolones (FQs)
Fluoroquinolones (FQs) are the most widely used broad-spectrum antibiotics in clinical practice and the first choice for the treatment of XDR tuberculosis.FQs commonly used in anti-tuberculosis treatment mainly include traditional ofloxacin, levofloxacin,moxifloxacin, ciprofloxacin and new sparfloxacin.In clinical practice, FQs are mainly used for the combined treatment of drugresistant tuberculosis due to the advantage that it is not easy to crossresistance to other anti-tuberculosis drugs.For example, it has a good therapeutic effect on patients with MDR infection resistant to first-line drugs INH and RIF.FQs act on MTB’ s DNA gyrase (type II DNASA topoisomerase) to inhibit its growth by inhibiting MTB transcription and translation[25].Therefore, FQs resistance loci are currently considered to be concentrated in the genes of quinolone resistance-determining regions (QRDRs)[26].Mutations in the gyrA and gyrB genes encoding DNA gyrase subunits result in increased MICs of FQs.In addition, the increase of drug efflux pump will also affect the resistance of FQs ; mTB cell wall thickening and permeability reduction are also the causes of FQs resistance, but the level of FQs resistance caused by cell wall thickening is low, and the molecular mechanism needs to be further clarified[27].
3.2 Para-aminosalicylic acid (PAS)
Para-aminosalicylic acid (PAS), a structural analogue of folic acid precursor p-aminobenzoic acid (PABA), is the most commonly used anti-tuberculosis drug in folic acid antagonist drugs.Other folic acid antagonists include dapsone, WR99210 and trimethoprim/ sulfamethoxazole (a compound of sulfamethoxazole and trimethoprim).Target studies have focused on several essential enzymes in the tetrahydrofolate de novo synthesis pathway:dihydropteroate synthase (DHFS, FolP1), dihydrofolate reductase(DHFR) and flavin-dependent thymidylate synthase (flavindependent thymidylate synthase)[28].In vivo, PAS can competitively bind to DHFS and inhibit the activity of DHFR, resulting in a significant decrease in tetrahydrofolic acid (THF) synthesis and a significant inhibition of the conversion of deoxyuridine 5-phosphate( dUMP ) to deoxythymidine 5-phosphate (dTMP), leading to DNA synthesis disorder and death of MTB[29].At present, more research on PAS resistance is Folc protein coding gene folC, and its mutation will increase the metabolic rate of MTB to PAS, resulting in PAS resistance[30].Riboflavin biosynthesis enzyme (RibD) catalyzes the conversion of dihydrofolic acid (DHF) to THF.Mutations in the gene encoding ribD lead to up-regulation of RibD expression in place of DHFR, resulting in PAS resistance[31].In addition, mutations in protein-coding genes such as acyltransferase PapA1, SigB and MmpL11 may also be related to PAS resistance, and the specific mechanism needs further study[32-33].
3.3 Aminoglycoside antibiotics and polypeptide drugs
Common second-line injection drugs in the treatment of tuberculosis include aminoglycosides such as kanamycin(Km), amikacin (Am) and viomycin, and polypeptides such as capreomycin.These drugs inhibit protein translation by binding to the 16S rRNA in the MTB ribosomal 30S subunit, triggering MTB death[34].Because of their similar drug action mechanisms, the drug resistance mechanisms of these drugs are often discussed together;moreover, more and more studies have begun to pay attention to the cross-resistance between these drugs.Studies have shown that the tlyA gene may encode a predicted rRNA methyltransferase,mutations in which can make MTB resistant to capreomycin and azithromycin[35].The mutation of rrs gene encoding 16S rRNA is related to the resistance of four drugs.Recent studies have shown that mutations in the promoter region of the eis gene encoding aminoglycoside acetyltransferase can affect the resistance of MTB to low levels of amikacin and kanamycin[36], and eis or whiB7 mutations can cause MTB to develop resistance to kanamycin[37].In clinical treatment, capreomycin has a good effect on stationary phase TB, and has less side effects than other second-line drugs.However,it is easy to produce drug resistance when used alone, so it is often combined with first-line drugs such as INH and EMB in clinical practice.
3.4 Linezolid (LZD)
Linezolid (LZD) was approved by FDA in the United States as the first fully synthetic oxazolidinone antibacterial drug in April 2 000,which can effectively treat various Gram-positive bacterial infections.LZD.It works on the 23 S rRNA of the ribosomal 50S subunit,inhibits the formation of the 70 S translation initiation complex by binding to the 50 S subunit, preventing protein translation and exerting anti-MTB effects[38].At present, most of the known genes related to LZD resistance are mainly focused onrplCgene and rrl gene.The rplC gene encodes the ribosomal protein L3 of the MTB ribosomal 50 S subunit.RplC mutations are often associated with varying degrees of LZD resistance, with the most common 460th Thr mutation to Cys[39].Gene rrl mainly encodes 23 S rRNA, and its mutation can also make MTB produce LZD resistance.Among them,G2061T is the most common mutation, and this mutation will make MTB strains produce high LZD resistance[40].In clinical treatment,LZD has strong side effects such as blood and bone marrow toxicity.About 40 % ~ 90 % of patients suffer from adverse reactions,and 6 % ~ 8 % of patients discontinue due to strong neurological lesions and bone marrow suppression[41].In the treatment of MTB,the clinical use of LZD is strictly limited by the large side effects.However, as the first fully synthesized oxazolidinone antibacterial drug, its application in tuberculosis treatment provides ideas for exploring new oxazolidinone anti-MTB drugs.
3.5 Ethylthioisonicotinamide (ETH) and propylthioisonicotinamide(PTH)
Both ethionamide (ETH) and protionamide (PTH) belong to thioamides.They are similar to INH in action, but their antibacterial activity is much lower than INH[42].The mechanism of action of ETH and PTH is similar to that of INH.The intracellular role of ETH and PTH requires the activation of the monooxygenase EthA protein to produce metabolites similar to the structure of isonicotinic acid, inhibit the activity of the target protein InhA enzyme, destroy the synthesis of mycolic acid in the cell wall of MTB, lead to the destruction of cell wall integrity, accelerate cell rupture and eventually lead to the death of MTB in the metabolically active or quiescent period[10].Thus, mutations associated with ETH and PTH resistance are primarily drug-activated ethA and drug targetinhAgene alterations[43].In addition, the transcriptional repressor EthR inhibits the expression of ethA by binding to the ethA-ethR intergenic region and decreases the activity of EthA protein, resulting in increased ETH resistance in Mycobacterium tuberculosis[44].Studies have found that in the clinical treatment of MDR-TB patients, ETH and PTH resistance is more serious.Therefore, some scholars suggest that patients with previous medication history of ETH and PTH should receive drug sensitivity tests before using the two drugs again to avoid insufficient therapeutic effect caused by drug resistance[37].
3.6 D-serine
D-cycloserine (DCS), like RIF, is a broad-spectrum antibiotic[45].D DCS is a structural analogue of D-alanine, which mainly affects the synthesis of peptidoglycan from D-alanine in MTB cell wall.It inhibits the activity of alanine racemase (encoded by thepncAgene) and alanine ligase (encoded by the ddlA gene), impairs cell wall synthesis, and reduces the acid resistance of bacteria to play a bactericidal or bacteriostatic role[46].By analyzing the known DCS resistant strains, it was found that the mutation genes related to DCS resistance were mainly concentrated on ald, pncA and ddlA genes.The genealdencodes L-alanine dehydrogenase and is involved in alanine metabolism.L-alanine compensation occurs in MTB due to the deletion of ald.The accumulation of L-alanine reduces the competitive inhibition effect of DCS, resulting in drug resistance in MTB[47].Gene pncA encodes alanine racemase and gene ddlA encodes alanine ligase.The overexpression of alanine racemase and alanine ligase caused by gene mutation may lead to DCS resistance in BCG strains.The latest study found that the mutations of pykA,cycA, ald and alr genes were found in 153 DCS-resistant strains of 513 XDR isolates, but the mechanism needs to be further studied[48].
3.7 Clofaziramine (CFZ)
Clofaziramine (CFZ), also known as chlorobenzene phenazine,is a kind of iminophenazine drug.Its key structural feature is phenazine nucleus, which makes it have high lipophilicity and high transmembrane permeability.Early CFZ was mainly used for the treatment of leprosy and has been used in recent years to treat tuberculosis[49].The detailed mechanism of action of CFZ is still unclear, which may inhibit the growth and reproduction of bacteria by inhibiting the killing effect of MTB-derived factor on phagocytic cells.May also be associated with intracellular oxidative stress such as hydrogen peroxide, superoxide or ROS[50].Studies have found that mutations in the transcription factor rv0678 of MTB can upregulate the expression of the efflux pump MmpL5, resulting in cross-resistance between CFZ and bedaquiline (BDQ)[51].
4.New drugs
4.1 Bedaquiline (BDQ)
Bedaquiline (BDQ) is a diarylquinoline approved by the FDA in December 2012.It binds to ATP synthase oligomer C in MTB,affect the activity of ATP synthase proton pump, specifically inhibit the activity of ATP synthase, so that ADP can not be converted to ATP, blocking the energy supply of MTB ATP to play an antituberculosis role[52].At present, BDQ has been used in MTB treatment in more than 50 countries and regions, and there have been reports of BDQ resistance cases and cross-resistance with CFZ[53].Current studies have found that MTB has a more complex BDQ resistance mechanism, including transcriptional regulation by overexpressing some transcription factors, reducing the number of TCA cycles to change ATP production and consumption, and changing metabolic pathways to maintain ATP levels[29].Studies on BDQ resistance genes also include mutations inatpE,rv0678and pepQ[54].In addition, Guo et al.found a new gene glpK related to BDQ resistance, but the specific resistance mechanism needs to be further elucidated[55].
4.2 Delamani (OPC-67683) and Putomani (PA-824)
Delamanib (OPC-67683) and Putomani (PA-824) were approved for marketing in 2014 and 2019, respectively.They are both prodrugs and belong to nitroimidazoles.In vitro and in vivo experiments have shown that they have strong antibacterial activity against MDR strains and bactericidal effect on MTB during dormancy and in host cells[56].As prodrugs, both require F420-dependent nitroreductase Ddn activation to alter the permeability of the cell wall by blocking the synthesis of mycolic acid in MTB cell wall, thereby exerting bactericidal effects.The activity of Ddn required for prodrug activation depends on the normal and reduced state of coenzyme F420.This morphological transformation is mediated by F420-dependent glucose-6-phosphate dehydrogenase (encoded by thefdg1gene), which is closely related to the synthesis and activation of fbiA, fbiB, and fbiC genes[56].At the same time, the fibD gene encoding guanosine monophosphate transferase is also necessary for the synthesis of F420[57].Therefore, the mutations ofddn,fgd1,fbiA, fbiB, fbiC and fbiD genes may be related to drug resistance[58].Among them,fbiDgene mutation is related to the high level of drug resistance of PA-824, but the relationship between fbiD gene mutation and OPC-67683 resistance needs further study[59].Other studies have shown that PA-824 activated by Ddn will produce NO and other active free radicals, inhibit MTB respiratory chain electron transfer, resulting in inhibited bacterial growth[60].In clinical practice, OPC-67683 is generally well tolerated, but when combined with other anti-TB drugs such as FQs, it may increase the probability of adverse events such as syncope and arrhythmia[61].
4.3 New drugs under development
Since 2021, in addition to the above three new drugs on the market,there are still 23 new chemical entity drugs entering clinical trials.Among them, there are four new drugs developed in China, namely:TBI-223 (oxazolidinones, phase I clinical trial stage), Auli-manid(Auli-manid, phase I clinical trial), Pyrfazimine (TBI-166, phase II clinical trial stage), and Shudapyridine (WX-081, phase II clinical trial stage)[5].
5.Prospects
In this paper, the mechanism of action and drug resistance of firstand second-line anti-tuberculosis drugs commonly used in clinical practice are systematically described, and the new drugs that have been marketed and are in the clinical trial stage are summarized (see table 1).
Humans have been fighting tuberculosis for centuries, and drugs to treat it have made good progress at this stage, but there are still many problems to be solved.From the current study, we still havelimited understanding of MTB resistance mechanisms, increasing the difficulty of new clinical anti-TB drug development.The launch of new drugs will undoubtedly bring more opportunities for tuberculosis patients to cure, but the price of new drugs is still very expensive.In addition, according to the latest WHO report statistics, 94 % of patients were cured for up to 2 years, and 4 %for up to 3 years[1].Long treatment cycle leads to poor patient compliance, further exacerbating the emergence of tuberculosis resistance.Therefore, it is urgent to develop new drugs and new preparation types to shorten the treatment time.Understanding the mechanisms of MTB resistance is critical to the development of new anti-tuberculosis drugs.The rapid development of molecular diagnosis and omics analysis has also provided a lot of help for the identification of drug resistance genes, but there are still many problems.For example, whether there is interaction between different drug resistance mechanisms, whether cross-resistance leads to the accumulation or antagonism of drug resistance mutations,and the competition between drugs under the same drug resistance mechanism are still topics worth studying.This review describes the antibacterial mechanism of anti-tuberculosis drugs and drug resistance mechanism of Mycobacterium tuberculosis, in order to provide reference for the development of more new drugs.

Tab1 Targets and drug resistance genes of anti-tuberculosis drugs
Authors’ contribution
Ge Sai: literature collection, collation and writing ; Song Xinyi,Jiang Huiyue, Li Zhaoyang: MS writing and drawing; Zhu Zhuangyan, Sun Manluan : MS design, review, guidance.
Author conflict of interest statement
All authors declare that there is no conflict of interest in the process of writing this article.
杂志排行
Journal of Hainan Medical College的其它文章
- Meta-analysis of the impact of hyperuricemia on contrast agent-related acute kidney injury after percutaneous coronary intervention
- Macrophage-derived exosomes mediate mir-222 targeting Caspase-10 to promote glioma proliferation
- Mechanism of action of verbascoside in mice with Parkinson's disease based on molecular docking, molecular dynamics and in vivo experiments
- Mechanism of Sanshi decoction inhibits macrophage pyroptosis by inhibiting BRD4/NF-κB/ NLRP3 pathway in the treatment of gouty arthritis
- Effects of female body mass index on embryo development and ART outcomes
- Prediction of potential targets and molecular mechanisms of Tripterygium hypoglaucum for oral lichen planus based on network pharmacology and molecular docking
