Transformer and CNN combined generative adversarial network for segmentation of placental tissue in MR images
2023-01-28YEZhengjieWANGYutaoXUJianJINWei
YE Zhengjie ,WANG Yutao ,XU Jian ,JIN Wei*
(1.Faculty of Electrical Engineering and Computer Science,Ningbo University,Ningbo 315211,China;2.Affiliated Hospital of Medical School,Ningbo University,Ningbo 315020,China;3.Ningbo Women &Children’s Hospital,Ningbo 315012,China)
Abstract: Accurate segmentation of placental tissue in magnetic resonance (MR) images is of great significance for the study of pregnancy and childbirth complications.However,manual annotation by radiologists is difficult to ensure the accuracy and objectivity of the results,and it is time-consuming and labor-intensive.To develop a deep learning model for automated placental tissue segmentation in MR images,we propose a generative adversarial network (TCGANet) that combines transformer and convolution neural network (CNN) to realize the segmentation purposes.It combines the feature embedding module with the skip connection which avoids the information loss caused by the traditional feature fusion method.On the basis of this technique,the context extractor module is introduced,and the self-attention mechanism of the transformer is adopted to capture the global dependencies,which can effectively represent the global and local information of MR images.In addition,to improve the precision of placental tissue edge segmentation,the generative adversarial architecture is introduced to supervise the generative network of placental tissue segmentation by the discriminator network.The results show that the proposed model is superior to other comparison methods in terms of quantitative metrics and boundary positioning accuracy,the accuracy of which reads 0.993±0.003,sensitivity reaches 0.903±0.093,specificity is 0.996±0.003,and Dice similarity coefficient is found to be 0.861±0.141.Ablation experiments on different structural units of the TCGANet have validated the network structure design,and most of the performance metrics are far better than those of the existing methods (P<0.05).The proposed TCGANet enables automated and accurate segmentation of placental tissue in MR images.
Key words: deep learning;placental tissue segmentation;adversarial network;MRI;attention mechanism
1 Introduction
The placenta is the critical organ for material exchange between the fetus and the mother,and the fetus relies on it to absorb nutrients from the mother.Placental abnormalities[1]are common obstetric diseases and may cause serious obstetric complications.Magnetic resonance (MR) imaging has the advantages of a large field of view and good soft tissue contrast and is an important complement to ultrasound[2].T2 weighted image (T2WI) clearly distinguish the placental tissue from the others,so the research in this paper is based on T2WI[3].Accurate segmentation of placental tissue is essential for diagnosing placental abnormalities[4].Traditionally,placental tissue segmentation mainly relies on manual labeling by radiologists,which is difficult to ensure the accuracy and objectivity of labeling results,and it is time-consuming and labor-intensive.Therefore,it is urgent to design an effective and reliable automatic segmentation method of placental tissue in MR images.
In recent years,convolution neural networks(CNNs) and other deep learning-based algorithms have developed rapidly,and have been widely used in the fields of medical image segmentation[5],classification[6-7],and detection[8].U-Net[9]is one of the most popular networks in the field of medical image segmentation,and subsequent improvements based on U-Net[10-11]are also widely used.Some studies have achieved great performance in medical image segmentation such as that of liver[12],brain[13],prostate[14],and other organs.
Although CNNs have been widely used in the field of medical image segmentation because of their excellent feature representation capabilities,they lack the ability to model long-range dependencies present in an image.More precisely,each convolution kernel only has a local subset of pixels in the whole image.Therefore,to obtain global information,these models stack multiple convolution layers and expand the receptive field through downsampling,which has limitations[15].Some works have been done to model the long-range dependencies of CNNs using image pyramids[16]and atrous convolution[17].Although these deep learning models could gain accurate segmentation masks,there are also varieties of challenges in placental tissue segmentation.Three major challenges to placental tissue segmentation in MR imaging are illustrated in Fig.1.
For placental tissue regions with discontinuous or large spans,as shown in Fig.1(a) and (b),learning longrange dependencies between the pixels corresponding to the mask is helpful to improve the performance of placental segmentation.For the small placental tissue area,as shown in Fig.1(c),more attention should be paid to local information.However,the placental tissue structures are irregular,complex,and multi-scale.Perceiving these placental tissue features with fixed regions could be inadequate.
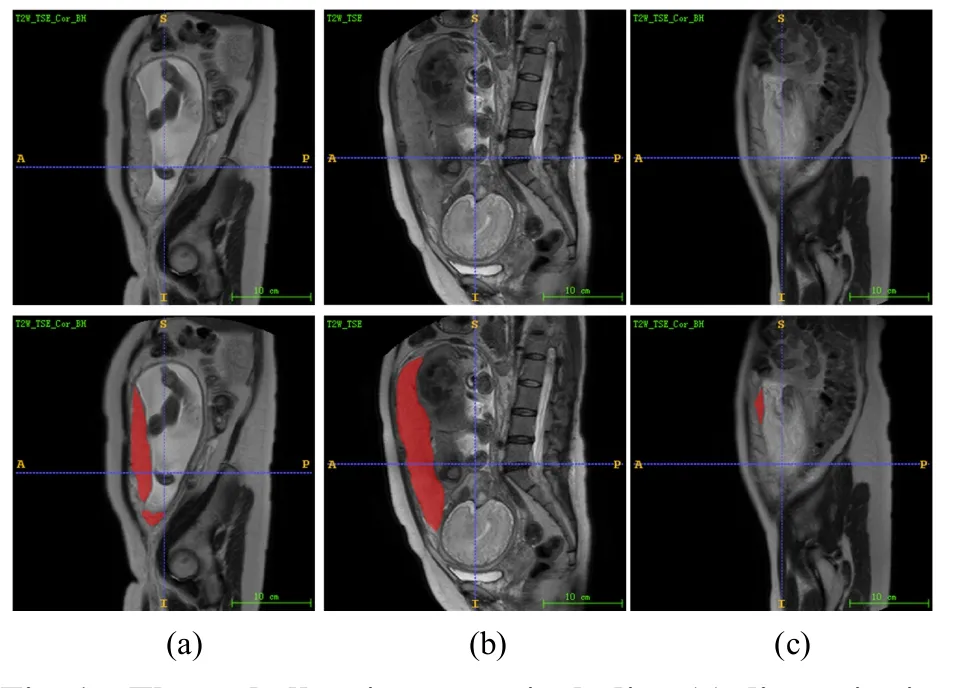
Fig.1 Three challenging cases,including (a) discontinuity of placental tissue area,(b) large span of placental tissue area,(c) small placental tissue area
Therefore,local and global information should be comprehensively considered in placental tissue segmentation,while most medical segmentation networks based on CNNs ignore the long-range contextual information.Transformer have recently revolutionized most of natural language processing(NLP) tasks like machine translation,question answering and document classification.The main reason for the success of transformers is their ability to learn long-range dependencies between the input tokens.Therefore,we believe that the combination of CNN and transformer is expected to achieve more accurate segmentation.
To address these challenges,this paper proposes TCGANet,which is based on a generative adversarial network.In this work,we aim to alleviate the aforementioned challenges from different perspectives.On the one hand,to better segment the placental tissues with different sizes and shapes,we made full use of global and local information.On the other hand,to accurately define the boundary of placental tissue,we used an adversarial network to monitor the generation network of placental tissue segmentation.The main contributions of this work can be summarized as follow:
(1) A deep network for automatic segmentation of placental tissue in MR images was constructed.Through the combination of the feature embedding module and skipping connection,the network embedded the highlevel features into the low-level features,alleviating the information loss caused by the traditional feature fusion method based on skip connection,and improving the accuracy of placental tissue segmentation.
(2) In the network architecture,a context feature extractor was introduced.This module can make full use of the ability of CNN to obtain local features and advantages of the transformer to learn long-term dependence,thus facilitating accurate segmentation of placental tissues with regional discontinuity and large size differences.
(3) Transformer and convolution neural network(CNN) were combined to form a generative adversarial network (TCGANet).Based on this architecture,TCGANet uses an adversarial network to supervise the generation network of placental tissue segmentation,thus improving the accuracy of placental edge localization.
The paper is organized as follows: Section 2 provides the related work.Section 3 describes the TCGANet network architecture proposed in this paper in detail.The experiment and results are introduced in section 4.Finally,the conclusive remarks and future work are given in sections 5-6.
2 Related Work
2.1 CNNs-based approaches
An end-to-end network based on CNN is widely used in image segmentation.Following the initial breakthrough made by FCN[18]in image segmentation,Li et al[19]designed a 3D multi-scale network for the segmentation of multimodal intervertebral disc MR images based on FCN.U-Net[9]is generally regarded as the baseline for medical image segmentation tasks,and various improvements have also been made for different segmentation tasks[20].Shahedi et al[21]proposed an improved U-Net for uterine and placental tissue segmentation in MR images.Despite the significant progress made by these methods,they are not sufficient to address the challenges of placental tissue segmentation.
2.2 GAN-based approaches
Generative adversarial networks (GAN) includes generator network and discriminator network[22].Luc et al[23]applied GAN to the field of semantic segmentation.GAN and its extensions solved many well-known challenging medical image problems[24].In the field of medical image segmentation,inspired by the classic GAN,SegAN was proposed by Xue et al[25]as an endto-end neural network for skin damage segmentation.Dai et al[26]proposed SCAN to segment lungs and hearts in chest X-ray images.Accurate segmentation results can also be obtained by using only limited training data.GAN-based methods are constantly being explored and improved,which provide the possibility to further improve the accuracy of segmentation.
2.3 Transformer-based approaches
Transformers[27]have become the model of choice in natural language processing (NLP).Enlightened by NLP successes,scholars began to explore the application of transformers in tasks in the field of computer vision,such as object detection in DETR[28].Vision transformer (ViT)[29]achieved excellent performance in image classification,and more than this,it laid the foundation for semantic segmentation.Valanarasu et al[15]designed a medical transformer(MedT) for medical image segmentation,which significantly improved the segmentation performance.In addition,Chen et al[30]used CNNs and transformer to build a more powerful hybrid encoder,confirming that the transformer’s strong long-range sequence modeling ability can make up for some of the deficiencies of CNNs.Many scholars have been combining transformer structure with convolution operation to achieve better performance[31-32]and exploring the performance of transformers in segmentation is one of current research trends.Several recent studies have introduced hybrid transformer-CNN models for MRI processing tasks such as reconstruction and synthesis that are highly related to the proposed architecture[33-34].
Therefore,this paper draws on the abovementioned existing state-of-the-art methods and proposes TCGANet for automatic placental tissue segmentation in MR images.
3 Materials and Methods
In this section,the proposed method and the related experimental setup and data preparation are described in detail here.
3.1 Dataset description and preprocessing
With the approval of the ethics committee,the data provided by the Affiliated Hospital of Medical School of Ningbo University and Ningbo Women and Children’s Hospital were used in this paper.The imaging equipment of the Affiliated Hospital of Medical School of Ningbo University is Ge signa twin speed 1.5T superconducting dual gradient magnetic resonance scanner with an 8-channel body phased array coil.The imaging equipment of Ningbo Women and Children’s Hospital is Philips Achieva Noval Dual 1.5T superconducting dual gradient magnetic resonance scanner,using a 16-channel body phased array coil.The scanned slice thickness is 4.5 mm,the slice spacing is 2 mm,and the matrix is 256×256.A total of 40 placental MR images cases were collected,of which 27 were from the Ningbo Women and Children’s Hospital and 13 were from the Affiliated Hospital of Medical School of Ningbo University.The placental tissue of each case is manually labeled by an experienced radiologist.According to the image quality,a total of 572 sagittal slices with T2-sequential containing placental tissues were selected.K-fold cross-validation can prevent overfitting problems caused by overly complex models and is a method that can improve the generalization ability of models.In order to maximize the training process of the model,improve the reliability of the test results and avoid over-fitting and under-fitting,this experiment uses the 5-fold cross-validation method.To avoid data leakage,all slices are divided into training and test sets by case.
Due to the scarcity of available data,data augmentation is critical for the analysis of medical images,and augmenting the dataset can also reduce the risk of overfitting.Standard data augmentation techniques are utilized,including image translation,rotation and scaling,vertical flipping,and horizontal flipping.
3.2 TCGANet architecture
The overall structure is shown in Fig.2,which is composed of the generator network (G) and discriminator network (D).

Fig.2 Architecture of proposed TCGANet,which includes generator network and discriminator network
During the training stage,G and D are trained alternately.When training the D,only the parameters of the D are updated,and the same scheme is applied to the G.After training,the segmentation results can be obtained from the model.
3.2.1 Generator network
The backbone of the generator network is constructed based on the basic U-Net,as shown in Fig.3.To mitigate the information loss caused by traditional skip connections,we are combining the feature embedding module and skip connections.The context extractor module is designed to improve the limitation that convolution can only obtain local features by capturing features of different scales and long-term context information.The details are as follows:
(1) Feature encoder module
In this paper,resnet[35]is chosen as the encoder.It was found that the pre-trained resnet[34]can achieve better performance through experiments.The first four feature extraction modules are retained,and each module is shown in Fig.3(a).The input of the first layer can be defined asH×W×C,H×Wis the input image size,Cis the number of channels.The output is a three-dimensional tensorh×w×c,h×wis the spatial dimension of the feature map,cis the number of channels of the feature map.Networks such as U-Net usually directly use skip connection to combine the low-level features obtained by the encoder with the upsampled high-level features.The fusion is as follows:
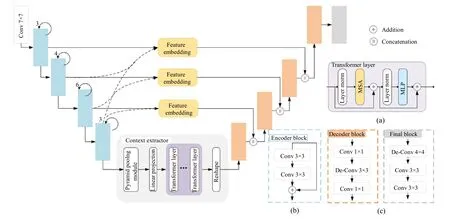
Fig.3 Architecture of proposed generator network,which consists of three parts: feature encoder module,context extractor module,and feature decoder module.Several key network blocks from different parts and some main notations are illustrated on the right

wherexiis the output feature of thei-th layer of the encoder.[·] denotes concatenation operation.This fusion method has drawbacks: First of all,due to significant differences in semantic information and resolution contained in low-level and high-level features,it is difficult to be fused directly.Secondly,a shallower network pays more attention to detailed information,including encoded clearer semantic boundaries.Instead,a deeper network pays more attention to semantic information and can describe the object more accurately.In short,a direct skip connection will cause some useful information loss.To solve the above problems,inspired by [36],this paper uses the feature embedding module and usesF(·) to express this operation.This feature fusion can be expressed as:

whereU(·) denotes the upsampling operation andndenotes the number of feature layers.Its detailed description is shown in Fig.4,where the “×” symbol represents the multiplication operation of elements.Embedding high-level features into low-level features are beneficial to supplementing low-level features.

Fig.4 The structure of the feature embedding module
As shown in Fig.3,the feature embedding module is applied to the first three layers of the encoder.
(2) Context extractor module
Context extractor module is designed,as shown in Fig.3,which is mainly composed of pyramid pooling module[16]andLlayers transformer.For placental tissues with large size differences,we used a pyramid pooling module to capture and aggregate features of different scales.For placental tissues with a large span,the transformer is introduced to capture long-term dependence using a self-attention mechanism.This module fully establishes the correlation of each pixel and integrates local features with global context information.
Context information is extracted from different perspectives and scales through the pyramid pooling module.Features of four different scales are merged,using the average pooling of sizes 1 ×1,2 ×2,3 ×3 and 6 ×6 .In this way,more spatial information can be obtained,which contributes to the segmentation of placental tissue with large differences in size.The output feature map can be expressed asf∈Rh×w×d.
Because the global information is beneficial to the segmentation of the placental tissue with a large area,the transformer is introduced to further capture the global information using self-attention mechanisms.The standard transformer expects a sequence as input.Thus,the feature mapF∈R h×w×dis divided into patches of sizeP×Pand flattened to obtain2N×Pmatrix,whereN=hw/P2is the input sequence length.Further mapping vectorized patches into a latentd-dimensional embedding space using a linear projection function.To encode the location information which is vital in placental tissue segmentation task,we add a 1D learnable position embeddingEpos∈R N×dto the patch embedding to retain positional information as follows:
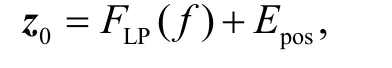
wherez0∈RN×drefers to the feature embeddings,andFLP(·) is the linear projection operation.The transformer encoder is composed ofLtransformer layers,each of them has a standard architecture as shown in Fig.3(b),which consists of a multihead selfattention (MSA) and multi-layer perceptron (MLP)blocks.Therefore the output of the ℓ-th(ℓ ∈[1,2,…,L])layer can be written as follows:

whereFLN(·) denotes the layer normalization operator andzℓis the output of ℓ-th transformer layer.MSA is composed ofmparallel self-attention (SA) heads,and the output is defined as:

whereWOrepresents the learnable weight matrices of different SA heads.SA is a parameterized function that learns the similarity between two elements in the input sequence (z) and their set of the query (q) and key (k) representations.SA is formulated as:

wherevdenotes the values in the input sequence anddh=d/mis a scaling factor.Finally,to fit the input dimension of the decoder,the sequence datazL∈RN×dis projected back to a standard 3D feature mapF∈Rh×w×dthrough the reshape operation.
(3) Feature decoder module
The feature decoder module is made up of four decoder blocks as shown in Fig.3(c),designed to recover high-resolution information and the feature map size from more abstract semantic features.The bottleneck structure is used to construct the decoder block.Each decoder block contains two pointwise convolutions to reduce the number of channels.Between the two pointwise convolutions,there is a layer of 3 ×3 deconvolution,which could learn a self-adaptive mapping to restore features with more detailed information.Ultimately,the feature decoder module outputs a mask of the same size as the original image.
3.2.2 Discriminator network
The edge definition of placental tissue is essential for placental tissue segmentation.The discriminative network can highlight the difference of edge distribution between segmented and real maps,enhance the perception of placental tissue edges,guide the segmentation network training,and further improve the accuracy of placental tissue boundary definition.The discriminator takes two input combinations,one set of original image and labeled image,and one set of original image and generator placenta segmentation image.The discriminator network distinguishes between ground truth and predicted mask,and provides a trainable loss function for the generator network,to supervise its training.Each block of the discriminator network includes 2 layers of convolution with a kernel size of 3 ×3,a stride of 1,and a padding of 1.The last layer of the discriminator network is activated using the sigmoid function.
3.3 Objective function
Assume thatxis an input image andyis the ground truth.G(x) is the segmentation result of generator network,D(x,y) are the probability that the input is ground truth,andD(x,G(x)) indicates the probability that the input is predicted mask.By using generator network to minimize the objective function,and discriminator network to maximize the objective function,the following function is optimized:

To better supervise the generator network and further improve the accuracy of placental tissue segmentation,a binary cross-entropy loss function is added to the generator network:

The final objective function is defined as:

4 Experiments and Results
4.1 Implementation details
The proposed method is based on the Windows 10 system and implemented by using Python 3.6 and Pytorch 1.7.1.The graphics card used is NVIDIA GeForce GTX 1080Ti GPU.Our experiments use a batch size of 8,use Adam optimization,the learning rate is 10-4,the number of layers of the transformer is set to 12,the embedding dim is 1 024,and the model is trained for 100 epochs.For all the comparison methods in this study,training and testing are carried out according to their papers and codes,and the other settings are the same as in this paper.
4.2 Evaluation metrics
In this paper,general indicators in the field of image segmentation are applied to measure the performance of different segmentation methods[37].In this study,True Positive (PT) is the correctly segmented placental tissue pixels,and False Positive (PF) is the wrongly segmented placental tissue pixels,also called over-segmented pixels.Similarly,True Negative (NT)means the correctly segmented non-placental tissue pixels,and False Negative (NF) is the pixels of placental tissue that are not segmented.Based on these definitions,the following evaluation indicators are introduced respectively.The mathematical expressions of the six metrics:MACC,MSEN,MSPE,MPPV,MDSC,andMJSCare presented below:

Accuracy (MACC) is the entire accuracy of segmentation of placental tissue pixels and nonplacental tissue pixels.Sensitivity (MSEN) is applied to evaluate the proportion of correctly segmented placental tissue pixels in the ground truth placental tissue pixels.Specificity (MSPE) aims to measure the ratio of how many non-placental tissue pixels from the non-placental tissue pixels ground truth are classified to be true.Positive predictive value (MPPV) indicates the proportion of placental tissue pixels in the prediction result that belong to the ground truth.Dice similarity coefficient(MDSC) is a commonly used evaluation index in medical image segmentation.Jaccard similarity coefficient (MJSC)is defined as the intersection of the segmentation result and ground truth divided by their union.
4.3 Results
In this section,the results of the ablation experiment are presented,and the qualitative and quantitative analysis results of the model proposed in this paper and the state-of-the-art methods are shown in detail.
4.3.1 Ablation study
To verify the effectiveness of each part of the proposed network in the task of segmenting the placental tissue,and to further discuss the contribution of each part,the following ablation experiments in this paper are performed.
We conducted the ablation study on placental tissue segmentation by evaluating three key components of TCGANet (discriminator,feature embedding module,and context extractor module).The detailed experimental results are presented in Table 1.

Table 1 Ablation study on different components of TCGANet
For ease of presentation,we signify discriminator,feature embedding module,and context extractor module as D,F,and C,respectively.As shown in Table 1,when TCGANet without (w/o) D,theMDSCis less than 80%,which shows that D effectively improves the overall segmentation performance.Similarly,F and C also contribute to placental tissue segmentation,because theMDSCof TCGANet w/o F,C is 82.56%.Compared with TCGANet,theMDSCof TCGANet w/o C dropped from 86.19% to 84.68%.C effectively utilizes global and local information,which shows that the designed C can help achieve better segmentation.TheMDSCof TCGANet w/o F is 85.03%,which proves that a suitable feature fusion strategy can further improve the segmentation performance of the placental tissue.It also means that correctly grasping more meaningful knowledge from different feature levels is helpful to the precise segmentation of placental tissue.
We further explore the effectiveness of each component of the context extractor module.The results are shown in Table 2.Both pyramid pooling module and transformer improve segmentation performance.

Table 2 Ablation study on different components of context extractor module
In Fig.5,we visualized the segmentation results and found that our proposed model was likely to obtain more continuous segmentation boundary maps,and could completely segment the placental region.In Fig.5(c),the result of segmentation without context extractor module is under segmentation,while in Fig.5(d),the result of segmentation without discriminator is poor boundary segmentation.The experimental results show that our method can segment the irregular placental tissue well.These visualizations further demonstrate the importance of gathering multi-scale context information and capturing long-term dependencies in placental tissue segmentation.

Fig.5 Segmentation results of different methods (a) the original images layered with ground truth labels,(b)-(d)segmentation results,(b) without context extractor module,(c) without discriminator,(d) proposed TCGANet
To verify the effect of transformers of different sizes on the performance of placental tissue segmentation,this paper conducts ablation experiments on different transformer scales.The scale of the transformer is determined by the level (L) of the transformer and the feature embedding dimension (d).As shown in Table 3,whend=1024 andL=12,the performance is the best.

Table 3 Ablation study on transformer
4.3.2 Comparison of state-of-the-art methods
From the perspective of quantitative analysis,various methods are compared with different segmentation evaluation indicators.Each evaluation indicator uses the mean and standard deviation to summarize the results,as shown in Table 4.In Table 4,the various indicators of the method in this paper are better than the existing medical image segmentation methods,MDSCis 0.903±0.093,MSENis 0.903±0.093,andMJSCis 0.778±0.169.TheMJSCof R2U-Net[10]is 0.664±0.258 slightly higher than that of U-Net[9],and other indexes are similar to U-Net.TheMSENandMDSCof Att-Unet[11]are 0.886±0.211 and 0.764±0.215,respectively,which are higher than those of U-Net,and it can be inferred that the attention-based network can improve the feature representation ability of placental tissue.

Table 4 Quantitative comparison with different methods in placental tissue segmentation
In addition,GAN,which uses U-Net as a generator,has achieved competitive performance in various indicators,theMPPVandMDSCare 0.829±0.179 and 0.747±0.188,respectively,which are slightly lower than the method in this paper.TheMPPVof TransUNet[30]is higher than the above networks,reaching 0.847±0.226.It shows that TransUNet can improve true positive by using the transformer for global modeling.The performance of our method in each metric was also compared with other methods using paired samplest-tests.The results indicated that most of the performance metrics of the proposed TCGANet were significantly better than the state-of-the-art method(P<0.05).
The qualitative analysis is performed,and the differences between these existing medical image segmentation methods for placental tissue segmentation tasks are provided in detail,as shown in Fig.6.
For the large placental tissue area,as shown in the first row of Fig.6,the results of R2U-Net are only partial placental tissue pixels,which may be related to its feature extraction strategy.Att-Unet and TransUNetare over-segmentation,and U-Net and our method can predict the placental tissue more accurately.In the small placental tissue area shown in the second row of Fig.6,U-Net,Att-Unet,GAN,and TransUNet did not completely predict the placental tissue.Only the segmentation result of our method is closest to the ground truth.It is also difficult for radiologists to distinguish these small placental tissues from other tissues on MR images,but the identification of these small placental tissues is also crucial for medical doctors to detect the condition of the fetus.For the placental tissue with irregular regional shapes,as shown in the third row of Fig.6,U-Net,Att-Unet,and R2U-Net all have over-segmentation.Over-segmentation will lead to higher false positives,which means that the shape of the placental tissue is missing in clinical applications.However,the method proposed in this paper can accurately segment the edge of the placental tissue.Fig.7 shows a visual comparison of the accuracy of edge localization in placental tissue segmentation by different methods.As can be seen,our results are much closer to the ground truth.This shows that the proposed method is able to capture more dependencies between pixels and thus better capture the contours of the placental tissue.
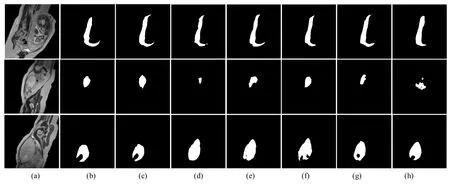
Fig.6 Qualitative comparison with different methods in placental tissue segmentation,(a) the original images,(b) the ground truth,(c)-(h) segmentation results using (c) proposed TCGANet,(d) U-Net,(e) Att-Unet,(f) R2U-Net,(g) GAN(U-Net+discriminator),(h) TransUNet

Fig.7 Segmentation results of the proposed TCGANet and other competing models,(a) the original images layered with ground truth labels,(b)-(g) segmentation results using (b) proposed TCGANet,(c) U-Net,(d) Att-Unet,(e) R2U-Net,(f) GAN(U-Net+discriminator),(g) TransUNet
5 Discussion
Automated placental tissue segmentation plays an important role in clinical diagnosis,as providing physicians with precise placental tissue localization and clear contour information can assist physicians in their diagnosis.Since placental tissue varies greatly in size in different slices and has a high similarity to surrounding tissue,there are still difficulties in segmenting placental tissue with large span differences and defining edges.Most of the existing medical segmentation models use an encoder-decoder structure,but this traditional skip connection method is prone to information loss.Generally,the shallow features contain more semantic information,such as the structure and boundary information of the placental tissue.On the contrary,the deep features contain more detailed information and are more capable to accurately describe the placental tissue.Therefore,these features should be fully exploited,and a feature embedding module is introduced in this paper.
Considering that the shortcoming of convolution is that it cannot capture long-distance pixel dependencies,it is not conducive to the segmentation of placental tissue with large spans.We designed the context extractor module,which uses the transformer to capture more global information to make up for the insufficiency of CNN and alleviate the limited receptive field of traditional segmentation networks.In addition,adversarial networks are used to supervise the generative network to improve the accuracy of placental edge definition.
Our proposed method still has some limitations.Fully supervised models rely on high-quality annotation data,so this method is suitable as an auxiliary tool for manual annotation,which can reduce the time of manual delineation,but cannot completely replace manual segmentation.
6 Conclusion
In this paper,a deep learning model for placental tissue segmentation in MR images was proposed to help radiologists further medical image-based diagnosis,anatomical structure modelling and surgical planning.
To validate the effectiveness of the proposed method,various ablation experiments were conducted and the results of the comparison experiments showed that the method can achieve better performance in the task of placental tissue segmentation in MR images,outperforming existing medical segmentation networks.In future work,we will explore the extension of the proposed network to other tasks of medical image segmentation.
