Single-molecular methodologies for the physical biology of protein machines
2022-12-28ShuangWang王爽YingLu陆颖andMingLi李明
Shuang Wang(王爽), Ying Lu(陆颖), and Ming Li(李明)
Beijing National Laboratory for Condensed Matter Physics,Institute of Physics,Chinese Academy of Sciences,Beijing 100190,China
Keywords: physical biology,single-molecules,research object reconstruction,precise measurements
1. Introduction
Physical biology is an interdisciplinary field that bridges biology with physical sciences and engineering. It focuses on research in which quantitative approaches lead to new insights into biological systems at all scales of space and time,and all levels of organizational complexity. Physical biology makes use of physical methodologies to gain quantitative understanding of dynamics of biological processes,often engaging reconstructed objects to avoid interference from unnecessary complications. It is enlightening to retrospect the early history of physics. Galileo revolutionized basic scientific principles by measuring balls rolling down ramps, along level tracks, and then up ramps (Fig. 1(a)). He discovered that a ball rolling down an incline and onto a horizontal surface would roll indefinitely.This property of a ball to roll on forever without changing speed along the level track is inertia. Galileo then used the rolling balls on inclined planes to develop his understanding of acceleration(Fig.1(b)). He did not have proper timing devices to measure the high speeds of free fall and used an inclined plane to lower the acceleration. The ball rolling down the plane strikes bells,allowing him to record the time taken to reach each bell. This allowed him to observe that the distancedcovered by the ball rolling down the inclined plane is proportional to the square of the timet,i.e.,d=(1/2)at2,whereais the acceleration of the system. Borrowing the concept of particles that had shaped the early development of physics,the physical biology of single molecules focuses on the dynamics of individual biological molecules that function as the basic elements in biology. Given the complexity of living systems in general, and cells in particular, it is hard to investigate the biomoleculesin situ, although progresses are being made in this direction. In physical biology, it is also desirable to reconstruct research objects in “clean” environments to avoid unnecessary interferences. This is much like to observe that a feather in a vacuum drop tube falls as fast as an iron ball,while the same feather falls much slower in air due to the resistance of air. Figure 1(c)gives an example of physical measurements on a molecular motor called myosin that consumes and converts chemical energy to its mechanical movements along an actin filament. A single actin filament was firstly assembledin vitroand held by an optical trap. A microbead connected to a myosin molecule was then monitored directly by an optical microscopy to measure its speed as a function of force,ATP concentration and temperature, providing insights into the mechanism of mechanochemical energy transduction.[1]

Fig. 1. Galileo’s rolling ball experiment (a) and inclined plane experiment (b). In physical biology, often an object is also reconstructed and investigated in the absence of unnecessary interferences. Depicted in (c)is a myosin moving along an actin filament held by an optical trap.
Physicists often give various stimuli to a system of interest and monitor its responses to decipher its properties.Following the same idea, researchers in the field of singlemolecule physical biology take advantages of their skills in mechanism, electricity, magnetism, optics and so on to study biomolecules. They not only observe the motions and functions of the molecules passively, but also manipulate the molecules to reveal their dynamic responses to stimuli. For instance, application of a stretching force can slow down the folding of a protein or even convert it to equilibrium, allowing one to measure precisely the folding dynamics.[2]The unfolding dynamics and energy landscape can be elucidated by measuring force-induced changes of the end-to-end distance of the protein[3–6](Fig. 2(a)). A more interesting example is about the organization of the so-called 30 nm chromatin. A segment of chromatin can be firstly producedin vitroand then connected to two long double-stranded DNA that function as handles for manipulation (Fig. 2(b)). A tension can then be exerted to the reconstituted structure to investigate the assembly by repetitively distorting the structure and then allowing it to reassemble.[7]
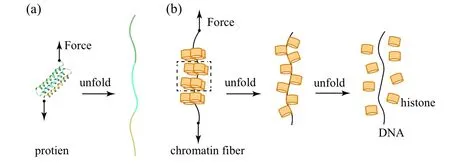
Fig.2. Investigation of dynamics of biological systems under force stimuli and manipulation. (a)Force-induced folding of a natural protein. (b)Disassembly of a chromatin fiber under force.
Protein machines participate in almost all biological processes that couple energy consumption with nanoscale conformational changes. Many proteins interact with nucleic acids to decode genetic information and work cooperatively to maintain genome integrity. On the other hand, membraneinteracting proteins are essential for cells to exchange matter,energy and information with their extracellular environment.Studying one protein at a time can provide us with extraordinarily clear and often surprising views of these molecules in action. In this review, we summarize recent advances in single-molecule physical biology in two aspects:nucleic acidinteracting proteins and membrane-interacting proteins.
2. Precise measurements in physical biology
Quantitative measurements lie in the core of physical biology. Atomic-resolution structures of biomolecules are the basis of all life sciences.A tremendous number of structures of biomolecules and their complexes have been solved thanks to the great efforts of structural biologists.[8–11]In physical biology,researchers are more concerned with motions and conformational transitions of individual biomolecules in various free energy landscapes.[7,12–14]Energy transduction and the correlated dynamics lie at the root of physical biology. What to measure and how to measure are essential questions in physical biology.
2.1. Single-molecular measurements of nucleic acid–protein interactions
Proteins that interact with nucleic acids are involved in many aspects of nucleic acid transactions such as DNA condensation,replication,transcription and so on.[10,15–21]These processes engage changes in contour lengths of DNA, separation of two strands of a DNA, and elongation of a singlestranded DNA or RNA. Researchers often construct special DNA structures as the substrates of the proteins of interest.For instance, a DNA hairpin can be used as the substrate of a helicase which unwinds a duplex DNA during DNA replication (Fig. 3(a)). F¨orster resonance energy transfer (FRET)has become a powerful and popular technique in recording conformational trajectories of single biomolecules.[22–24]Detailed description on single-molecule FRET techniques can be referred to Refs. [25–27]. When a pair of dyes are attached to the two overhangs of the hairpin, the distance between the two dyes increases when the helicase unwinds the hairpin,and the decrease in efficiency of FRET between the two dyes as a function of time yields the unwinding rate of the helicase.
FRET is based on the dipole–dipole interaction, whose sensitivity is of angstrom scale, but its measurable range is usually limited to 2–8 nm.[22,23]The reconstructed DNA structures can be tethered to single-molecule manipulation tools such as magnetic traps or optical traps to overcome the limitation.[28–31]The single-molecule manipulation tools can also be used to study force-induced effects of DNA enzymes by measuring the end-to-end distance as a function of time(Fig. 3(b)). Detailed description on single-molecule manipulation techniques can be referred to Refs. [29,32–34]. Some nucleic acid-interacting proteins such as RNA polymerase and topoisomerase change the topology of DNA rather than its contour length.[35]During transcription, an RNA polymerase generates a transcription bubble in which genetic information is exposed and transmitted to an RNA sequence. Taking advantage of the ability of rotational manipulation of magnetic traps, the reconstructed linear DNA can be supercoiled by rotating the magnets,with which transcriptional dynamics of an RNA polymerase can be monitored via the topological changes of the DNA[36–39](Fig. 3(c)). The translocation dynamics of an RNA polymerase can be studied by firstly reconstructing an RNA polymerase on DNA and then monitoring the transcription elongation via end-to-end distance changes under optical trap with a high tempo-spatial resolution[13,40,41](Fig. 3(d)). Additionally, various protein machineries interacting with RNA polymerases can be studied via fluorescence method. In such studies, the RNA polymerases are fluorescently labeled and their interactions can be imaged by colocalization of fluorescent spots with different colors whose durations imply kinetics of these interactions[42–44](Fig.3(e)).
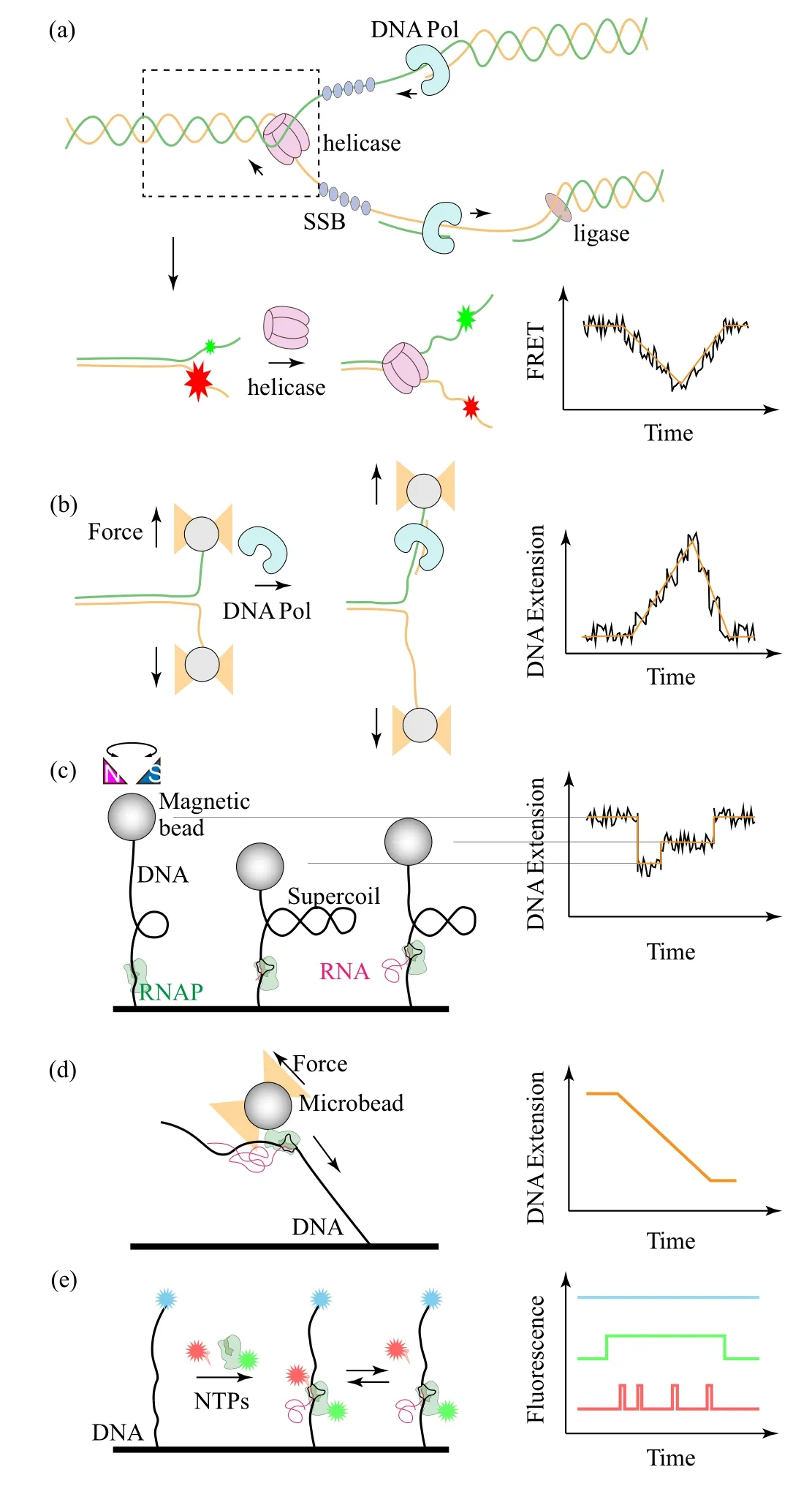
Fig.3. Examples of quantitative measurements in single-molecule physical biology. (a)Scheme of the DNA replication process at a DNA fork(upper panel) which can be reconstructed for studying DNA unwinding by DNA helicases via single-molecule FRET (lower panel). (b) A similar structure can be connected via long handles to an optical trap to study polymerases.(c)Torsional manipulation of a DNA and measurements of RNA polymerase dynamics via single-molecule magnetic trap. (d) Transcription dynamics can also be studied via optical trap by monitoring the end-to-end distance changed by RNA polymerase or(e)via single-molecule fluorescence colocalization of various components during transcription.
2.2. Single-molecular measurements of protein–membrane interactions
Cell membranes play important roles in many metabolic processes. They not only serve as a naturally protective barrier for the cell, but also allow cells to cooperatively form organisms of higher orders with various functions.[45]A variety of proteins are embedded in cell membranes, involved in various cellular functions such as cell adhesion, ion conductivity and signaling.[46–48]Key parameters to characterize the protein–membrane interactions include positions and orientations of the protein inside the membrane, conformational changes upon activation,and the number of proteins in a cluster formed with other proteins or with themselves.
Proteins are extremely crowded in cell membranes. A protein of interest is often(fluorescently)labeled so that its dynamics can be distinguished from others(Fig.4(a)). One can engineer genes of the proteins of interest by flanking a gene that can co-express a fluorescent protein to study the dynamics of the proteins in live cells.[49,50]Different proteins can be distinguished and simultaneously tracked via their fluorescence in the cells,[51]reflecting their assembly and organization in response to the environmental stimuli.[52]Interactions of cells with their environments engage interactions between membrane proteins, such as antigens and receptors which are crucial for target recognition and immune cell activation.[53–55]One can modify a ligand to biotinylate it for tethering a streptavidin coated magnetic bead and let them to interact with the receptors on the cell surfaces. An force can be applied by adding a magnet,and the value of force of the antigen/receptor interaction can be precisely characterized[56](Fig. 4(a)). By using a pair of micropipettes to separately manipulate a cell and a microsphere on which proteins of interest are coated,and then separating the cell and the microsphere in fine steps,a slowly increasing force will rupture the interaction, quantifying the strength of the interactions[57](Fig. 4(b)). Taking the feature that a short DNA hairpin can be opened at a force around 10 pN (1 pN = 10−12N), researchers reconstructed DNA hairpins with labeled fluorophores on the overhangs to construct tension sensors[58](Fig. 4(c)). These tension sensors can quantitate the interaction between the antigens and receptors on the cell. The involved force regulation is highly relevant to immune cell activation and target recognition.
In many situations, however, direct observation of the proteins in living cells does not provide real information about the intrinsic dynamics due to unnecessary interference from other biomolecules in the cellular environment. As a result,materials mimicking cell membranes would have many advantages,and have been desired for a long time.[59–61]“Clean systems” in the view of physicists such solid supported lipid membranes can be produced.Such two-dimensional platforms are compatible with numerous single-molecule techniques for manipulation or fluorescence imaging.[62–65]To mimic natural cell membranes, many different artificial lipid vesicles spanning a large diameter range from 20 nm to 2µm have been fabricated using different methods.[66]Proteins of interest can be reconstituted into the membranes to mimic their natural environment,where a controllable number of proteins which interact with the membrane proteins of interest can also be engaged(Fig.4(d)).Such artificial lipid vesicles were exploited as platforms for reconstituting ion channels and transporters and used for single-molecule FRET measurements,opening a new door for investigating this important class of proteins in their native membrane environment.[67]On the other hand, extracellular vesicles spontaneously secreted by cells or giant plasma membrane vesicles (GPMV) released by cells upon chemical treatments are also model systems for membrane proteins[68](Fig. 4(e)). These objects are more like cells than man-made vesicles. Observations on vesicles can therefore be doublechecked on them to avoid artificial effects. Finally, if necessary, the observations can be confirmed on real cells to see if the dynamics and functions of the proteins of interest would be modified by other proteins, by which corporation of proteins could be unveiled.[69]
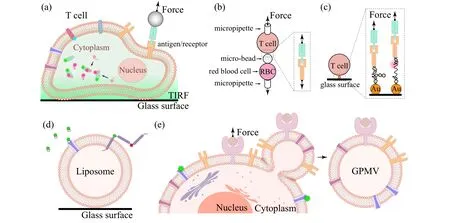
Fig. 4. (a) A cell with fluorescently labeled proteins and with a receptor connected to a magnetic trap through an antibody. (b) Cells hold by micropipettes are used as sensors for cell–cell interactions. (c) DNA hairpins connecting cell surface and glass surface can be used as tension sensors to study cell–matrix interactions. (d) A man-made giant unilamellar vesicle (GUV) with reconstituted membrane proteins. (e) A giant plasma membrane vesicle(GPMV)released by cells. Both GUV and GPMV are model systems for studying protein–membrane interactions.
2.3. Other single-molecular measurement methods
In addition to optical measurements,magnetic resonance is also potentially useful to measure the composition and structure of molecules containing spins via detecting their absorbance in alternating magnetic field. This method has been recently developed for precise measurements of single biological molecules. Nanodiamond with a nitrogen vacancy combined with a recently developed atomic-sized magnetic field[70,71]allows detection of the magnetic resonance spectrum of single proteins under ambient conditions or in aqueous solution.[72,73]Ionic pumps transduce materials and related information between inside and outside of a cell. An ionic pump can be reconstructed onto lipid bilayers whose dynamics can be measured via current or fluorescent signals.[74–76]Such phenomena have recently resulted in nanopore assays where a nanopore protein is reconstructed on an artificial membrane allowing ionic current measurements when different sized molecules pass through the channel. The ionic current detection can be converted from fluorescent signals by using an indicator dye Fluo-8 and thus allows a high throughput optical recording at a density of∼104nanopores per mm2.[77,78]Other membrane proteins are continuously found as nanopore candidates exhibiting high sensitivities for single-molecule detections because of its two sensing spots.[79]
3. Nucleic acids transactions
Proteins engage in all nucleic acid transactions, such as replication, transcription and so on.[80,81]How a DNA polymer is organized by histone proteins in assistance with related chaperons is a key question that must be answered at first. Due to the highly dynamic and heterogeneous properties of chromatin fibers,it is technically challenging to investigate the packing density and structural dynamics of chromatin fibers.[7]By using magnetic tweezers, Liet al.was able to manipulate chromatin fiber and trace its extension during the folding and unfolding process under tension to investigate the dynamic structural transition at single-molecule level. They investigated the hierarchical organization and structural dynamics of chromatin fibers reconstitutedin vitrowith purified histones and man-made DNA template in the presence of H1 chaperones (Fig. 5(a)). They measured the force extension curves of the chromatin fibers and observed structural transitions near 3 pN, which could be attributed to the disruption of nucleosome–nucleosome interactions in the higherorder chromatins because the nucleosomes remain intact at such low forces(Fig.5(b)). The details of the folding and unfolding dynamics of the chromatin fiber at∼3.5 pN indicate that tetra-nucleosome units exist as a stable structural intermediate of the 30-nm chromatin fiber and further unfold to a more extended “beads-on-a-string” conformation by disrupting the nucleosome–nucleosome interactions[7](Fig.5(c)).
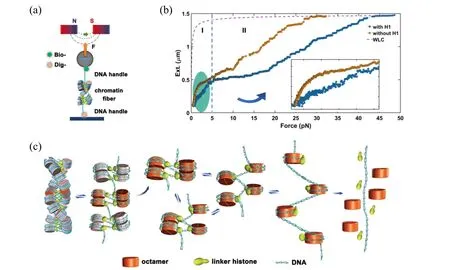
Fig. 5. (a) Schematic representation of chromatin fiber studied with magnetic tweezers. (b) Force–extension curves of the chromatin fibers.(c)Model of the dynamic organization of a chromatin fiber. Reproduced with permission.[7]
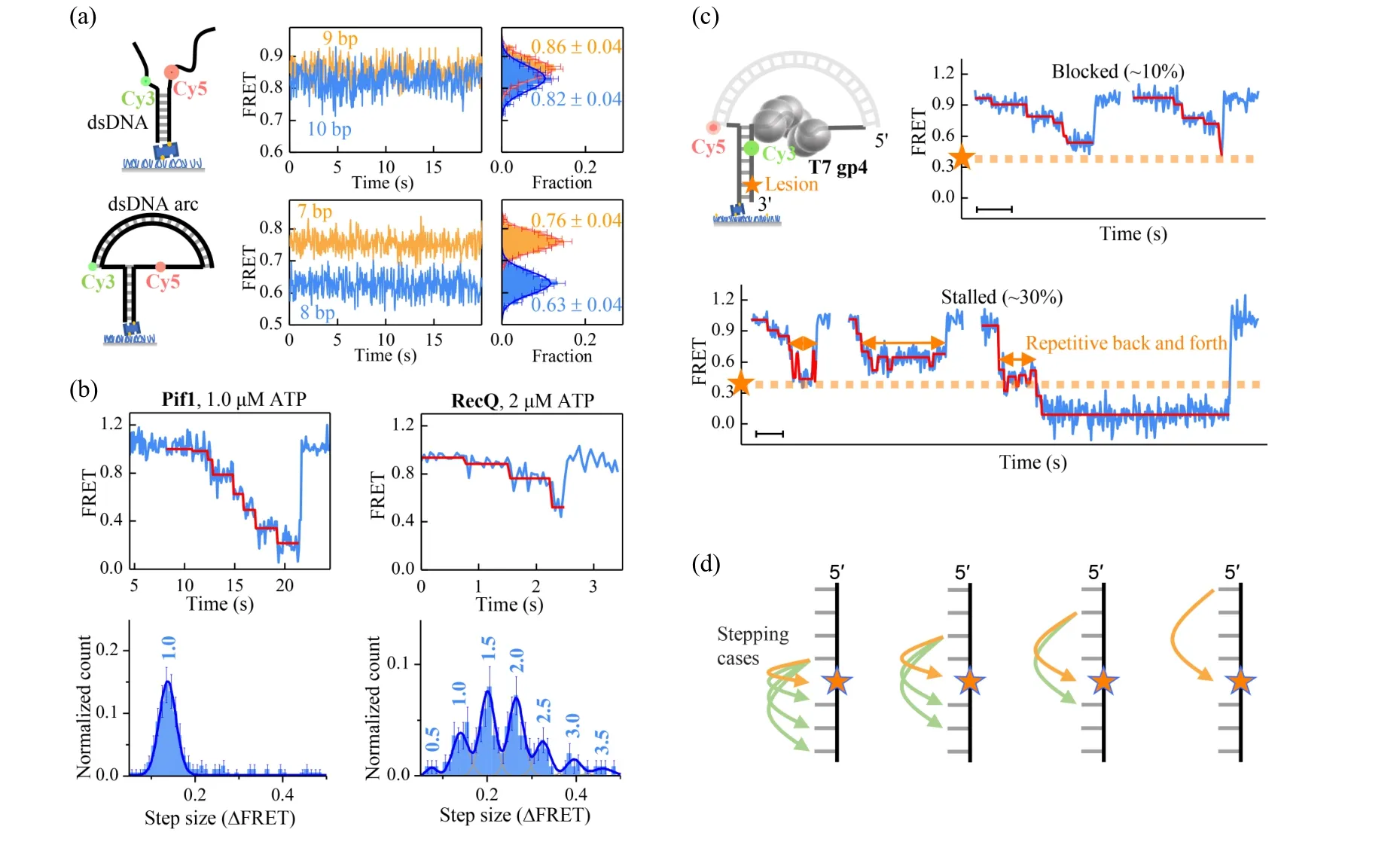
Fig.6. (a)The FRET efficiency changes in a nanotensioner is significantly enlarged when the Cy3–Cy5 distance is increased by 1 bp. (b)Stepping of two non-ring helicases in the assay with nanotensioner(top panel). Histograms of step sizes(∆FRET)in the FRET range of[0.2,0.8](bottom panel). The step sizes in units of bp are indicated on top of each peak. Reproduced with permission.[83] (c) DNA unwinding patterns of the ring-shaped T7 gp4 helicase on a DNA containing an abasic lesion. The dashed orange lines represent the position of the lesion. Scale bars, 1 s.(d) Nonuniform stepping of the ring-shaped gp4 helicase explains the unwinding patterns. If DNA contains an abasic lesion, gp4 would not be affected in 6 cases(green arrows),while it would be affected in another 4 cases(orange arrows). Reproduced with permission.[84]
Single-molecular FRET can detect angstrom scale changes in distances, which is the length scale of 1 bp in dsDNA.[23]However, with forked DNA substrates, the resolution is often limited to 2–3 bp because thermal fluctuations of the ssDNA overhangs disturb the inter-dye distances significantly[82](top panel in Fig. 6(a)). By using a duplex DNA as a nanotensioner to stretch the overhangs, Linet al.was able to exert force∼6 pN on the fork and resolve 1-bp length difference with FRET[83](bottom panel in Fig. 6(a)).Using the DNA nanotensioner, they investigated the stepping of yeast helicase Pif1 and theE.colihelicase RecQ at low ATP concentrations(Fig.6(b)). The step size distribution of Pif1 is narrow and has a single peak at 1.0 bp,while the step size distribution of RecQ has several peaks at 0.5 bp, 1.0 bp, 1,5 bp,2.0 bp, 2.5 bp, and 3.0 bp. Although significantly different,the stepping of the two non-ring helicases can be generally explained by a simple model,i.e.,a helicase breaks 1 bp with the energy of 1 ATP hydrolysis and may sequester or release the nascent nucleotides randomly[83](Fig.6(c)). Maet al.studied the stepping of the ring-shaped T7 gp4 helicase when encountering an abasic lesion.[84]They found that the T7 helicase may be blocked(∼10%)or stalled(∼30%)for a while at the lesion,or unblocked(∼60%)by the lesion. In the stalled pattern(Fig.6(c)),the helicase may move back and forth repetitively before the lesion. They also found that the stepping of the ring-shaped T7 helicase is randomly uneven from 1 nt to 4 nt, which explains the random block of the helicase in the context of hand-over-hand mechanism. In that work,they also predicted an intermediate conformation of the DNA-helicase complex.
4. Dynamics of single molecules in live cells
Biologists desire to watch biomolecules in live cells. A protein performs various functions synergistically with many other proteins. Detections of single molecules in live cells provide more realistic results and a more direct view of the dynamics of biological molecules.[85–87]The single-molecule tracking(SMT)in living cells has greatly promoted our understanding of intracellular transport dynamics. Different from simple liquid environments,the intracellular transport is more complex due to the influence of active fluctuations and diverse cyto-architectures. With the single-molecule fluorescence imaging and the trajectory reconstruction, the SMT method provides the highly spatiotemporal information (millisecond,nanometer)of individual molecules,which is generally hidden by ensemble-averaged methods before. Recently, by labelingβ1-integrin proteins with quantum dots,the on-membrane diffusion dynamics of integrins during the epithelial to mesenchymal transition(EMT)is first revealed[88](Fig.7(a)).The enhanced integrin diffusion may represent a new physical hallmark for detecting EMT cells. Liet al.introduced a method based on SMT to rapidly map intracellular diffusion, revealing heterogeneous and compartmentalized diffusion resulting from restriction of the endoplasmic reticulum[89](Fig. 7(b)).In addition to the diffusion, directed motion driven by motor proteins along cytoskeletons is another important way for intracellular transport(Fig.7(c)). For example,endocytic transport of the EGFR and NGFR complex in real time has been revealed.[90–92]A more interesting study showed that the intracellular transport of endocytic vesicles is accelerated in the early apoptosis cells as a result of the elevated intracellular ATP concentrations, which is found to be important in the apoptotic progression.[93]Recently, with the development of 3D SMT technique, it was newly discovered that intracellular diffusion is anisotropic quasi-2D rather than isotropic 3D in adherent cells.[94,95]As for the endocytic vesicles, the intracellular transport changes from thermal-dominated 3Dconstrained motion to active-dominated quasi-2D motion with varying timescale,and inside the vesicles,it was observed that endocytic nanoparticles make diffusive motions on their inner membranes.[96]

Fig.7. Intracellular transport dynamics revealed by single-molecule tracking. (a)On-membrane diffusion of integrins labeled with quantum dots.The β1-integrins exhibit significantly enhanced diffusion during EMT.Reproduced with permission.[88](b)Intracellular diffusion of single quantum dots(red)confined by the endoplasmic reticulum(green). With the diffusion trajectories,the heterogenous and compartmentalized diffusion map of a cell is generated. Reprinted with permission.[89] (c) Intracellular transport of endocytic vesicles contains directed motion and nondirected diffusive motion. In early apoptotic cells,the velocity and run length of the directed motion increase by over 30%. Reprinted with permission.[93]
To fully uncover the working mechanisms of the membrane-associated biomolecules, it is optimal to develop high-precision single-molecule methods to monitor their conformational changes on live cell membranes. Houet al.developed a FRET-based single molecule fluorescent method named QueenFRET (quenchers in extracellular environment FRET)[69]to probe the movement of the fluorescence-labeled site of the biomolecule in the direction normal to live cell membrane with sub-nanometer precision. QueenFRET takes the principle of FRET from one donor to multiple acceptors(Fig. 8(a)). Biocompatible quenchers are added to the extracellular culture medium to serve as FRET acceptors. As a result,the movements of the labeled site in normal direction are visualized by the intensity changes of the donor fluorophore induced by the changes of the FRET efficiency(Fig.8(b)).[97]The application of QueenFRET has uncovered the motion patterns of human host defensive peptide LL-37. The LL-37 oligomers are found to be dynamic on live cell membrane,in which the N-terminal of LL-37 switches among four different semi-stable depths in the lipid bilayer (Figs. 8(c) and 8(d)).Another example is the mixed lineage kinase domain-like protein (MLKL), which is a key protein in necroptosis.[98]The combination of QueenFRET and fluorescence lifetime imaging (FLIM) clearly showed that the short form (residues 2-123) MLKL inserted deeply in the hydrophobic core of the cell membrane,whereas that of the long form(residues 2-154)of MLKL tended to stay at the intracellular surface of the cell membrane.

Fig.8. Study of dynamics of membrane proteins with the QueenFRET technique. (a) Principle of QueenFRET. (b) Distance dependence of the donor(rhodamine-B here) intensity in different concentrations of the quencher(blue dextran) outside the cell. Reprinted with permission.[97] (c) and(d)Dynamics of the N-terminus of oligomeric LL-37 molecules. Reprinted with permission.[69]
5. Perspective
Single-molecule physical biology is a relatively new frontier and has been undergoing explosive growth. It makes good use of numerous single-molecule tools and gained remarkable new findings in biological processes. Single-molecule physical biology often involves reconstructed “clean systems” to investigate the mechanisms of life processes. Normally, the research objects are specifically reconstructed with purified nucleic acids or protein machineries. Obviously,complicated biological systems which really function in live cells will be the next focus of single-molecule physical biology.In vitroreconstruction of these complicated objects is the primary barrier,challenging the current techniques in both biophysics and biochemistry. Improvements on resolution of current tools or developments of new tools capable of detecting extra physical or biological properties are badly required to visit the multiple aspects of the biological systems. Integrated uses of singlemolecule tools have been accomplished showing remarkable insights into the dynamics and mechanisms of complicated biological systems. Applications of several tools to gain different information about the same research object will allow multiple physical views of the same biological system. Integration of multiple single-molecule tools or incorporating other tools into the single-molecule tools are highly encouraged. Efficient extraction of raw data and coupling of these data with theoretical calculations or new mathematical models will further improve our quantitative understanding of the biological systems.Computer modeling is hence an indispensable supplement to physical measurements of single molecules in physical biology.[99–104]
Acknowledgements
This work is supported by the National Key Research and Development Program of China (Grant No. 2019YFA0709304), the National Natural Science Foundation of China (Grant Nos. 12090051 and 12022409), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB37000000), and the Youth Innovation Promotion Association of CAS (Grant Nos. 2021009 and Y2021003). We sincerely apologize to the authors whose work could not be included because of the scope of this review. We thank Prof. Wei Li (Institute of Physics, Chinese Academy of Sciences),Prof. Hui Li(Beijing Normal University),Dr. Jianbing Ma(Institute of Physics,Chinese Academy of Sciences) and Dr. Dongfei Ma (Songshan Lake Materials Laboratory)for their kind discussion and figure preparation.
猜你喜欢
杂志排行
Chinese Physics B的其它文章
- Fault-tolerant finite-time dynamical consensus of double-integrator multi-agent systems with partial agents subject to synchronous self-sensing function failure
- Nano Ag-enhanced photoelectric conversion efficiency in all-inorganic,hole-transporting-layer-free CsPbIBr2 perovskite solar cells
- Low-voltage soft robots based on carbon nanotube/polymer electrothermal composites
- Parkinsonian oscillations and their suppression by closed-loop deep brain stimulation based on fuzzy concept
- Temperature dependence of spin pumping in YIG/NiO(x)/W multilayer
- Interface effect on superlattice quality and optical properties of InAs/GaSb type-II superlattices grown by molecular beam epitaxy
