Halophyte Vegetation Influences Soil Microbial Community of Coastal Salt Marsh
2022-12-27GUChenSHIJiyanRUIJianliangYUYanmingHUANGWeibinLUZhinaiCHENYaoCHENXiaojunDONGShudiHUZhijunandYEChenghua
GU Chen, SHI Jiyan, RUI Jianliang, YU Yanming, HUANG Weibin, LU Zhinai,CHEN Yao, CHEN Xiaojun, DONG Shudi, HU Zhijun, and YE Chenghua
1) Power China Huadong Engineering Corporation Limited, Hangzhou 311122, China
2) MOE Key Laboratory of Environmental Remediation and Ecosystem Health, and Institute of Soil and Water Resources and Environmental Science, College of Environmental and Resource Sciences, Zhejiang University, Hangzhou 310058, China
Abstract Coastal wetlands are the most productive ecosystems worldwide and can provide important ecosystem services, yet the characteristics of microbial community within these systems remain poorly understood. Microbial community of salt marsh vegetation and the associated soil physio-chemical properties were investigated in this study. Three typical Suaeda australis, Phragmites australis, Spartina alterniflora wetlands, and non-vegetated bare mudflats in the Zhoushan Islands were studied to advance the understanding of the characteristics of soil bacterial communities in coastal wetlands. Results showed that the bare mudflats exhibited high pH value and soil moisture content compared with the vegetated samples. In different vegetation types, the organic matter content, total nitrogen, and total potassium content decreased in the order: S. alterniflora wetland > P. australis wetland > S. australis wetland, and there was no obvious difference in total phosphorous content. The halophytes could decrease soil salinity compared with bare mudflats. Proteobacteria, Nitrospinae, Bacteroidetes, Acidobacteria, and Nitrospirae were the predominant level across all samples. Functional prediction showed that SPA-covered soil might play vital roles in sulphur cycling, while SUA and PHR covered soils were involved in nitrogen cycling. This study could provide the first insight into the microbial community of this study area and contribute to a better understanding of vegetation microbiota and bioremediation in coastal wetland ecosystem.
Key words illumina sequencing; salt marsh; vegetation type; soil bacterial community; function prediction
1 Introduction
The coastal wetland is a transition zone between ocean and land, possessing the dual environmental function(Huaet al., 2017). However, the increased human activities along coastlines and sea-level rise have led to the loss and degradation of coastal wetlands (Deeganet al., 2012;Tianet al., 2016). Coastal salt marshes are intertidal wetland ecosystems that provide habitat to halophytes, and it is ecologically and economically vulnerable (Chaudharyet al., 2018). Halophytes are representative vegetation of salt marshes with special morphological and physiological features in adaptation to saline environments (Glennet al., 1999).
It had been emphasized by many studies the importance of the bacteria in the soil of wetlands in regulating the biogeochemical processes of coastal ecosystems (Zhanget al., 2019a; Chiet al., 2020). Soil bacteria play vital roles in the recycling and transformation of major nutrients and maintain ecosystem stabilities (Huet al., 2016).It has been demonstrated that the soil property and the halophyte species exert important effect on soil bacterial community (Chaudharyet al., 2015). For instance, salinity, dissolved, pH, nutrients, and contaminants have been confirmed to directly shape the structure and composition of bacterial communities in coastal wetlands (Guoet al.,2017; Xieet al., 2017; Wanget al., 2018a). The microbial community composition and diversity varied under the vegetated wetland soils (such asPhragmites australis)(Chiet al., 2020). Moreover, salt marsh sediments are sinks for various anthropogenic pollutants (Reboreda and Caçador, 2007). Once contaminants enter the salt marsh,they become distributed in the sediments and plants. The ubiquitous occurrence of microorganisms in salt marsh sediments may play an important role in the degradation of pollutants.
Zhoushan is the largest archipelago in China. Zhoushan Island is located in the southern flank of Yangtze River Estuary and outer edge of Hangzhou Bay. The coastline type is mainly composed of tidal flat wetlands.Spartina alterniflora(smooth cordgrass), an invasive species,which experienced a rapid expansion over Chinese coasts(Zhouet al., 2009).S. alterniflorais one of the local representative species in the Zhoushan Island with an area of 83.07 hm2(Luet al., 2013; Liet al., 2020). Moreover, the secondary indigenous salt marsh plant communities in this region dominated bySuaeda australisandPhragmites australis. Other plant species, such asKandeliacandel, has sporadic distribution in the tidal flat, which has been introduced in the Zhoushan Island since 2020.Although many reports on the distribution of pollutions in coastal seawater and sediment of Zhoushan Island (Wanget al., 2016; Zhaiet al., 2021), relatively less effort is directed to understand the characteristics of microbial community. Here, we hypothesized that bacterial community composition is varied distinctly owing to the differences of soil physiochemical properties and vegetation types compared with non-vegetated mudflats. The heterogeneity of different wetlands might shape the microbial community and further lead to changes in metabolic functions.To test the hypothesis, high-throughput sequencing technology was applied to analyse the genetic data of wetland soils.
Therefore, this study aims to: i)investigate the structure and diversity of bacterial community under vegetated wetland soils and bare mudflat areas; ii)predict the function profiles of bacterial communities. The findings are fundamental for understanding the bacterial community in coastal saline-alkali soil of the Zhoushan Island and further provide bioremediation strategies for salt marshes.
2 Materials and Methods
2.1 Site Description and Soil Sample Collection
Soil samples were collected in December 2020 from Dinghai District, located in the Zhoushan Island (29˚32´–30˚04´N, 121˚30´–123˚25´E). The island has a subtropical monsoon climate with an annual average temperature of approximately 16℃, and the annual average sunshine duration is about 1941–2257 h (Linet al., 2018; Wanget al., 2018b). Three wetland sites dominated bySuaeda australis(defined as ‘SUA’ hereafter),Phragmites australis(PHR), andSpartina alterniflora(SPA)were chosen as the sampling points, bare mudflat (MUD)without plants was also included as reference. The triplicate surface soil samples at a depth of 0 – 20 cm were collected with polyethylene container and then mixed to create a single sample for each site. Samples were transported to the laboratory on ice and frozen at -20℃ in darkness on the same day. Subsamples were air-dried at room temperature and sieved through a 2-mm sieve before physico-chemical properties analysis. Others were stored at-80℃ for high throughput sequencing analyses.
2.2 Soil Chemical Analysis
Several parameters were measured to get the physicochemical characteristics of the soil samples. Soil pH and electrical conductivity (EC)were detected using a digital pH meter. Soil salinity was determined after drying residue method. Soil moisture content was analysed by the weight loss of the soil after drying at 105℃ until weight stability. Soil organic matter (SOM)was measured using the dichromate titration method (Schumacher, 2002). Total phosphorus (TP), available phosphorus (AP), total nitrogen (TN), ammonium nitrogen (NH4+-N), nitrate nitrogen (NO3--N), and available nitrogen were determined by standard methods (Zhanget al., 2019a). Soil total potassium (STK)and available potassium (SAK)were measured using graphite furnace atomic absorption spectrometry.
2.3 Soil DNA Extraction and High Throughput Sequencing
Soil total DNA was extracted using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA)according to the manufacturer’s instruction. The concentration and purity of the DNA were quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc.,Wilmington, Delaware, USA). The V4-V5 region of the bacteria16S ribosomal RNAgene of the samples were amplified by PCR. The16S rRNAgenes were amplified using the primers 515F (5’-GTGCCAGCMGCCGCGG-3’)and 907R (5’-CCGTCAATTCMTTTRAGTTT-3’).PCR amplification products were extracted from 2%agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA,USA)according to the manufacturer’s instructions and quantified using QuantiFluor™-ST (Promega, USA). The amplicons were sequenced using IlluminaNovaSeq platform (Nanjing GenePioneer Co., Ltd.)with double-ended(paird-end)sequencing. The PCR products were performed in the software package QIIME V1.17 to according to specific filtering criteria to obtain the high-quality clean tags. The resulting16S rDNAgenesequences were selected and compared for relative abundance of bacterial taxa. The sequences with 97% similarity were assigned to the same operational taxonomic units (OTUs). Taxonomic classification of OTUs was performed using UPARSE V7.1 and chimeric sequences were identified and removed using UCHIME. The phylogenetic affiliation of each16S rRNAgenesequence was analyzed by uclust against the SILVA (SSU123)16S rRNAdatabase using confidence threshold of 50%. The alpha-diversity indices and rarefaction curves were calculated by MothurV1.21.1.The beta-diversity using both weighted and unweighted UniFrac was calculated by QIIME software. Principal coordinate analysis (PCoA)was conducted using the WGCNA package in R (Version 2.15.3). The functional annotation of the bacterial taxa was performed using the Functional Annotation of Prokaryotic Taxa (FAPROTAX)database (Loucaet al., 2016).
3 Results and Discussion
3.1 Soil Characterization
The physicochemical characteristics of soils are summarized in Table 1. The properties of soils showed differences among the four wetland types. The soil pH of samples ranged from 8.17 – 8.30, indicating the study area presented alkaline environment (pH > 8). It was found that the MUD exhibited the highest pH value and soil moisture content compared with the vegetated samples. In different vegetation types, the SOM content, TN, and STK content decreased in the order: SPA wetland > PHR wetland > SUA wetland, and there was no obvious difference in TP content across the three plant covers. The NH4+-N and NO3--N were relatively abundant under PHR-covered soil, whereas the SUA area was relatively low. SPA wetlands exhibited higher amount of AP and SAK contents than the other wetland types. The SAK contents of PHR and SUA wetlands were almost similar,and AP contents in PHR wetland was more enriched in SUA wetland. The MUD exhibited the highest EC (581 Ms m-1)and dramatically declined in the halophyte-covered wetland soils (334 – 353 Ms m-1). It was found that soil salinization was associated with the increase of EC.Similar EC results were also reported by Yin and Yan(2020)and Wanget al.(2020), suggesting that EC was a crucial environmental factor in separation of the bacterial communities between the halophyte-covered soils and bare mudflats, and halophyte could decrease soil EC.Collectively, the physicochemical properties extensively varied among halophyte vegetation types.
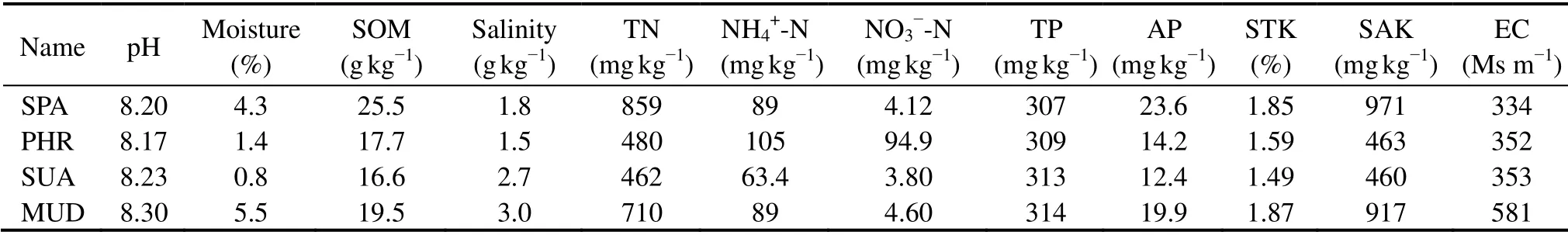
Table 1 Soil physicochemical parameters of different wetland types
3.2 Bacterial Community Diversity and Composition
Halophytes play central roles in coastal ecosystems.Microorganisms are abundant and diverse in soils, and they play important roles in biogeochemical cycling of nutrients. However, few reports have reported the bacterial communities related to halophytes. Illumina sequencing of16S rRNAgene amplicons was performed to profile the complex bacterial communities of halophytescovered and bare mudflat coastal soils. A total of 368353 clean tags from the 4 samples with average 46914 highquality sequences per sample were obtained, with a range of 42354 to 50193 sequences in each sample. As shown in Fig.1, the rarefaction curves for the samples were reached plateau, indicating that the bacterial diversity was covered sufficiently at the sequencing depth.
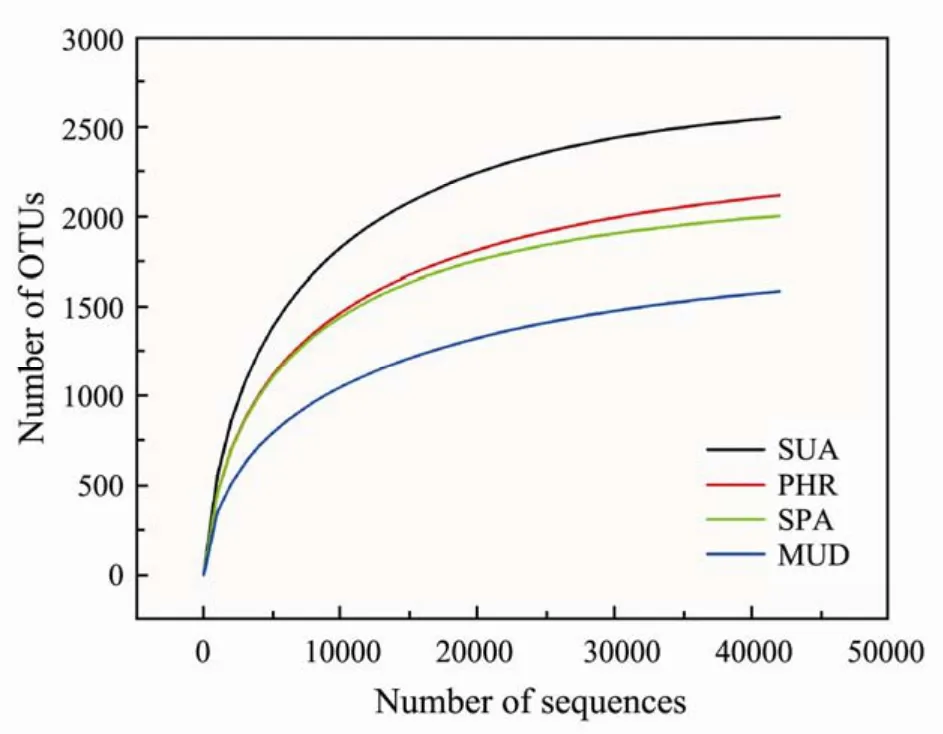
Fig.1 Rarefaction curves of all samples.
3.3 Alpha-Diversity Estimation
The α-diversity indices, including Chao1, observed species, PD whole tree, Shannon, and Simpson index, are tabulated in Table 2. Among the α-diversity indices, Chao1 and observed species estimate the species richness, while Shannon and Simpson indices estimate the bacterial community diversity. Alpha-diversity analysis in Fig.2 showed that the bacterial diversity and richness of YSGT sample had the lowest values, indicating that the vegetated wetland soils were richer and more diverse when compared with the bare mudflat. This is consistent with a previous study demonstrating elevated bacterial diversity inLycium chinense,Tamarix chinensis, andGossypiumhirsutumcovered saline soils (Wanget al., 2020). The flourishing of bacterial communities in the halophytecovered soils could be attributed to the decrease in soil salinity. In vegetated soils, the HHMC sample showed the highest richness and diversity values compared with the others.
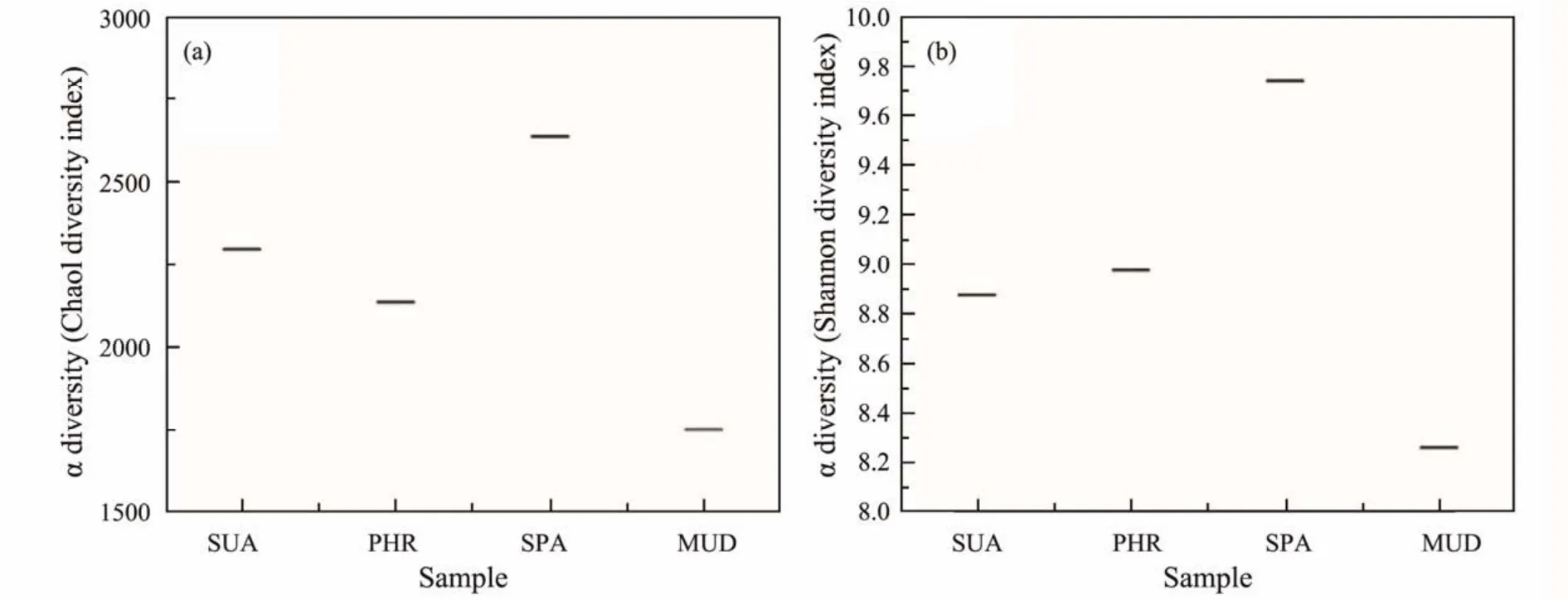
Fig.2 Alpha-diversity of all samples.
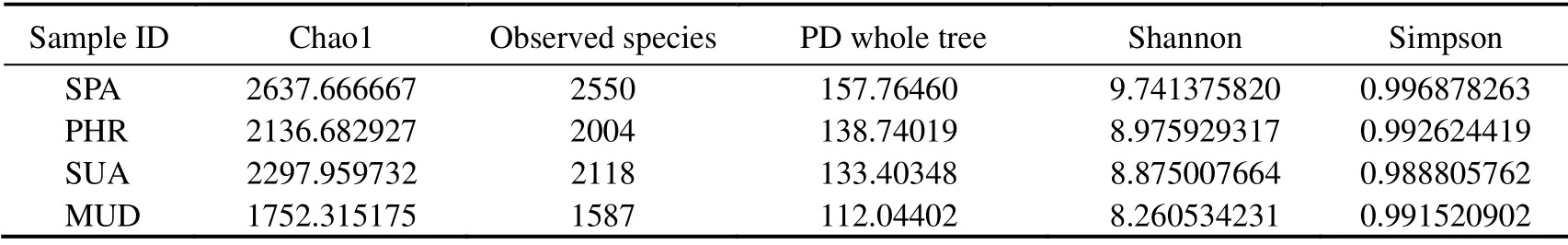
Table 2 Diversity indices of the bacterial community in different wetland types
3.4 Beta-Diversity Estimation
The sequence data were subjected to PCoA based on the weighted UniFrac distance analysis to reveal the similarity of microbial communities across the wetland samples at the genus level (Fig.3a). The first two PCs explained a total of 82.51% variance (PC1 = 60.54% and PC2 = 21.97%)of bacterial communities. The composition and structure of bacterial community were clearly clustered into two groups. The soil samples of SPA and MUD were closer to each other and well separated from those of others, suggesting these two sites exhibited similar bacterial community structure. Furthermore, the SUA and PHR samples were grouped together and distantly separated from the samples of SPA and MUD, indicating these samples showed similar community compositions.Additionally, UPGMA tree based on weighted UniFrac distance also revealed the same clustering among all samples (Fig.3b). Collectively, these results showed that the halophyte plant species played an important role in shaping microbial community composition.
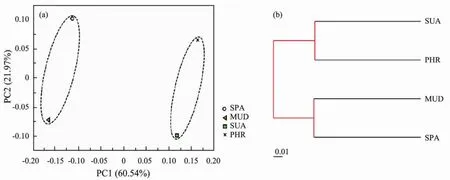
Fig.3 (a)PCoA based on sequence data of 16S rRNA genes of all samples; (b)UPGMA clustering of all samples based on weighted UniFrac distance.
3.5 Soil Bacterial Community Under Different Conditions
The Venn diagram in Fig.4 showed that the SPA sample had 832 unique OTUs, PHR had 371 unique OTUs, SUA had 392 unique OTUs, while MUD had 210 exclusive OTUs. The number of 455 OTUs were shared among the four groups. These results indicated that SPA had the highest number of unique bacterial OTUs, whereas MUD had the lowest number of unique bacterial OTUs.
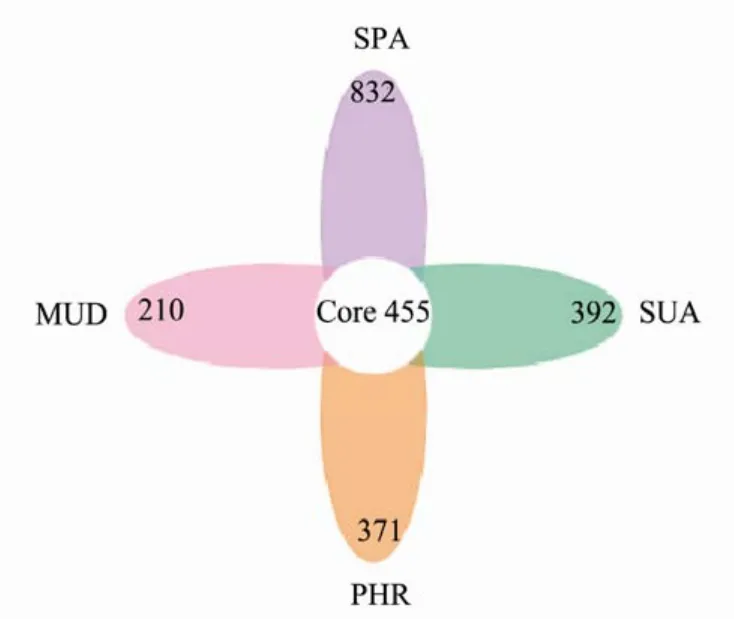
Fig.4 Venn diagram showing the number of operational taxonomic units excluded and shared in different wetland soil samples.
The prevalence of the top 10 bacterial genera in the soil samples are presented in Fig.5. At the genus level, the community structure varied among the soil samples. The generaThiobacillus,MND1, andLuteimonashad high relative abundance in SUA soil sample. The genusNitrospinahad the highest relative abundance in PHR soil.Woeseiawas the most dominant genus in SPA and MUD soil samples. The most abundant bacterial phyla were Proteobacteria, Nitrospirae, Bacteroidetes, Acidobacteria, and Nitrospirae. The enrichment of bacteria of these phyla were mainly involved in the biological nitrogen cycle(Chiet al., 2020). A study of wetland soil samples from China revealed that Proteobacteria, Chloroflexi, Acidobacteria and Bacteroidetes were the dominant bacterial phyla (Anet al., 2019). Similar, Jinget al.(2019)found the phyla Acidobacteria, Bacteroidetes, Chloroflexi were dominated in Suaeda glauca covered soil, while Proteobacteria, Bacteroidetes, Chloroflexi, Actinobacteria, Acidobacteria were the major bacterial phyla inAtriplextriangulariscultivated soils. Generally, Proteobacteria is a broad phylum of the bacteria domain in marine environments, while the high abundance of this phylum might be due to its diverse nitrogen fixation and nutrient cycling(Sunet al., 2019). It has been demonstrated that the phylum Bacteroidetes were prevalent in high-salinity environments due to their strong salinity adaptability (Zhaoet al., 2020). Acidobacteria might play essential role in ecological functions of coastal soils (Rampelottoet al.,2013). The relative abundance of Acidobacteria might be related to the higher root exudate (Jinget al., 2019). As an aerobic chemolitho autotrophic nitrite-oxidizing bacterium in the soil, Nitrospiraeis mainly participate in the ammonia and ammonium salts transformation into nitrate and increase the nitrogen nutrition utilization by plants(Aguilaret al., 2019). Nitrospinae is known as a widespread, ecologically significant phylum in the marine environments (Bemanet al., 2013).
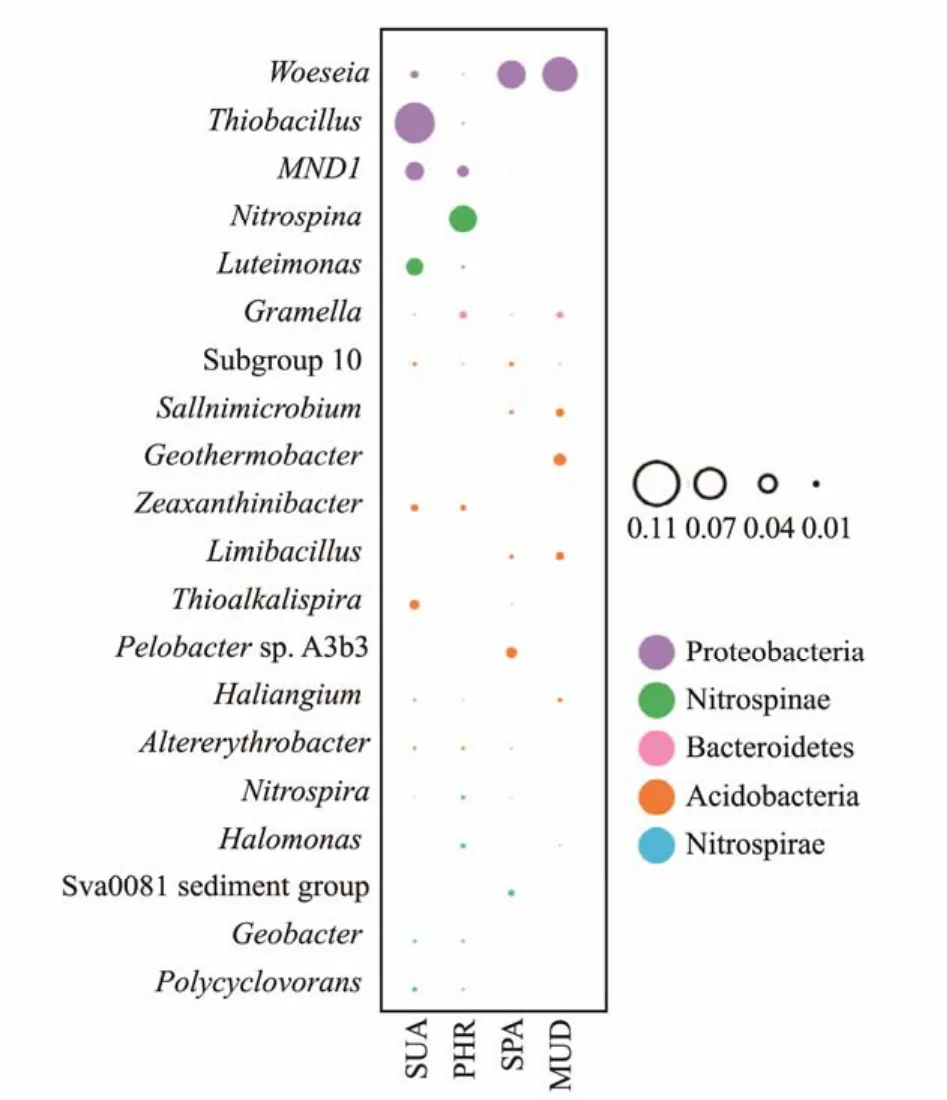
Fig.5 Composition of the bacterial community in all samples.
As indicated in Fig.6, the relative abundance of aerobic and anaerobic bacteria in SPA soil sample were almost similar. In MUD, SUA, and PHR samples, the relative abundances of aerobic bacteria were much higher than that of anaerobic bacteria, suggesting the O2-dominated biological reactions were more abundant in these wetland soils.
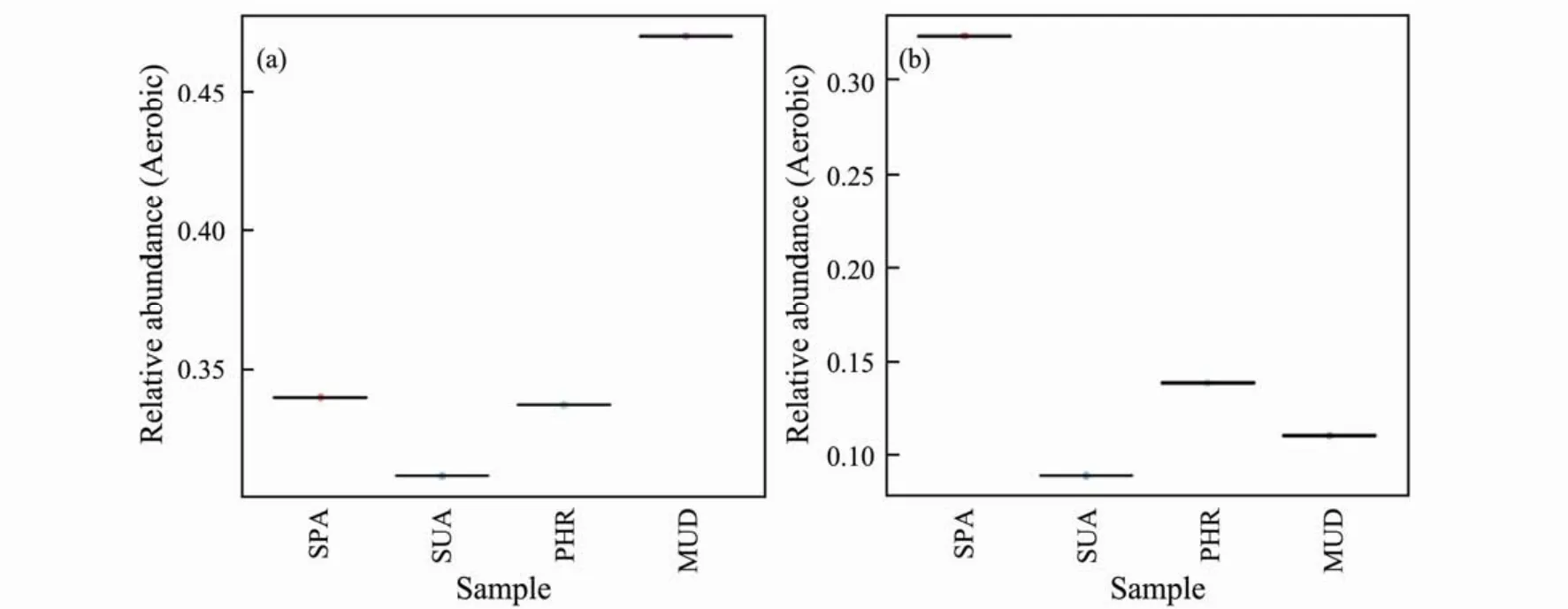
Fig.6 Relative abundance of aerobic and anaerobic bacteria.
3.6 Predictive Functional Profiling
The ecological functions were predicted by using FAPROTAX for the bacterial communities in different soil samples (Fig.7). The FAPROTAX analysis showed knallgas was the dominant predicted function in MUD sample. The chemoautotrophic knallgas bacteria can utilize H2/CO2for growth as the terminal electron acceptorunder aerobic conditions (Mülleret al., 2013).
The functions of chitinolysis, xylanolysis, dark sulfide oxidation, nitrogen fixation, aerobic anoxygenicphototroph, dark hydrogen oxidation, photoheterotrophy, ureolysis, manganese oxidation, aromatic compound degradation, phototrophy were predicted in SUA sample. It has been reported that the marine chitinolytic bacteria play a fundamental role in the nutrient cycling in the oceans viachitinous compound biodegradation (Jahromi and Barzkar, 2018). Microbial xylanases (1,4-β-D-xylan-hydrolase)play a key role in the biodegradation of xylan (Dimitrovet al., 1997). It has been suggested that the aerobic anoxygenic photophic bacteria is widely distributed in the oceanic environment, which could significantly contribute to the carbon cycle in the oceans (Lamiet al., 2007).Photo heterotrophic microbes are capable of utilizing dissolved organic materials and harvesting light energy in the ocean (Cottrell and Kirchman, 2009). Natural and man-made aromatic compounds are widely distributed in the environment, and the degradation of these compounds is accomplished by marine microbes (Guet al., 2018).
In SPA sample, dark sulfite oxidation, fermentation,thiosulfate respiration, sulfite and sulfate respiration, respiration of sulfur compounds, human gut, mammal gut,fumarate respiration, aromatic hydrocarbon degradation,dark iron oxidation functions were predicted. These results suggested that the diversity of respiratory substrates(thiosulfate, sulphite, sulphate, fumarate compounds)of marine bacteria in SPA wetland soil. Diaset al.(2016)demonstrated that Fe(II)/Fe(III)cycling was occurredviareactive oxygen species in the rhizosphere of aSpartinadominated salt marsh systems.
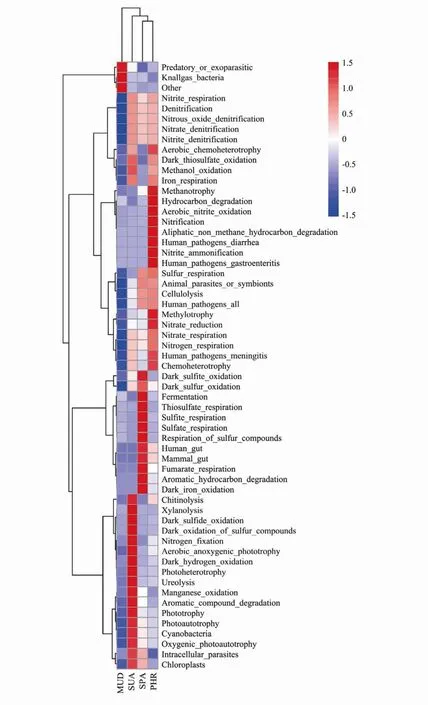
Fig.7 Functional predictions using FAPROTAX of bacterial community in all wetland soil samples.
The aerobic chemoheterotrophy, methanotrophy, hydrocarbon degradation, aerobic nitrite oxidation, nitrification,aliphatic non methane hydrocarbon degradation, human pathogens diarrhea, nitrite ammonification, human pathogens gastroenteritis, methylotrophy, nitrate reduction functions were plentiful in PHR samples. These results suggested that functional bacteria involved in nitrogen cycling was predominated in PHR wetland soils. The aerobic chemoheterotrophy function was also found to be dominant in Bohai Sea sediments (Zhanget al., 2019b).Wanget al.(2020)suggested that the prevalence of this function was probably due to the carbon released from the plants enhanced the growth of the chemoheterotrophic microorganism. It has been indicated that the methanotrophy process could prevent the escape of methane produced within marine sediments into the atmosphere (Sivanet al., 2014).
Overall, halophyte vegetation in salt marsh system played an important role in ecological functions, such as nutrient cycling and biodegradation of pollutants. In particular, the above results indicated that soil bacterial communities under SUA and SPA vegetated study areas might be capable of biodegrading aromatic compounds,such as polycyclic aromatic hydrocarbons. Sulphur cycling was found in SPA covered soil, while SUA and PHR covered soils were involved in nitrogen cycling. The findings highlight the importance of bacteria in the phytoremediation of coastal soil and also indicate that the bacteria-mediated functions involved are halophyte species specific.
4 Conclusions
In summary, halophyte-covered and un-vegetated wetland soil bacterial community structure and diversity were analysed in a salt marsh of the Zhoushan Island, China.The soil bacterial communities inS. australis,P. australis,S. alterniflorawetland soils were different from that in bare mudflats, suggesting that halophyte-covered coastal soil greatly altered the soil bacterial communities. The ecological functions of bacterial OTUs were predicated and could be potentially used as bioindicators. This study could contribute to a better understanding of soil microbial community in coastal wetland ecosystems. In particular, it is of great importance when considering these species for phytoremediation of polluted coastal wetland ecosystem.
Acknowledgements
This work was supported by the grant from the Postdoctoral Advance Programs of Zhejiang Province and Scientific Project Establishment of Huadong Engineering Corporation Limited (No. KY2020-SD-11). The authors gra- tefully acknowledge Mr. Zelong Zhao and Dr. Zhaojing Zhang for their great helps and valuable suggestions.
杂志排行
Journal of Ocean University of China的其它文章
- A Theoretical Model for the Microwave Emissivity of Rough Sea Surfaces
- Mechanism of Regional Subseasonal Precipitation in the Strongest and Weakest East Asian Summer Monsoon Subseasonal Variation Years
- Analytical and Experimental Studies on Wave Scattering by a Horizontal Perforated Plate at the Still Water Level
- Theoretical Prediction of the Bending Stiffness of Reinforced Thermoplastic Pipes Using a Homogenization Method
- Penetration Resistance of Composite Bucket Foundation with Eccentric Load for Offshore Wind Turbines
- Application of Converted Displacement for Modal Parameter Identification of Offshore Wind Turbines with High-Pile Foundation
