栽培稻芽期耐低温全基因组关联分析
2022-12-01逄洪波程露于茗兰陈强李玥莹吴隆坤王泽潘孝武郑晓明
逄洪波,程露,于茗兰,陈强,李玥莹,吴隆坤,王泽,潘孝武,郑晓明
栽培稻芽期耐低温全基因组关联分析
逄洪波1,程露1,于茗兰1,陈强2,李玥莹1,吴隆坤3,王泽1,潘孝武4,郑晓明5,6
1沈阳师范大学生命科学学院,沈阳 110034;2沈阳师范大学实验教学中心,沈阳 110034;3沈阳师范大学粮食学院,沈阳 110034;4湖南省农业科学院水稻研究所,长沙 410125;5中国农业科学院作物科学研究所,北京 100081;6海南三亚中国农业科学院国家南繁研究院,海南三亚 571700
【目的】水稻是重要的粮食作物,芽期是水稻生长发育过程中最脆弱的时期,直播稻遭遇冷害时发芽率大幅降低,减产严重。深入了解耐冷性的遗传机制,为培育芽期强耐受性水稻品种奠定基础。【方法】以世界范围内14个国家代表性的238份水稻种质资源为试验材料,于2021和2022年在沈阳开展表型鉴定试验,统计不同水稻品种在人工气候培养箱15℃低温条件下第1—10天的发芽率和相对发芽率,利用R语言绘制5—10 d的频率直方图,通过表型丰富度Hill值选择宜作关联分析的天数,将发芽率和相对发芽率表型数据与重测序数据相结合,进行基于混合线性模型MLM(QK)的全基因组关联分析,并对所获得的SNP位点进行耐冷候选基因的预测。【结果】发芽率频数分布直方图和表型丰富度计算结果显示第8天发芽率多态性最好,其Hill值为0.84,高于其他几天发芽率(0.48—0.83),可用于全基因组关联分析;主成分分析结果显示,这些水稻品种可以分为、、、和5个亚群;2个指标进行的GWAS分析检测到3个相同的显著性SNP位点,均位于第4染色体,解释表型的11.9%—25.4%;在上下游各50 kb进行基因搜索,共发现24个相关候选基因,进一步开展LD和单倍型分析,发现和的不同单倍型耐冷性之间存在极显著差异。被编码区SNP分为5个单倍型,且Hap_3的耐冷性显著强于Hap_1;被编码区SNP分为18个单倍型,且77 bp处的氨基酸变异(S>L)存在籼粳差异。结果表明,编码糖基转移酶的基因和编码F-box蛋白基因可能与水稻芽期耐冷性密切相关。【结论】在238份水稻种质资源中共检测到3个与芽期耐冷性显著关联的SNP位点,筛选出2个与水稻芽期耐冷性相关的候选基因。
水稻;芽期;耐冷性;发芽率;全基因组关联分析
0 引言
【研究意义】水稻(L.)是一种重要的粮食作物,世界上60%以上人口以水稻作为主食[1-3]。与其他作物不同,水稻对温度较为敏感[4-5]。据统计,全球每年约1 500万hm2水稻种植区遭受低温冷害,造成大面积减产,尤其是日本、韩国以及中国东北和西南地区,严重影响世界粮食安全[6-9]。因此,提高水稻耐冷性已成为水稻育种的重要目标。低温对水稻各个发育阶段均会产生影响[10-13]。萌发期是确保水稻幼苗稳定生长、直接影响最终产量的重要阶段[9]。直播稻可以大量减少劳动力和种植成本[14],随着直播稻在亚洲温带国家的大面积应用,芽期耐冷性已经成为水稻种植所面临的一个重要挑战[15]。因此,挖掘芽期耐冷基因对培育耐冷品种、扩大水稻播种面积乃至提高水稻产量至关重要。【前人研究进展】培育耐冷品种一直受育种专家的关注。水稻芽期耐冷性遗传结构复杂,属于数量性状,受多个基因共同控制,已有对水稻芽期耐冷性的相关研究,定位了一系列与芽期耐冷性相关的QTL[16],同时,研究表明,发芽率是衡量水稻芽期耐冷性强弱的重要指标[17-21]。如LI等[22]以发芽率作为芽期耐冷指标,对粳稻和籼稻品种的重组自交系群体进行芽期表型耐冷评价,最终选择前四代品种发芽率用于后续QTL分析;FUJINO等[11]以2份不同粳稻品种的重组自交系为研究对象,测定其发芽率并用于QTL分析,共检测到、和3个与低温萌发相关的QTL。全基因组关联分析(genome-wide association studies,GWAS)是以基因组重测序为背景,以连锁不平衡(linkage disequilibrium,LD)为基础,利用自然群体在全基因组范围内的遗传变异多态性,深入探讨基因型与表型变化之间的相关性,进而获得最有可能影响特征性状的相关候选基因或基因组区域的一种方法[23-27]。GWAS分析突破了传统的单个数量性状基因定位的局限性,不需构建专门群体,利用现有的自然群体就可进行分析,且所关联的位点多态性等位基因数量庞大,一次可关联多个性状[28-29]。目前,全基因组关联分析已经广泛应用于大豆[30-31]、拟南芥[32]、玉米[33]、水稻[34]和棉花[35]等众多作物发育性状以及农艺性状等研究中,并确定了多种性状的许多QTL。该方法已成功应用于探究水稻芽期耐冷性相关的遗传基因座[36-37]。如FUJINO等[36]以63份日本水稻种质资源为材料,进行水稻低温发芽率的筛选,鉴定到115个SSR标记和另外2种标记,并确定了17个与芽期耐冷性相关联的QTL;PAN等[37]以174份中国水稻品种为材料展开全基因组关联分析,鉴定出51个与水稻芽期和孕穗期耐冷相关的QTL;ZHANG等[6]对249份籼稻品种进行全基因组关联分析,在第1染色体上定位到与水稻芽期耐冷性显著相关的QTL,并鉴定3种芽期耐冷性候选基因;HAN等[38]对200份水稻品种进行全基因组关联分析,共在7条染色体上定位到9个与水稻低温发芽率相关的QTL。【本研究切入点】随着年轻人员的外出打工和农业生产的机械化,直播稻面积不断增大。尽管低温发芽性在水稻栽培中具有重要意义,但其遗传机制尚不清楚。【拟解决的关键问题】本研究选取来自14个国家的238份水稻种质资源为材料,利用人工气候培养箱进行低温胁迫,将重测序数据与芽期发芽率表型数据相结合,进行全基因组关联分析。旨在通过全基因组关联技术筛选出与水稻芽期耐冷性状显著相关的单核苷酸多态性位点,进而挖掘与芽期耐冷性相关的基因,为培育优良耐冷性品种水稻提供基因资源。
1 材料与方法
1.1 试验材料
238份水稻品种包括55个温带粳稻()、26个热带粳稻()、87个籼稻()、66个奥斯稻()和4个香稻(),分别属于中国、印度和日本等14个不同国家,代表世界范围内具有代表性的水稻种植区域和品种类型,由中国农业科学院作物科学研究所提供。所有水稻材料2021年种植于中国农业科学院三亚南繁基地(中国海南,108.37°E,18.10°N),收获干燥后-40℃保存。材料具体信息详见电子附表1。
1.2 发芽率和相对发芽率指标的测定
2020—2021年,在沈阳进行表型鉴定试验。将供试材料置于50℃烘箱中处理72 h,以打破休眠。参考FUJINO等[11]和LI等[22]方法测定发芽率,每份试验材料挑选40粒饱满成熟的种子,以8行5列(8×5)用已灭菌镊子整齐地铺在有润湿的双层滤纸的玻璃培养皿(直径9 cm)中,加入10 mL无菌水,分别放置于低温15℃和对照30℃(12 h光照/12 h黑暗),光强为8 000 lx,相对湿度60%的人工气候培养箱(RDN-1000E-4,宁波乐电仪器制造有限公司)中低温处理10 d。以种子露白作为萌发标准,每天统计发芽种数,用于发芽率指标的计算,每个品种3次重复。计算公式如下:
发芽率(%)=(15℃低温发芽的种子数量/种子总数)×100;
相对发芽率(%)=(15℃处理发芽率/对照发芽率30℃)×100。
1.3 数据处理与分析
1.3.1 表型数据整理 利用R语言(v3.5.0)绘制发芽率频数分布直方图。
1.3.2 数据质量控制 使用Illumina HiSeq 2500 Sequencing Systems Platform(Illumina Inc., San Diego, CA,USA)对238份水稻材料进行全基因组测序,通过SICKLE(https://github.com/najoshi/sickle)修剪reads的3’末端,处理raw reads以获得per read≤20的average quality score(QS)。将high-quality reads与水稻参考基因组序列日本晴(MSU 7.0)进行比对(日本晴序列下载自http://rice.plantbiology.msu.edu/ pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0/all.dir/)。利用Perl脚本结合plink2软件,对所有原始标记位点进行处理,去除稀有变异位点、高缺失率位点、高杂合率位点和严重偏离哈迪-温伯格平衡的位点,最终筛选出高质量的SNP显著性位点,经数据质量控制后用于后续全基因组关联分析的高质量SNP数量为7 143 225个。
1.3.3 群体遗传结构分析 利用Admixture软件进行群体结构分析,得到Q矩阵用于后续线性模型构建。利用GCTA软件(v1.93.2)进行主成分分析(principal component analysis,PCA),获得各个PC的方差解释率及样本在各个PC中的得分矩阵。在GWAS中,为避免可能存在的小家系导致假阳性,往往会把亲缘关系矩阵作为随机效应协变量矩阵(K矩阵)加入GWAS模型。
1.3.4 全基因组关联分析 利用GEMMA软件(v0.98.1)比较全基因组关联测序4种分析模型(GLM、GLM(Q)、MLM(K)和MLM(QK))所得结果,最终采用混合线性模型(MLM(QK))模型进行GWAS分析,即将群体结构(Q)和个体遗传关系(K)加入混合线性模型中,对Q和K的影响进行预测评估,从而控制和减少对目标基因关联定位的影响[39]。Admixture最优K值对应的群体结构矩阵作为相应模型的Q矩阵,GCTA(v1.93.2)软件计算的样品间亲缘关系矩阵作为相应模型的K矩阵。GWAS分析结果采用曼哈顿图和QQ图进行展示。
1.3.5 LD分析和单倍型分析 使用LDBlockShow软件的默认参数进行LD分析。单倍型分析所用165个栽培稻(56个粳稻和109个籼稻,具体信息见电子附表2)的编码区序列是从FRGB数据库(https://www. rmbreeding.cn/)下载,使用ClustalX 1.81进行序列比对,DnaSP 6进行核苷酸多态性和单倍型分析。
2 结果
2.1 水稻芽期耐冷性表型数据分析
共统计238个水稻供试品种在15℃低温条件下第1—10天的发芽率,由于前4 d发芽品种很少,故在此只展示第5—10天发芽率的频数直方图(图1)。根据遗传学中的表型丰富度Hill多样性指标[40]进行5— 10 d的发芽率多样性计算,计算公式为:
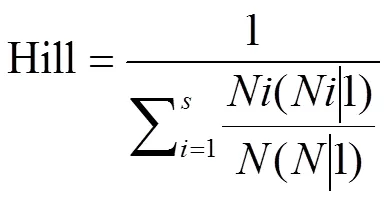
第5—10天的发芽率丰富度依次为0.48、0.65、0.78、0.84、0.83和0.80,第8天的发芽率丰富度最高,为0.84,故选择第8天的发芽率数据用于后续的全基因组关联分析。
2.2 水稻芽期耐冷性的全基因组关联分析
2.2.1 群体遗传结构分析 基于筛选后的SNP标记,利用GCTA软件(v1.93.2)对238供试材料进行主成分分析,获得各个PC的方差解释率及样本在各个PC中的得分矩阵。经计算,将数据矩阵转化成图片(图2),前3个主成分的方差解释率分别为10.21%、6.82%和3.43%,可以解释群体遗传结构20.46%变异。根据PCA分析结果,可将238水稻品种划分为5个类群,分别为、、、和。
基于筛选后的SNP标记,利用GCTA软件(v1.93.2)对238供试材料进行亲缘关系分析,即两特定材料之间的遗传相似度与任意材料之间的遗传相似度的相对值。根据亲缘关系kinship频率分布图(图3-A),可以清晰展示样本间亲缘关系值的主要分布图范围,从而推断样本间遗传差异的大小。其中,图中的负值可能是由于标准化的结果,值的大小表示两样本之间关系的相似性即相对远近,值越大,说明两样本之间相似性远大,即亲缘关系越近;值越小,说明两样本之间相似性越小,即亲缘关系越远。由图3可见,绝大多数试验材料的亲缘关系位于-0.5,说明大部分试验材料之间的相似性较小,亲缘关系相对较远。因此,可以避免因家系群体对全基因组关系分析结果产生假阳性。根据亲缘关系矩阵绘制热图(图3-B),可将供试材料分为5个亚群,与PCA分析结果一致。

图2 水稻全基因组单核苷酸多态性数据的主成分分析
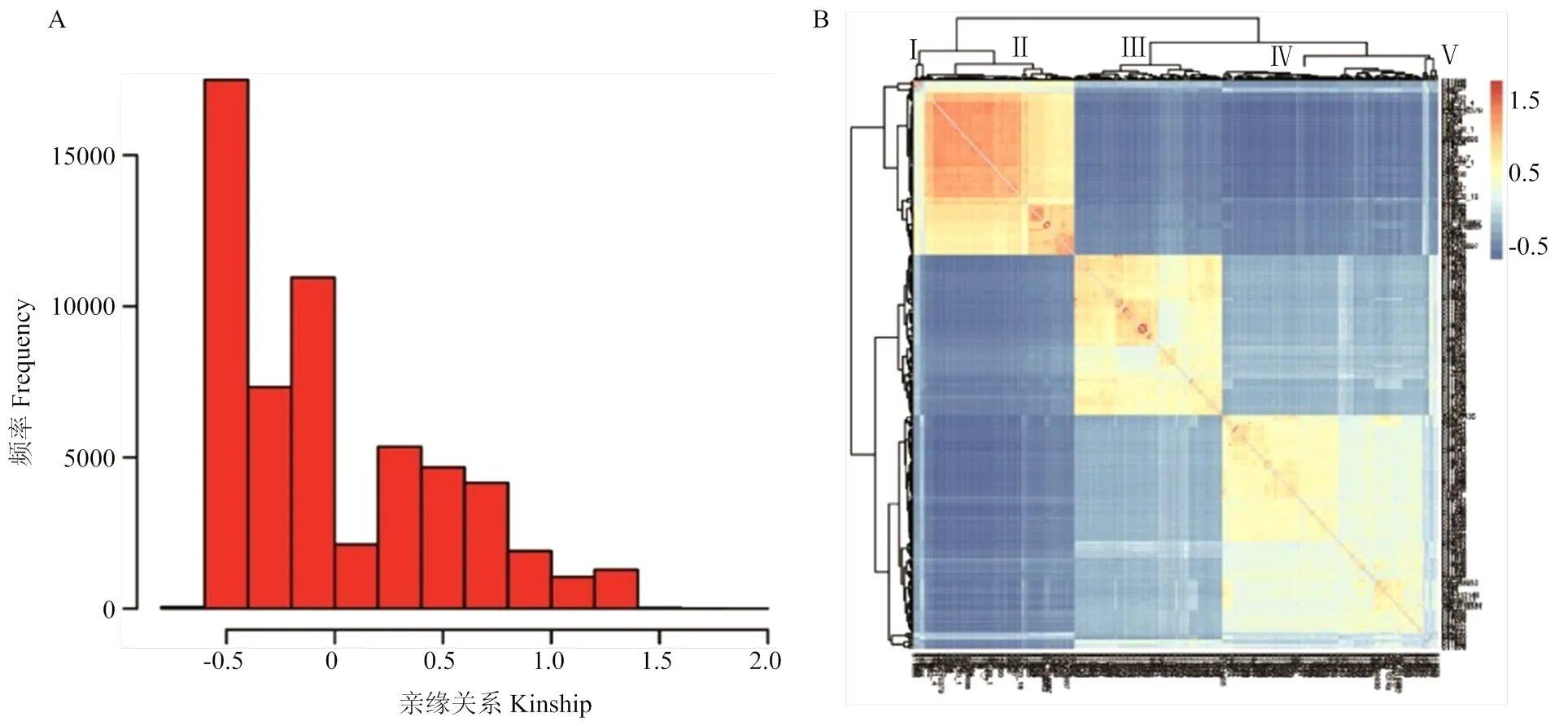
A:样本间亲缘关系直方图;B:样本间亲缘关系热图
2.2.2 GWAS定位目标性状SNP位点 采用混合线性模型MLM(QK)进行GWAS分析(图4),阈值设为-log10()=7,利用发芽率指标仅在第4染色体上关联到3个显著性SNP位点,分别是Chr.4:14334616、Chr.4:14333724和Chr.4:14528782,可以解释形态学性状的11.9%—25.4%,其中,Chr.4:14334616 与发芽率的关联程度最高,为8.69E-08。利用相对发芽率共关联到3个显著性SNP,均位于第4染色体上,且这3个位点与利用发芽率关联到的位点完全相同,可以解释形态学性状的13.1%—25.6%,与相对发芽率性状关联程度最高的SNP位点(Chr.4:14334616)为9.23E-08。

图4 238个水稻品种发芽率和相对发芽率的全基因组关联分析
2.2.3 筛选水稻芽期耐冷候选基因 根据发芽率和相对发芽率2个指标的关联结果,第4染色体Chr.4:14333724、Chr.4:14334616和Chr.4:14528782重复出现,故后续对这3个显著性SNP进一步深入分析。搜索每个显著性SNP位点上、下游各50 kb区间,预测可能与芽期耐冷性状相关联的基因,共关联到24个基因(—),通过国家水稻数据中心(www.ricedata.com)对关联的基因进行Interpro蛋白功能预测(表1)。将显著性SNP位点所在区域的曼哈顿图放大,同关联到的24个基因进行共线分析,同时绘制了LD单倍型块图(图5),结合基因注释结果,推测含有CCHC结构域的锌指蛋白基因()、编码糖基转移酶的基因(和)和含有F-box结构域的基因()可能与芽期耐冷性相关。
2.2.4 候选基因的核苷酸多样性和单倍型分析 为了进一步分析4个候选基因是否与芽期耐冷性相关,对3K数据库中165个栽培稻品种的编码区序列进行核苷酸比对,结果显示,和不存在SNP位点(结果未展示)。中鉴定出4个SNP(图6-A),分别为164 bp(R>H)、194 bp(Q>R)、206 bp(L>P)和235 bp(C>T)。其中235 bp位置的突变C>T引入了终止密码子。进一步的单倍型分析结果显示,存在5种单倍型;Hap_1为主要单倍型,有145个品种(占84.8%);其次是具有22个籼稻品种的Hap_3。只有单倍型Hap_1中同时包含粳稻和籼稻(图6-A)。为进一步分析这些单倍型之间的差异,比较核苷酸多态性与相对发芽率之间的相关性。Hap_1(43.23%)和Hap_3(61.51%)间存在显著差异(曼-惠特尼秩和检验,=0.005),说明Hap_3是优异单倍型。存在9个SNP和1个Indel(图6-B)。其中,6个SNP为有义突变,分别为77 bp(S>L)、166 bp(K>D)、466 bp(H>D)、563 bp(R>H)、567 bp(K>N)和1 018 bp(C>R)。这些SNP将分成18个单倍型(Hap_1—Hap_18);只有Hap_4、Hap_7和Hap_8中同时含有粳稻和籼稻。单倍型中品种数量最多的是Hap_4,有86个(52.12%),其次是Hap_3,有26个品种(15.76%)。有趣的是,77 bp处的基因变异(S>L)在籼稻和粳稻亚种之间是不同的。据此,可以将这些单倍型分为A组(平均发芽率=40.25%)和B组(平均发芽率=54.80),结合相对发芽率的表型数据,发现这两组之间存在显著差异(曼-惠特尼秩和检验,=0.004)。同样,Hap_4(40.75%)和Hap_2(75.45%)(曼-惠特尼秩和检验,=0.001)、Hap_4(40.75%)和Hap_3(62.66%)之间也存在显著性差异(曼-惠特尼秩和检验,=0.002),Hap_2(75.45%)和Hap_3(62.66%)之间没有显著性差异(Mann-Whitney秩和检验,=0.295),说明77 bp(S>L)是关键的核苷酸变异位点。同时,Hap_4—Hap_7的两两比较分析,均没有显著性差异,说明609和1 344 bp的无义突变可能不会影响其耐冷性,也可能与Hap_5—Hap_7的样本量小有关。而其他单倍型的数量太少,无法进行相关的统计分析。

图中间部分24个黑色方框依次为24个候选基因,红色竖线代表3个显著性SNP所在位置,4个灰色箭头分别指LOC_Os04g24830、LOC_Os04g24840、LOC_Os04g24850和LOC_Os04g25140
3 讨论
3.1 水稻芽期耐冷指标筛选
芽期低温胁迫是导致中国南部水稻种植区,特别是直接稻种植区减产的重要因素[41-42]。耐冷性是多个基因共同控制的复杂性状[43-44],表型鉴定是研究耐冷性状的重要手段[6, 45]。水稻芽期各形态指标测定时间短、简便易行,在进行大规模水稻耐冷种质资源的筛选和鉴定方面具有一定的优势。种子在低温条件下的发芽率是具有遗传学性状的表现之一,其基因型很大程度上决定了种子芽期的耐冷性,因此,种子的低温发芽率可以作为调查水稻芽期耐冷性的重要指标[46-47]。目前,已有前人在水稻芽期耐冷筛选和鉴定方面开展了一些相应的研究,如CRUZ等[48]以种子萌发指数百分比作为芽期耐冷指标对不同起源的24份水稻种质资源进行评价,共筛选出4份强耐冷品种;JI等[18]以发芽率作为一个芽期耐冷指标来评估低温萌发能力的遗传控制,并在此基础上使用重组置换系进行水稻耐冷QTL分析;YE等[49]以17份水稻种质资源为材料,以发芽率和平均发芽速度为指标对其萌发阶段进行耐冷评价。故本研究以水稻芽期发芽率为耐冷评价指标对耐冷性进行快速鉴定。

表1 24个候选基因其基因注释
3.2 水稻芽期耐冷基因位点鉴定方法
GWAS通过重测序识别高分辨率SNP,以鉴定复杂数量性状的靶基因区域,精确度更高,且可以一次关联定位到多个性状[28-29],已经被广泛用于识别作物性状相关的遗传基因座,并为其遗传基础提供新的见解。如对非生物胁迫的响应[50-51]、生物应激[52-53]以及许多其他农艺性状特征[54-55]等。HUANG等[24]以517个水稻地方品种为材料,第一次证实将GWAS和重测序数据相结合可以用于水稻复杂性状的研究。FUJINO等[36]以63个水稻品种为材料展开全基因组关联分析,鉴定出6个与水稻抽穗期和17个与水稻芽期耐冷相关的QTL;MIURA等[45]以98个粳稻和籼稻品种的回交自交系为研究对象,测定其发芽率并用于QTL分析,共检测到、、、和5个与低温萌发相关的QTL;JIANG等[56]对2个不同粳、籼稻品系杂交的F2进行全基因组关联分析,共在7条染色体上定位到了11个与水稻低温发芽率相关的QTL。故本研究采用全基因组关联分析法对238份水稻种质资源进行耐冷性相关基因位点检测。

红色代表粳稻和籼稻共有的单倍型,蓝色代表基因编码区SNP中的有义突变;绿色表示235 bp处核苷酸变异(C>T)导致基因提前出现终止子;黄色表示粳稻和籼稻在77 bp处存在碱基差异(C>T)
3.3 与水稻芽期耐冷相关的候选基因
基于发芽率的全基因组关联分析共得到了3个显著性SNP,均位于第4染色体,关联到24个基因。进一步对其进行LD和单倍型分析,结果发现,和各自具有的SNP将其分成了5个和18个单倍型,的Hap_3是优异单倍型,在77 bp处的变异(S>L)可能与粳稻和籼稻的分化有关。编码糖基转移酶,与之前发现的调控孕穗期耐冷基因Ctb2位点相距12.3 Mb[57],且CTB2也是个编码葡糖基转移酶基因。低温胁迫下,Ctb2通过影响甾醇糖苷和乙酰化甾醇糖苷的含量维持细胞膜的渗透性,保护花粉粒及花粉外壁结构,最终提高水稻的耐冷性。无独有偶,LI等[58]发现拟南芥中的UDP-糖基转移酶UGT79B2/3过表达植株耐冷性增强,且其表达受CBF1直接调控。SHI等[59]鉴定的苗期耐冷基因OsUGT90A1也属于糖基转移酶家族成员,而糖基转移酶属于GT家族中最大的一个分支,参与很多代谢过程,故推测很有可能在水稻芽期低温胁迫的响应中发挥着重要作用。编码的是含有F-box结构域的蛋白质基因,与SAITO等[60]分离的开花期耐冷基因相距约17 Mb,也属于F-box 蛋白,以往研究表明,F-box蛋白整合了几乎所有的植物激素信号通路,在调节各种发育过程和应激反应中发挥重要作用[61-63]。水稻中耐冷相关基因MAIF1编码的也是F-box蛋白[62],因此,本研究筛选编码F-box蛋白的基因有很大可能参与芽期耐冷调控,当然还需要进一步功能分析加以验证。
4 结论
在第4染色体上检测到3个完全一致的显著性SNP,筛选出2个与水稻芽期耐冷性密切相关基因(和)。
[1] ZHENG S, LIU S, FENG J, WANG W, WANG Y, YU Q, LIAO Y, MO Y, XU Z, LI L, GAO X, JIA X, ZHU J, CHEN R. Overexpression of a stress response membrane protein geneenhances rice tolerance to salt, cold and heavy metal stress. Environmental and Experimental Botany, 2021, 182: 104327.
[2] WANG H, LEE A R, PARK S Y, JIN S H, LEE J, HAM T H, PARK Y, ZHAO W G, KWON S W. Genome-wide association study reveals candidate genes related to low temperature tolerance in rice () during germination. 3 Biotech, 2018, 8(5): 1-13.
[3] CHENG S H, CAO L Y, ZHUANG J Y, CHEN S G, ZHAN X D, FAN Y Y, ZHU D F, MIN S K. Super hybrid rice breeding in china: achievements and prospects. Journal of Integrative Plant Biology, 2007, 49(6): 805-810.
[4] VILAS J M, CORIGLIANO M G, CLEMENTE M, MAIALE S J, RODRIGUEZ A A. Close relationship between the state of the oxygen evolving complex and rice cold stress tolerance. Plant Science, 2020, 296: 110488.
[5] SIPASEUTH, BASNAYAKE J, FUKAI S, FARRELL T C, SENTHONGHAE M, SENGKEO, PHAMIXAY S, LINQUIST B, CHANPHENGSAY M. Opportunities to increasing dry season rice productivity in low temperature affected areas. Field Crops Research, 2007, 102(2): 87-97.
[6] ZHANG M, YE J, XU Q, FENG Y, YUAN X, YU H, WANG Y, WEI X, YANG Y. Genome-wide association study of cold tolerance of Chineserice varieties at the bud burst stage. Plant Cell Reports, 2018, 37(3): 529-539.
[7] SUH J P, JEUNG J U, LEE J I, CHOI Y H, YEA J D, VIRK P S, MACKILL D J, JENA K K. Identification and analysis of QTLs controlling cold tolerance at the reproductive stage and validation of effective QTLs in cold-tolerant genotypes of rice (L.). Theoretical and Applied Genetics, 2010, 120(5): 985-995.
[8] FUJINO K, MATSUDA Y. Genome-wide analysis of genes targeted by qLTG3-1 controlling low-temperature germinability in rice. Plant Molecular Biology, 2010, 72(1): 137-152.
[9] FUJINO K, SEKIGUCHI H, MATSUDA Y, SUGIMOTO K, ONO K, YANO M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(34): 12623-12628.
[10] SAITO K, HAYANO-SAITO Y, MARUYAMA-FUNATSUKI W, SATO Y, KATO A. Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theoretical and Applied Genetics, 2004, 109(3): 515-522.
[11] FUJINO K, SEKIGUCHI H, SATO T, KIUCHI H, NONOUE Y, TAKEUCHI Y, ANDO T, LIN S Y, YANO M. Mapping of quantitative trait loci controlling low-temperature germinability in rice (L.). Theoretical and Applied Genetics, 2004, 108(5): 794-799.
[12] ANDAYA V C, TAI T H. Fine mapping of thelocus, a major QTL for seedling cold tolerance in rice. Theoretical and Applied Genetics, 2006, 113(3): 467-475.
[13] LOU Q, CHEN L, SUN Z, XING Y, LI J, XU X, MEI H, LUO L. A major QTL associated with cold tolerance at seedling stage in rice (L.). Euphytica, 2007, 158(1): 87-94.
[14] KUMAR V, LADHA J K. Direct seeding of rice. recent developments and future research needs. Advances in Agronomy, 2011, 111: 297-413.
[15] IWATA N, SHINADA H, KIUCHI H, SATO T, FUJINO K. Mapping of QTLs controlling seedling establishment using a direct seeding method in rice. Breeding Science, 2010, 60(4): 353-360.
[16] ZHANG Z H, QU X S, WAN S, CHEN L H, ZHU Y G. Comparison of QTL controlling seedling vigour under different temperature conditions using recombinant inbred lines in rice (). Annals of Botany, 2005, 95(3): 423-429.
[17] CHEN L, LOU Q J, SUN Z X, XING Y Z, XIN-QIAO Y U, LUO L J. Qtl mapping of low temperature on germination rate of rice. Rice Science, 2006, 13(2): 93-98.
[18] JI S L, JIANG L, WANG Y H, ZHANG W W, LIU X, LIU S J, CHEN L M, ZHAI H Q, WAN J M. Quantitative trait loci mapping and stability for low temperature germination ability of rice. Plant Breeding, 2009, 128(4): 387-392.
[19] WAN J M, JIANG L, TANG J Y, WANG C M, HOU M Y, JING W, ZHANG L X. Genetic dissection of the seed dormancy trait in cultivated rice (L.). Plant Science, 2006, 170(4): 786-792.
[20] SUGIMOTO K, TAKEUCHI Y, EBANA K, MIYAO A, HIROCHIKA H, HARA N, ISHIYAMA K, KOBAYASHI M, BAN Y, HATTORI T, YANO M. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(13): 5792-5797.
[21] SHARIFI P. Evaluation on sixty-eight rice germplasms in cold tolerance at germination stage. Rice Science, 2010, 17(1): 77-81.
[22] LI L, LIU X, XIE K, WANG Y, LIU F, LIN Q, WANG W, YANG C, LU B, LIU S, CHEN L, JIANG L, WAN J. qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (L.). Theoretical and Applied Genetics, 2013, 126(9): 2313-2322.
[23] ALBINANA C, GROVE J, MCGRATH J J, AGERBO E, WRAY N R, BULIK C M, NORDENTOFT M, HOUGAARD D M, WERGE T, BORGLUM A D, MORTENSEN P B, PRIVE F, VILHJALMSSON B J. Leveraging both individual-level genetic data and GWAS summary statistics increases polygenic prediction. American Journal of Human Genetics, 2021, 108(6): 1001-1011.
[24] HUANG X, WEI X, SANG T, ZHAO Q, FENG Q, ZHAO Y, LI C, ZHU C, LU T, ZHANG Z, LI M, FAN D, GUO Y, WANG A, WANG L, DENG L, LI W, LU Y, WENG Q, LIU K, HUANG T, ZHOU T, JING Y, LI W, LIN Z, BUCKLER E S, QIAN Q, ZHANG Q F, LI J, HAN B. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature Genetics, 2010, 42(11): 961-967.
[25] HUANG X, ZHAO Y, WEI X, LI C, WANG A, ZHAO Q, LI W, GUO Y, DENG L, ZHU C, FAN D, LU Y, WENG Q, LIU K, ZHOU T, JING Y, SI L, DONG G, HUANG T, LU T, FENG Q, QIAN Q, LI J, HAN B. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nature Genetics, 2011, 44(1): 32-39.
[26] ZHAO K, TUNG C W, EIZENGA G C, WRIGHT M H, ALI M L, PRICE A H, NORTON G J, ISLAM M R, REYNOLDS A, MEZEY J, MCCLUNG A M, BUSTAMANTE C D, MCCOUCH S R. Genome-wide association mapping reveals a rich genetic architecture of complex traits in. Nature Communications, 2011, 2(1): 1-10.
[27] HSIAO C F, CHIU Y F, CHIANG F T, HO L T, LEE W J, HUNG Y J, CHEN Y D, DONLON T A, JORGENSON E, CURB D, RISCH N, HSIUNG C A, GROUP S A S. Genome-wide linkage analysis of lipids in nondiabetic Chinese and Japanese from the SAPPHIRe family study. American Journal of Hypertension, 2006, 19(12): 1270-1277.
[28] NICOLAE D L, GAMAZON E, ZHANG W, DUAN S, DOLAN M E, COX N J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genetics, 2010, 6(4): e1000888.
[29] YANG J, FERREIRA T, MORRIS A P, MEDLAND S E, GENETIC INVESTIGATION OF A T C, REPLICATION D I G, META ANALYSIS C, MADDEN P A, HEATH A C, MARTIN N G, MONTGOMERY G W, WEEDON M N, LOOS R J, FRAYLING T M, MCCARTHY M I, HIRSCHHORN J N, GODDARD M E, VISSCHER P M. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature Genetics, 2012, 44(4): 369-375.
[30] LI Y H, LI D, JIAO Y Q, SCHNABLE J C, LI Y F, LI H H, CHEN H Z, HONG H L, ZHANG T, LIU B, LIU Z X, YOU Q B, TIAN Y, GUO Y, GUAN R X, ZHANG L J, CHANG R Z, ZHANG Z, REIF J, ZHOU X A, SCHNABLE P S, QIU L J. Identification of loci controlling adaptation in Chinese soya bean landraces via a combination of conventional and bioclimatic GWAS. Plant Biotechnology Journal, 2020, 18(2): 389-401.
[31] HWANG E Y, SONG Q, JIA G, SPECHT J E, HYTEN D L, COSTA J, CREGAN P B. A genome-wide association study of seed protein and oil content in soybean. BMC Genomics, 2014, 15(1): 1-12.
[32] WU S, ALSEEKH S, CUADROS-INOSTROZA A, FUSARI C M, MUTWIL M, KOOKE R, KEURENTJES J B, FERNIE A R, WILLMITZER L, BROTMAN Y. Combined use of genome-wide association data and correlation networks unravels key regulators of primary metabolism in. PLoS Genetics, 2016, 12(10): e1006363.
[33] WENG J, XIE C, HAO Z, WANG J, LIU C, LI M, ZHANG D, BAI L, ZHANG S, LI X. Genome-wide association study identifies candidate genes that affect plant height in Chinese elite maize (L.) inbred lines. PLoS One, 2011, 6(12): e29229.
[34] WU J, FENG F, LIAN X, TENG X, WEI H, YU H, XIE W, YAN M, FAN P, LI Y, MA X, LIU H, YU S, WANG G, ZHOU F, LUO L, MEI H. Genome-wide association study (GWAS) of mesocotyl elongation based on re-sequencing approach in rice. BMC Plant Biology, 2015, 15: 218.
[35] LI Z, WANG X, CUI Y, QIAO K, ZHU L, FAN S, MA Q. Comprehensive genome-wide analysis of thaumatin-like gene family in four cotton species and functional identification ofinvolved in regulating tolerance toand drought. Frontiers in Plant Science, 2020, 11: 575015.
[36] FUJINO K, OBARA M, SHIMIZU T, KOYANAGI K O, IKEGAYA T. Genome-wide association mapping focusing on a rice population derived from rice breeding programs in a region. Breeding Science, 2015, 65(5): 403-410.
[37] PAN Y, ZHANG H, ZHANG D, LI J, XIONG H, YU J, LI J, RASHID M A, LI G, MA X, CAO G, HAN L, LI Z. Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS One, 2015, 10(3): e0120590.
[38] HAN L Z, ZHANG Y Y, QIAO Y L, CAO G L, ZHANG S Y, KIM J H, KOH H J. Genetic and QTL analysis for low-temperature vigor of germination in rice. Acta Genetica Sinica, 2006, 33(11): 998-1006.
[39] ZHOU X, STEPHENS M. Genome-wide efficient mixed-model analysis for association studies. Nature Genetics, 2012, 44(7): 821-824.
[40] PEET R K. The measurement of species diversity. Annual Review of Ecology and Systematics, 1974, 5(1): 285-307.
[41] YANG J, YANG M, SU L, ZHOU D, HUANG C, WANG H, GUO T, CHEN Z. Genome-wide association study reveals novel genetic loci contributing to cold tolerance at the germination stage in indica rice. Plant Science, 2020, 301: 110669.
[42] LIN J, ZHU W Y, ZHANG Y D, ZHU Z, ZHAO L, CHEN T, ZHAO Q Y, ZHOU L H, FANG X W, WANG Y P, WANG C L. Detection of QTL for cold tolerance at bud bursting stage using chromosome segment substitution lines in rice (). Rice Science, 2011, 18(1): 71-74.
[43] XIAO H, CHEN J F, ZHANG Z X. Influence of deposition temperature on the structure of Si3N4 thin film prepared by MWECR-PECVD. Plasma Science & Technology, 2004, 6(5): 2485-2488.
[44] 纪素兰, 江玲, 王益华, 刘世家, 刘喜, 翟虎渠, 吉村醇, 万建民. 水稻种子耐低温发芽力的QTL定位及上位性分析. 作物学报, 2008, 34(4): 551-556.
JI S L, JIANG L, WANG Y H, LIU S J, LIU X, ZHAI H Q, YOSHIMURA A, WAN J M. QTL and epistasis for low temperature germinability in rice. Acta Agronomica Sinica, 2008, 34(4): 551-556. (in Chinese)
[45] MIURA K, LIN S Y, YANO M, NAGAMINE T. Mapping quantitative trait loci controlling low temperature germinability in rice (L.). Breeding Science, 2001, 51(4): 293-299.
[46] HYUN D Y, OH M, CHOI Y M, LEE S, LEE M C, OH S. Morphological and molecular evaluation for germinability in rice varieties under low-temperature and anaerobic conditions. Journal of Crop Science and Biotechnology, 2017, 20(1): 21-27.
[47] ANGAJI S A, SEPTININGSIH E M, MACKILL D J, ISMAIL A M. QTLs associated with tolerance of flooding during germination in rice (L.). Euphytica, 2009, 172(2): 159-168.
[48] CRUZ R P, MILACH S C K. Cold tolerance at the germination stage of rice: methods of evaluation and characterization of genotypes. Scientia Agricola, 2004, 61(1): 1-8.
[49] YE C, FUKAI S, GODWIN I, REINKE R, SNELL P, SCHILLER J, BASNAYAKE J. Cold tolerance in rice varieties at different growth stages. Crop and Pasture Science, 2009, 60(4): 328-338.
[50] WANG X, WANG H, LIU S, FERJANI A, LI J, YAN J, YANG X, QIN F. Genetic variation incontributes to drought tolerance in maize seedlings. Nature Genetics, 2016, 48(10): 1233-1241.
[51] WAN H, CHEN L, GUO J, LI Q, WEN J, YI B, MA C, TU J, FU T, SHEN J. Genome-wide association study reveals the genetic architecture underlying salt tolerance-related traits in rapeseed (L.). Frontiers in Plant Science, 2017, 8: 593.
[52] JIA L, YAN W, ZHU C, AGRAMA H A, JACKSON A, YEATER K, LI X, HUANG B, HU B, MCCLUNG A, WU D. Allelic analysis of sheath blight resistance with association mapping in rice. PLoS One, 2012, 7(3): e32703.
[53] KANG H, WANG Y, PENG S, ZHANG Y, XIAO Y, WANG D, QU S, LI Z, YAN S, WANG Z, LIU W, NING Y, KORNILIEV P, LEUNG H, MEZEY J, MCCOUCH S R, WANG G L. Dissection of the genetic architecture of rice resistance to the blast fungus. Molecular Plant Pathology, 2016, 17(6): 959-972.
[54] YANG W, GUO Z, HUANG C, DUAN L, CHEN G, JIANG N, FANG W, FENG H, XIE W, LIAN X, WANG G, LUO Q, ZHANG Q, LIU Q, XIONG L. Combining high-throughput phenotyping and genome- wide association studies to reveal natural genetic variation in rice. Nature Communications, 2014, 5(1): 1-9.
[55] WANG D, LIU J, LI C, KANG H, WANG Y, TAN X, LIU M, DENG Y, WANG Z, LIU Y, ZHANG D, XIAO Y, WANG G L. Genome-wide association mapping of cold tolerance genes at the seedling stage in rice. Rice, 2016, 9(1): 61.
[56] JIANG L, LIU S, HOU M, TANG J, CHEN L, ZHAI H, WAN J. Analysis of QTLs for seed low temperature germinability and anoxia germinability in rice (L.). Field Crops Research, 2006, 98(1): 68-75.
[57] LI J, ZENG Y, PAN Y, ZHOU L, ZHANG Z, GUO H, LOU Q, SHUI G, HUANG H, TIAN H, GUO Y, YUAN P, YANG H, PAN G, WANG R, ZHANG H, YANG S, GUO Y, GE S, LI J, LI Z. Stepwise selection of natural variations atandimproves cold adaptation during domestication ofrice. New Phytologist, 2021, 231(3): 1056-1072.
[58] LI P, LI Y J, ZHANG F J, ZHANG G Z, JIANG X Y, YU H M, HOU B K.UDP‐glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. The Plant Journal, 2017, 89(1): 85-103.
[59] SHI Y, HUY P, LIU Y, CAO S, ZHANG Z, CHU C, SCHLPPI M R. Glycosyltransferase OsUGT90A1 helps protect the plasma membrane during chilling stress in rice. Journal of Experimental Botany, 2020, 71(9): 2723-2739.
[60] SAITO K, HAYANO-SAITO Y, KUROKI M, SATO Y. Map-based cloning of the rice cold tolerance gene. Plant Science, 2010, 179(1/2): 97-102.
[61] CALLIS D J. Ubiquitin, hormones and biotic stress in plants. Annals of Botany, 2007, 99(5): 787-822.
[62] YAN Y S, CHEN X Y, YANG K., SUN Z X, FU Y P, ZHANG Y M, FANG R X. Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Molecular Plant, 2011, 4(1): 190-197.
[63] KOCH K. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Current Opinion in Plant Biology, 2004, 7(3): 235-246.
Genome-wide association study of cold tolerance atthe germination stage of rice
PANG HongBo1, CHENG Lu1, Yu MingLan1, CHEN Qiang2, LI YueYing1, WU LongKun3, WANG Ze1, PAN XiaoWu4, ZHENG XiaoMing5,6
1College of Life Science, Shenyang Normal University, Shenyang 110034;2Experiment Teaching Center, Shenyang Normal University, Shenyang 110034;3College of Grain Science and Technology, Shenyang Normal University, Shenyang 110034;4Rice Research Institute, Hunan Academy of Agricultural Sciences, Changsha 410125;5Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081;6Sanya National Research Institute of Breeding in Hainan, Chinese Academy of Agricultural Sciences, Sanya 571700, Hainan
【Objective】Rice is an important food crop, and its growth and development are most vulnerable at the germination stage. Under cold stress, direct-seeded rice exhibited significantly reduced germination rates (GRs) and yield compared with normally grown plants. Thus, a better understanding of genetic mechanisms regulating cold tolerance will enable to develop rice varieties with improved tolerance during germination. 【Method】238 representative rice germplasm resources from 14 countries worldwide were tested in phenotypic identification in Shenyang in 2021 and 2022; the low-temperature germination rate and relative low-temperature germination rate (LTGR and relative LTGR; 1-10 days under 15℃) were evaluated in an artificial climate incubator, and a 5-10 day LTGR histogram was constructed using R. The day suitable for GWAS was determined by phenotypic variation (Hill) and a mixed linear model combining LTGR and relative LTGR phenotype data with resequencing data. 【Result】LTGR histogram and phenotypic variation showed optimal GR on day 8 (Hill=0.84), i.e., it was higher than on other days (Hill=0.48-0.83), which could be used for GWAS. The principal component analysis results divided all germplasms into five groups—,,,, and. GWAS analysisof two indicators detected three identical significant single nucleotide polymorphisms (SNPs) related to cold tolerance in rice at the germination stage. These were located on chromosome 4, which could explain 11.9%-25.4% of the phenotype. In addition, 24 candidate genes were screened in the 50-kb region upstream and downstream of these three SNPs. Further linkage disequilibrium analysis and haplotype analysis were carried out and highly significant differences were found between different haplotypes of theandgenes for cold tolerance.was divided into five haplotypes by the coding region SNP, and Hap_3 was significantly more cold tolerant than Hap_1;was divided into 18 haplotypes by the coding region SNP and the amino acid variation (S>L) at 77 bp was different inandrice. These results showed that the genes encoding glycosyltransferases () and F-box protein () might be closely related to cold tolerance in rice.【Conclusion】 A total of three SNP loci were detected in 238 rice germplasm resources, and two candidate genes were screened for their association with cold tolerance during germination in rice.
L.; seed germinability; cold tolerance;germination rate; GWAS

10.3864/j.issn.0578-1752.2022.21.001
2022-07-15;
2022-08-17
国家自然科学基金(31970237)、辽宁省自然科学基金面上项目(2022-MS-309)、沈阳市中青年科技创新人才计划(RC190223)、沈阳师范大学“百人计划”拔尖人才项目(SSDBRJH2002012)、沈阳师范大学重大项目孵化工程(ZD202104)
逄洪波(通信作者),Tel:024-86593335;E-mail:panghb@synu.edu.cn。通信作者郑晓明,E-mail:zhengxiaoming@caas.cn
(责任编辑 李莉)
