Mutations of the brpR and brpS genes affect biofilm formation in Staphylococcus aureus
2022-06-07AllisonZankLillianSchulteXavierBrandonLaurenCarstensenAmyWescottWilliamSchwan
Allison Zank, Lillian Schulte, Xavier Brandon, Lauren Carstensen, Amy Wescott, William R Schwan
Allison Zank, Lillian Schulte, Xavier Brandon, Lauren Carstensen, Amy Wescott, William R Schwan, Department of Microbiology, University of Wisconsin-La Crosse, La Crosse, WI 54601, United States
Abstract BACKGROUND In the United States, Staphylococcus aureus (S. aureus) kills tens of thousands of individuals each year and the formation of a biofilm contributes to lethality.Biofilm-associated infections are hard to treat once the biofilm has formed. A new stilbene drug, labeled SK-03-92, was shown to kill S. aureus and affected transcription of two genes tied to a putative two-component system (TCS) we have named brpR (biofilm regulating protein regulator) and brpS (biofilm regulating protein sensor).AIM To determine if BrpR and BrpS regulate biofilm formation, brpR and brpS mutants were assessed using biofilm assays compared to wild-type S. aureus.METHODS A combination of biofilm and quantitative real-time-polymerase chain reaction assays were used. In addition, bioinformatic software tools were also utilized.RESULTS Significantly more biofilm was created in the brpR and brpS mutants vs wild-type cells. Quantitative real-time polymerase chain reactions showed the brpS mutant had differences in transcription of biofilm associated genes that were eight-fold higher for srtA, two-fold lower for lrgA, and 1.6-fold higher for cidA compared to wild-type. Bioinformatic analysis demonstrated that the S. aureus brpR/brpS TCS had homology to streptococcal late-stage competence proteins involved in celldeath, increased biofilm production, and the development of persister cells.CONCLUSION Our study suggests that brpR/brpS is a TCS that may repress S. aureus biofilm production and be linked to late-stage competence in S. aureus.
Key Words: Biofilm; Two-component system; Stilbene; Staphylococcus aureus; Late-stage competence; SK-03-92
lNTRODUCTlON
Staphylococcus aureus(S. aureus) is a significant pathogen of humans, causing more than 700000 skin/soft tissue infections, nearly 120000 bloodstream infections, and close to 20000 deathsperyear in the United States[1-3]. Because drug resistance within this species continues to increase, new drugs are needed to treat human infections. Our research group has developed a new stilbene drug labeled SK-03-92 with efficacy against all Gram-positive bacteria that were tested, including methicillin-resistantS. aureus[4].An mRNA microarray was performed on SK-03-92 treatedvsuntreatedS. aureuscells to try to elucidate the mechanism of action of the drug[5]. From this microarray, the genes for a putative two-component system (TCS) (annotated asMW2284/MW2285) were the most downregulated at the transcriptional level. Moreover, transcription of thesrtAgene (encoding sortase A) was upregulated and thelrgAgene encoding an anti-holin was downregulated following SK-03-92 treatment. Additionally, SK-03-92 treatment led to a high degree of persister cells and greater biofilm formation. Because of the effect on biofilm formation, theMW22284gene was labeledbrpR(biofilm regulating protein regulator) and theMW2285gene was labeledbrpS(biofilm regulating protein sensor).
Transcriptional changes of thesrtAandlrgAgenes as well as high numbers of persister cells suggested that SK-03-92 treatment may induce late-stage competence inS. aureus. Although competence allows DNA uptake to occur in heavily stressed cells, transformation is only one effect of bacterial competence. During early competence, which occurs prior to transformation, a large proportion of the stressed bacterial population dieviaholin-induced autolysis[6]. It is this phenomenon that supplies environmental DNA (eDNA) to the remaining cells for DNA uptake. Additionally, the surplus eDNA provides scaffolding for the rapid formation of a biofilm[7]. The final stage of natural competence is metabolic dormancy[8]. Current estimates show that only the youngest 1% of the original population survive to become a dormant cell. Thus, when faced with resource competition, a thriving bacterial colony has the ability to rapidly transform itself into a small group of latent (i.e.persister) cells living within a biofilm. These surviving cells re-emerge once environmental resources again become plentiful.This is one strategy used by bacterial cells to survive antibiotic challenge and re-infect the host[9].
The initiation of competence has been shown to rely on a symphony of genetic switches that begin to harmonize when short-sequence amino acids, known as competence stimulating pheromones (CSPs)bind to certain specific membrane proteins. The initiation of the CSP alarmone response has been well characterized in streptococcal species[10]. These membrane proteins are autoinducers that comprise one half of a specific TCS[11]. InStreptococcus pneumoniae(S. pneumoniae) andStreptococcus mutans(S. mutans), this response is initiated following the interaction with self-produced autoinducing pheromones known as CSPs, which are short, 14 residue peptides. The CSP is then received by the membrane bound sensor kinase ComD (S. pneumoniae)[12] or BrsM (S. mutans)[13]. Next, ComD or BrsM phosphorylate the cytoplasmic response regulator ComE (S. pneumoniae)[12] or BrsR (S. mutans)[13], which ultimately controls programmed cell death and persistence. InS. aureus, neither the CSP nor the TCS by which CSPs are received have yet been identified.
In this study, we have shown that mutations of thebrpRandbrpSgenes inS. aureusstrain Newman showed greater biofilm formation and transcriptional changes of thesrtAandlrgAgenes than wild-typeS. aureus. Furthermore, we have used bioinformatic tools to show that thebrpR/brpSTCS has homology to the BrsR/BrsM[13] and ComE/ComD[12] late-stage competence TCSs inS. mutansandS. pneumoniae,respectively. These findings suggest that thebrpR/brpSTCS may be specific for the reception and resultant signal cascade of a molecule that induces late-stage competence inS. aureus.
MATERlALS AND METHODS
Bacterial strains, plasmids, and growth conditions
All of the bacterial strains and plasmids used in this study are shown in Table 1. TheS. aureusparent strain Newman was isolated from a human infection[14]. The JE2 strain, created by the University of Nebraska Medical Center, is theS. aureusparent strain USA300 LAC CA-MRSA cured of its plasmids[15]. Strains NE272 (brpS mutant), NE671 (brpR mutant), and NE1787 (srtA mutant) are erythromycinresistant (EmR) mutants representing part of the Nebraska Transposon Mutant Library created by the University of Nebraska Medical Center bymarinertransposon mutagenesis[15] and obtained from the Network on Antimicrobial Resistance inS. aureus(NARSA) strain repository (Table 1). TheE. colistrain DH5α is a cloning strain with mutations that enable high-efficiency transformation[16].S. aureusstrain RN4220 is a transformation efficient strain ofS. aureus[17].

Table 1 Bacterial strains and plasmids used in this study
To clone thebrpRandbrpSgenes for complementation studies, plasmid pALC2073 was used. This plasmid carries ampicillin and chloramphenicol resistance genes,E. coliand Gram-positive origins of replication, and axyl/tetOtetracycline inducible promoter[18].
All media was purchased from Thermo Fisher Scientific (Thermo Fisher Scientific, Pittsburgh, PA,United States). All antibiotics were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO,United States).E. colistrains were grown in Luria (LB) broth shaken at 250 rpm at 37 °C or on Luria agar(LA) incubated at 37 °C. TheE. colistrains carrying the pALC2073 plasmid were selected for in media containing 100 μg/mL ampicillin.
AllS. aureusstrains were grown in brain heart infusion (BHI) broth with 1% (wt/vol) glucose (BHI-G)or trypticase soy broth shaken at 250 rpm at 37 °C. Agar grownS. aureuscultures were passaged on BHI agar at 37 °C. Themarinertransposon mutant strains were grown in media with 5 μg/mL oferythromycin (Em5).S. aureusstrains carrying the pALC2073, pXB3-1, or pAMZ1-3 plasmid were selected for in media containing 10 μg/mL of chloramphenicol (Cm10). To induce thexyl/tetOpromoter on the pXB3-1 and pAMZ1-3 plasmids, tetracycline at 0.25 μg/mL was added to the growth medium.
Transduction of S. aureus
ThebrpR::mariner,brpS::mariner, andsrtA::marinermutations were transduced into strain Newman using the a φ80a bacteriophage[19]. The transductants were then selected for on BHI agar containing Em5. All mutations were verified by polymerase chain reaction (PCR) and biofilm assays.
Construction of brpS and brpR complementing plasmids
ThebrpRcomplementing plasmid was constructed using the pALC2073 backbone[18]. Isolation of pALC2073 plasmid DNA followed the manufacturer’s instructions for the Qiagen QiaPrep plasmid isolation kit (Qiagen, Germantown, MD, United States). The full-length coding region of theS. aureusstrain MW2brpRgene was PCR amplified using the MW2284I/MW2284M primers (Integrated DNA Technologies, Coralville, IA, United States; Table 2) and the following PCR conditions: 35 cycles, 94 °C 1 min, 57 °C 1 min, 72 °C 1 min.S. aureusstrain MW2 chromosomal DNA served as a template. ThebrpRDNA was amplified to have aKpnI site on the 5’ end and anEcoRI site on the 3’ end. PCR amplifiedbrpRgene product was digested withKpnI andEcoRI (New England Biolabs, Ipswich, MA, United States), and then ligated withKpnI/EcoRI cut pALC2073 plasmid DNA using T4DNA ligase (New England Biolabs). Ligated DNA was transformed intoE. colistrain DH5α cells[16]. Transformants were selected on LA containing 100 μg/mL ampicillin, and one resulting plasmid, plasmid pXB3-1, was used for further experiments.

Table 2 Primers used in this study
A recombinantbrpScomplementing plasmid was also constructed. Isolation of pALC2073 plasmid DNA used the same plasmid isolation kit described above. The full-length coding region of theS. aureusstrain MW2brpSgene was PCR amplified using the GEX-5XB/GEX-5XC primers (Table 2) and the following PCR conditions: 35 cycles, 94 °C 1 min, 57 °C 1 min, 72 °C 1 min.S. aureusstrain MW2 chromosomal DNA was used as the template. ThebrpSDNA was amplified to have aKpnI site on the 5’end and anEcoRI site on the 3’ end. PCR amplifiedbrpSgene product was digested withKpnI andEcoRI(New England Biolabs), and then ligated withKpnI/EcoRI cut pALC2073 plasmid DNA using T4DNA ligase (New England Biolabs) immediately downstream from the tetracycline-induciblexyl/tetOpromoter on pALC2073. Ligated DNA was transformed intoE. colistrain DH5α cells[16]. Transformants were selected on LA containing 100 mg/mL ampicillin, and one resulting plasmid, plasmid pAMZ1-3,was used for further experiments.
Plasmid DNA fromE. coliwas purified with a Qiagen Plasmid Miniprep Kit (Qiagen) and electroporated into theS. aureusstrain RN4220[20] using a GenePulser, (Bio-Rad, Hercules, CA, United States)under the following conditions: 100 W capacitance, 25 mF resistance, 2.5 kV charge voltage, 4 s.Transformants were selected for on BHI agar containing Cm10after one hour of expression in BHI broth.Finally, plasmid DNA was re-isolated from oneS. aureusstrain RN4200 transformant carrying either the pXB3-1 or pAMZ1-3 plasmid using the method noted above with one alteration. TheS. aureuscells were incubated with 50 μL of lysostaphin (10 mg/mL; Remel, San Diego, CA, United States) for 60 min at 37°C prior to the first step to facilitate lysis of the staphylococcal cells. Each isolated plasmid DNA sample was then cut with theKpnI andEcoRI restriction endonucleases to verify the insertion. TheS. aureusstrain Newman was then transformed with 10 mL of pXB3-1 or pAMZ1-3 plasmid DNA using electroporation as outlined above and transformants selected for on BHI agar containing Cm10.
Biofilm assays
To determine the effect of thebrpRandbrpSmutations on the ability ofS. aureusNewman to form a biofilm, biofilm assays were performed[21]. Briefly, cultures of theS. aureuswere grown at 37 °C with shaking (250 rpm) overnight in BHIG broth with the appropriate antibiotic(s). Each strain was then diluted 1:100 in BHI-G and 220 μL of the solution was placed in microtiter wells in triplicate in a 96-well microtiter plate. The microtiter plates were statically incubated for 24 h at 37 °C to allow a biofilm to form. Each well was then rinsed three times with sterile water. The biofilms were then allowed to settle(10 min), stained with crystal violet dye (0.1% wt/vol) for 10 min, and then washed with sterile water.After allowing the well contents to dry fully in a sterile hood, the dried contents were incubated in 33%acetic acid at room temperature for 30 min. The contents of the well were vigorously curettaged. The optical densities were measured on a SpectraMax M3 96-well microtiter plate reader (Molecular Devices, San Jose, CA, United States) at an optical density of 570 nm. In addition to wild-type Newman cells, NewmanbrpRand NewmanbrpSmutant strains as well asbrpRandbrpSmutants containing the pXB3-1 or pAMZ1-3 plasmids were tested and compared with a NewmansrtAtransposon mutant strain that served as a negative control. To achieve statistical significance, the biofilm assays were performed a minimum of five times in triplicate for each strain.
Quantitative real-time PCR
Total RNA was isolated fromS. aureusstrains grown to early logarithmic phase in BHI broth with shaking (250 rpm) incubated at 37 °C using a High Pure RNA Isolation kit ΔΔ(Roche Diagnostics,Indianapolis, IN, United States) with an additional lysostaphin treatment step to help lyse theS. aureuscell walls and a DNase I digestion to digest contaminating DNA. To confirm RNA concentration and ensure the integrity of each RNA sample, an aliquot of each RNA sample was analyzed on a Nanodrop machine (Thermo Scientific, Waltham, MA, United States) and electrophoresed through 0.8% agarose gels. The cDNA for each strain was then synthesized from 2 μg of total RNA according to manufacturer’s instruction using a First-Strand Synthesis kit (Life Technologies, Carlsbad, CA, United States). All quantitative real-time PCR (qRT-PCR) trials were performed according to manufacturer’s instruction using the iTaq Universal SYBR Supermix kit (BioRad, Hercules, CA, United States).Oligonucleotide primers that targeted theftsZ,srtA,lrgA, andcidAgenes were synthesized (Table 2) by Integrated DNA Technologies. To perform qRT-PCR, the minimum information for publication of quantitative real-time PCR experiments guidelines were followed and theftsZhousekeeping gene was used as a standardization control[22]. All replicates were performed at least three times on a CFX96 qPCR instrument (BioRad, Hercules, CA, United States) under the following conditions: 94 °C, 20 s; 55°C, 30 s; and 72 °C, 1 min for 35 cycles. The level of target gene transcript from each strain was estimated against theftsZgene standard curve. Additionally, crossover points for all genes were standardized to the crossover points forftsZin each sample using the 2-ΔΔCTformula[23].
Bioinformatic tools
The sequenced genomes ofS. aureusstrains MW2 and Newman used in this study are publicly available on GenBank (NCBI, genome assembly ASM1126v1)[24-26]. The protein annotations for all of the bacterial strains included in this study were found on BioCyc or GenBank[26,27]. BioCyc was also used to search for brpRS homologs downstream of themqo2gene. The UniProt Consortium was used to obtain amino acid FASTA sequences[28]. Domain motifs were sought using NetPHOS, ExPASy, Prosite,and GenomeNet[28-30]. Protter was used to two-dimensionally visualize brpS and brsM[31]. I-TASSER and PyMOL were used together to three dimensionally visualize BrpR and BrpS[32,33]. I-TASSER and PyMol were also used to visually verify DNA binding in residues predicted by DP-Bind[34]. Finally,protein sequence homology analyses were performed by BLASTp (NCBI) with the following parameters: Max target sequences = 100, automatically adjusted parameters for short input sequences,expect threshold = 10, word size = 3, max matches in a query range = 0, matrix = BLOSUM62, gap costs= 11 existence and 1 extension, and a conditional compositional score matrix adjustment[35].
Statistical analysis
Calculation of the means, standard deviations, and paired Student’s t-tests were performed using Microsoft Excel.P< 0.05 were considered significant.
RESULTS
Alignment of the brpS/brpR genes
An alignment of thebrpRandbrpSgenes that encode the BrpR and BrpS proteins is seen on theS. aureusMW2 genome sequence (Figure 1)[24]. These genes overlap in a unidirectional in-tandem sequence. The overlappingbrpRSgenes lie just 66 base pairs upstream from themqo2gene, encoding one of the two malate: Quinone oxidoreductases (MQO2) produced byS. aureus. The bi-functional MQO2 protein is able to generate oxaloacetate through an oxidation of malate as well as donate electrons to the electron transport chain[36].
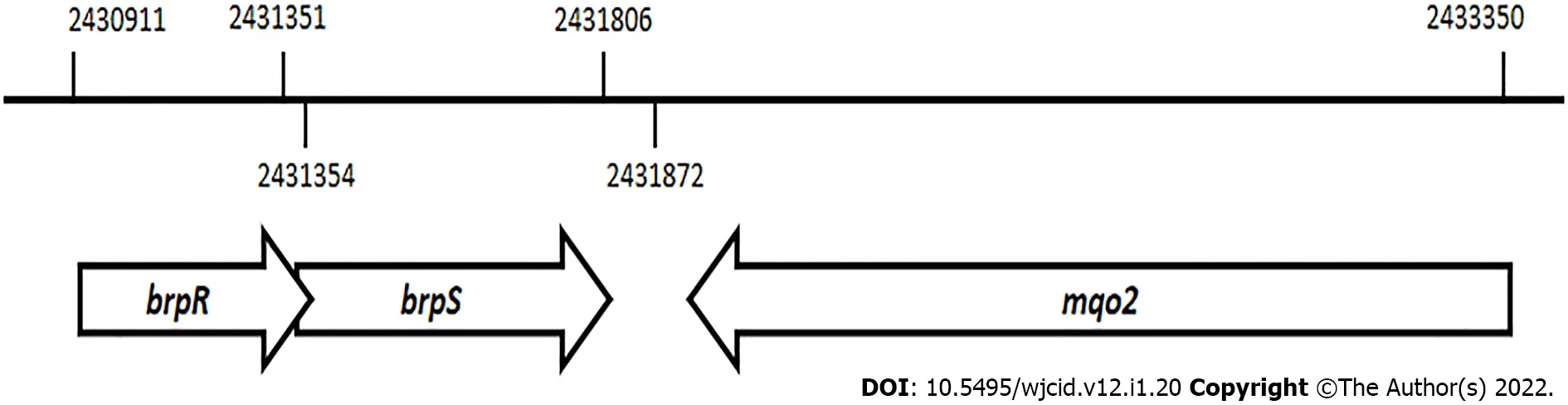
Figure 1 Schematic representation of the chromosomal position and organization of the brpR, brpS, and mqo2 genes in the Staphylococcus aureus strain MW2 genome.
Mutations in the brpR and brpS genes cause greater biofilm formation in S. aureus
To confirm biofilm formation was linked to BrpR and BrpS, individualbrpRandbrpSmutations were moved to theS. aureusstrain JE2 background[15] to theS. aureusstrain Newmanviatransduction.Biofilm production of both mutants was compared to the unmutated wild-typeS. aureusNewman strain. Significantly more biofilm material was produced by thebrpSandbrpRmutants (1.8-fold and 1.73-fold higher, respectively,P< 0.001) compared to wild-type. Complementation of thebrpRandbrpSmutants caused biofilm expression to either return to wild-type levels or there was less biofilm material formed (Figure 2). ThesrtAmutant displayed a 1.73-fold decline in the biofilm forming ability compared to the wild-type strain (P< 0.001). This suggested that the putative BrpRS TCS may repressS.aureusbiofilm production.
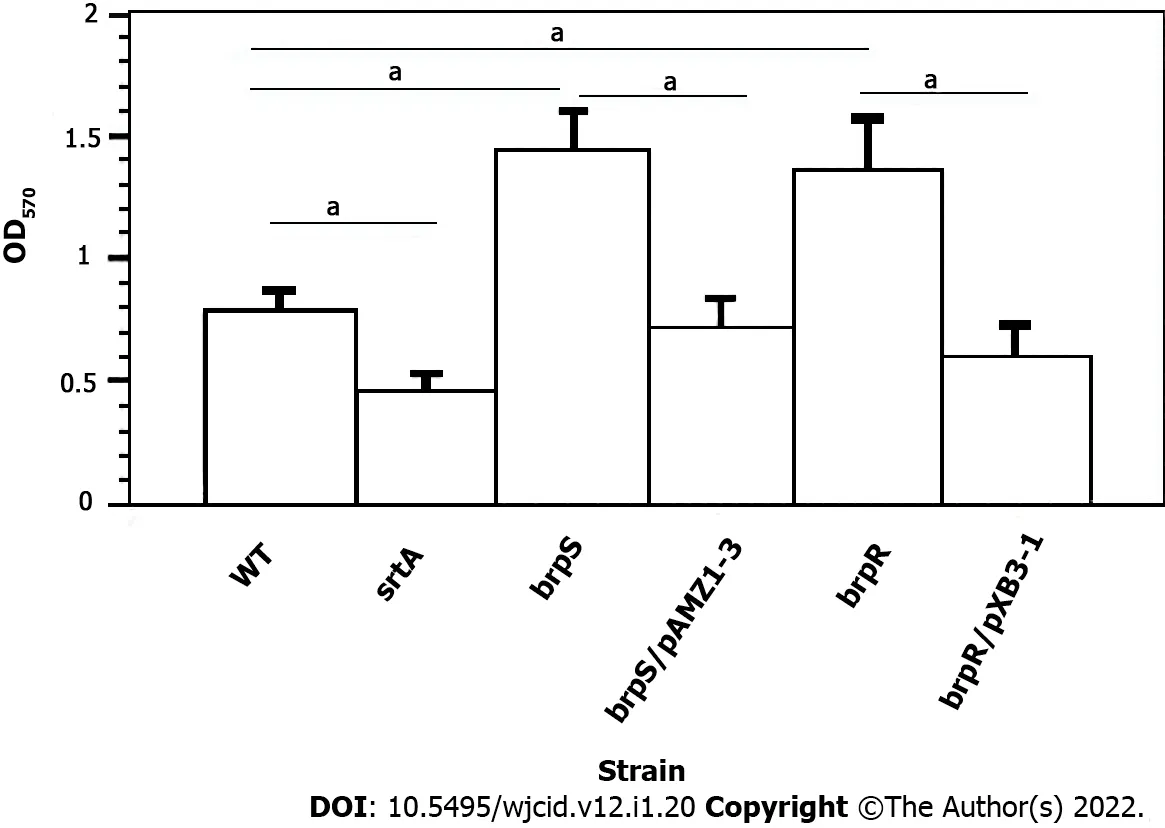
Figure 2 Effect of brpR and brpS mutations and complementation on Staphylococcus aureus biofilm formation. All experiments represent the mean ± SD from five different runs done in triplicate for each strain. Biofilm formation was done on wild-type Staphylococcus aureus (S. aureus) strain Newman (WT,open column), S. aureus Newman srtA mutant (black column), S. aureus Newman brpS mutant (right striped column), S. aureus Newman brpS mutant/pAMZ1-3 (left striped column), S. aureus Newman brpR mutant (dark right striped column), and S. aureus Newman brpR mutant/pXB31 (dark left striped column). Differences were statistically compared by analysis of variance where aP < 0.001.
Transcription of srtA and lrgA are regulated by a brpS mutation
Previously, we showedsrtAtranscription was elevated andlrgAtranscription was lower after SK-03-92 treatment ofS. aureuscells compared to untreated cells[5]. ThesrtAgene encodes sortase A[37] and thelrgAgene encodes an anti-holin[38] that are important for the formation of biofilms[39,40]. Total RNA was collected during the mid-exponential growth phase from theS. aureusNewmanbrpSmutant,S.aureusNewmanbrpSmutant containing the pAMZ1-3 plasmid, and wild-type S. aureusNewman cells Each RNA sample was converted to cDNA for qRT-PCRs analysis. ThebrpSmutant displayed 8.5-fold highersrtAtranscription (P< 0.008), 2-fold lowerlrgAtranscription (P< 0.016), and 1.6-fold highercidAtranscription (P< 0.43)vsthe wild-type strain (Figure 3). Complementation of thebrpSmutation causedsrtAtranscription to drop to a 2.2-fold increase, a 1.3-fold increase inlrgAtranscription, and a 1.2-fold increase incidAtranscription. These results demonstrated that transcription of some biofilm-associated genes was regulated by a mutation in thebrpSgene.

Figure 3 Quantitative reverse transcribed-polymerase chain reaction results of Staphylococcus aureus Newman cidA, lrgA, and srtA transcription in wild-type bacteria (standardized to 0) compared to a Staphylococcus aureus Newman brpS mutant (black column) and the complemented brpS mutant (white column). The data represents the mean ± SD from three separate runs.
BrpR/BrpS homology to other TCS proteins
BrpR and BrpS homologs were identified by BLAST analyses in multiple Gram-positive bacterial pathogens, includingBacillus cereus, Clostridioides difficile, Enterococcus faecalis, Lactobacillus species,Staphylococcus haemolyticus, Streptococcus pneumoniaeComD/ComE, and S. mutansBrsR/BrsM as well as three other bacterial species (Escherichia coliYehT/YehU, Mycobacterium tuberculosisYehT/YehU, andChlamydia trachomatis)[26,35]. Of these, theS. mutansBrsR/BrsM TCS that senses CSP and then induces late-stage competence showed the highest homology (Figure 4)[12].
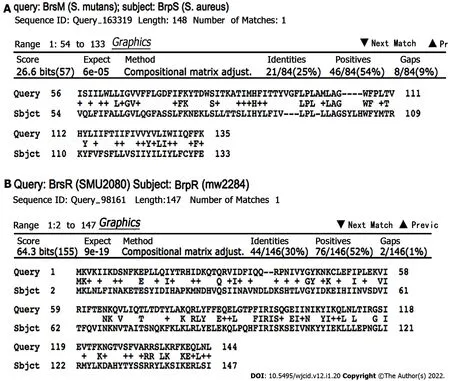
Figure 4 Bioinformatic comparison of the BrsRM proteins of Streptococcus mutans with the BrpRS proteins of Staphylococcus aureus.A: BrsM, a two-component system membrane protein responsible for activating competence in response to sensing competitor organisms within a niche, shares sequence similarity with BrpS; B: BrsR, which is the cognate response regulator to BrsM, shares sequence similarity with BrpR. BLASTp NCBI. Algorithm parameters:Max target sequences = 100, automatically adjusted parameters for short input sequences, expect threshold = 10, word size = 3, max matches in a query range = 0,matrix = BLOSUM62, gap costs = 11 existence and 1 extension, and a conditional compositional score matrix adjustment.
Putative structures of the BrpS protein
S. aureusBrpS andS. mutansBrsM have a similar arrangement of reactive residues. Lysine, serine,threonine, histidine, tyrosine, and glutamic acid residues were illuminated on a 2-dimensional Prottergenerated image of each protein (Figure 5)[29]. This mapping suggested that BrpS and BrsM are partitioned into distinct functional domains separated by the membrane. Functionality appears to occur at the intercellular loop (staphylococcal N’-76-KYTDWSITKAT-86-C’), at the extracellular loop (staphylococcal N’-108-PLTVHY-113-C’), and within a single reactive residue near the membrane at K135(staphylococcal) within the C’-terminal tail region.

Figure 5 Highlight of amino acid residues shared by competence stimulating pheromone, competence stimulating pheromone-2, and BrpS. The predicted exterior segment of BrpS, which spans N’-1-MKNLKNSLFISLIIGLSLSLFFSMLFADGKYYPLNPQSTIGILYYTHFT-50-C’, was compared to competence stimulating pheromone (CSP) and CSP-2 by BLASTp. The competence stimulating peptides of Streptococcus mutans (1-SGSLSTFFRLFNRSFTQA-18,CSP) and Streptococcus pneumoniae (1-EMRISRIILDFLFLRKK-17, CSP-2) has 56% similarity to CSP and 30% similarity to CSP-2. CSP: Competence stimulating pheromone.
Additionally, the region at the N’-terminus of brpS displays sequence homology with the secretedS.mutansandS. pneumoniaeCSPs (Figure 6). The segment of brpS spanning the regions from N’-1-MKNLKNSLFISLIIGLSLSLFFSMLFADGKYYPLNPQSTIGILYYTHFT-50-C’ showed 56% similarity with CSP (S. mutans, 1-SGSLSTFFRLFNRSFTQ A-18) and 30% similarity to CSP-2 (S. pneumoniae,1-EMRISRIILDFLFLRKK-17).
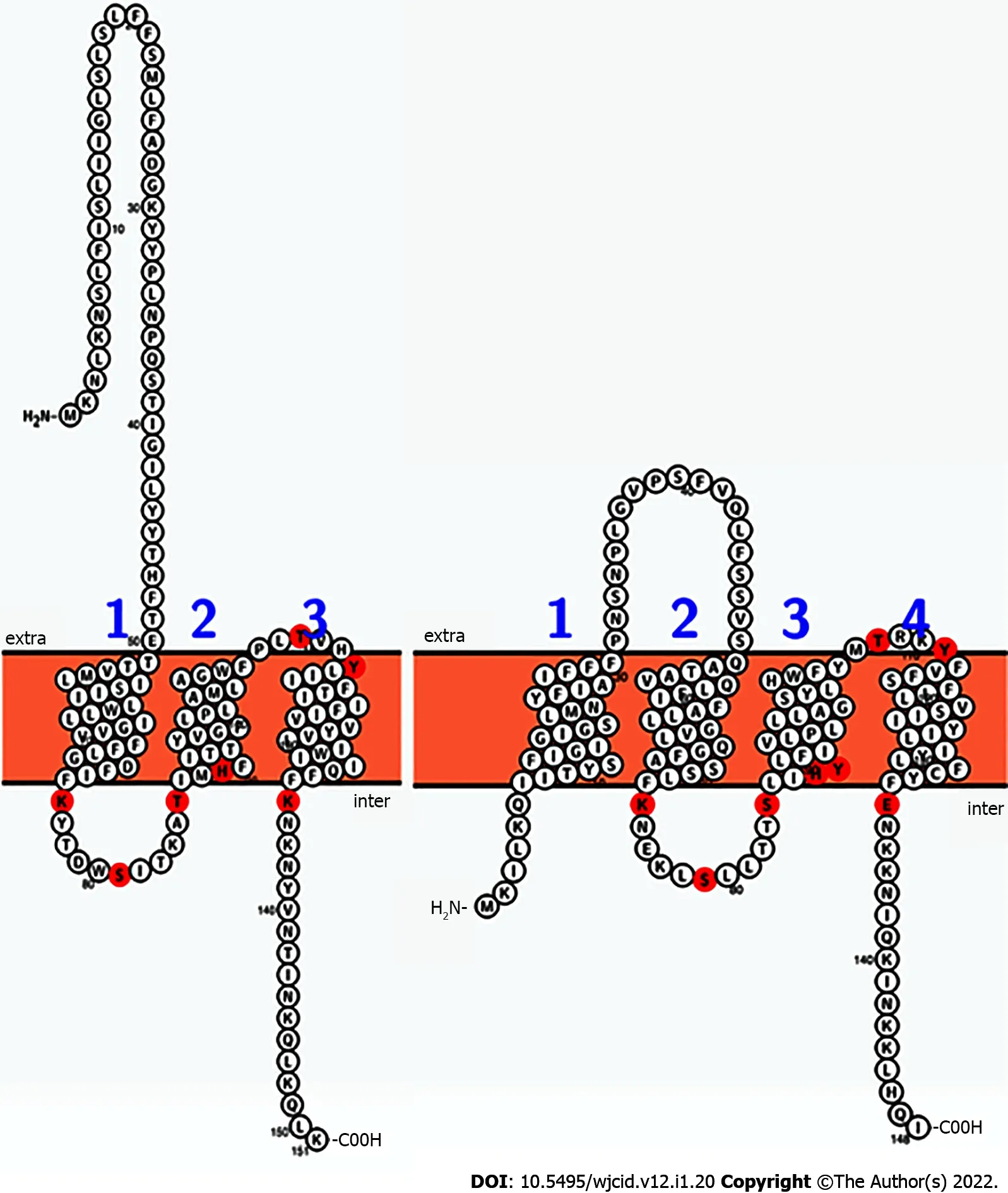
Figure 6 Comparison of the sequences and predicted topologies of the putative two-component system membrane sensor BrpS (left,Staphylococcus aureus) and BrsM (right, Streptococcus mutans). According to this prediction, residues likely to be reactive (red) are topologically arranged in similar loci among both proteins. Intra is proposed to correspond with the cytoplasmic space, and extra is proposed to correspond with the extracellular milieu of the cell. Figures generated by Protter.
DlSCUSSlON
S. aureuscauses 65% of biofilm-associated infectionsperyear[40,41]. Biofilms provide a defense against host immune defenses as well as most antibiotics. An understanding of what regulatesS. aureusbiofilm formation could lead to treatment options that target this process inS. aureus.
Both sortase A (SrtA) and antiholin (LrgA) are importantS. aureusproteins needed for creation and maintenance of biofilms. Sortase A promotes the covalent anchoring of surface proteins to the cell wall ofS. aureus[42] that are important in the first stage of biofilm formation. Cell death releases eDNA that is tied to holin/antiholin action. An integral part of matureS. aureusbiofilms is eDNA[7]. The function of the antiholin LrgA is to prevent cell autolysis by complexing with CidA holins[38,43].
In this study, we have shown that mutations in either thebrpRorbrpSgene cause an increase in biofilm formation as well as transcriptional changes of thesrtAandlrgAgenes, which are linked events.Previous studies with transposon mutants of what was an uncharacterized gene, that we have namedbrpS,displayed better biofilm formation than the wild-type strain[44,45]. Strains with a mutatedlrgAgene have also been shown to produce increased levels of biofilms[46]. Another study has shown that cell lysis caused eDNA to be rapidly produced that could act as a scaffolding for newly forming biofilms[10].
Thein silicodata; biofilm results with thebrpRandbrpSmutants; and the data from the transcript abundance changes of thelrgA, srtA, brpR,andbrpSgenes suggest that BrpR/BrpS comprise a TCS that may be involved in late-stage competence. From thein silicoanalysis, we speculate that the BrpR protein(that possesses an apparent LytTR DNA binding-motif[47]) may represssrtAtranscription. Other proteins that have LytTR motifs, such as BrsR and ComE, have been shown to have multifunctional activities tied to activation and repression[47-49]. Further analysis is required to show that BrpR is capable of binding to this region.
From the data presented, we speculate that BrpS is a receptor for a CSP-like pheromone secreted byS.aureusas a response to competition for resources. The leader peptide of BrpS may function to competitively antagonize the extracellular receptor portion of BrpS from the CSPs of competitive species, such asS. mutansandS. pneumoniae,that inhabit the human upper respiratory tract.
A number of previously completed studies focused on biofilm production and bacterial cell viability due to interactions with CSP-like pheromones. A study by Zhanget al[49] demonstrated that withinS.mutansthere was a 76.3% decline in cell viability and biofilm mass increased by 89.3% following the addition of CSP to bacterial growth media[49]. In addition, supernatant collected fromS. mutansthat was co-cultured withAggregatibacter actinomycetemcomitanscaused a 1.3-fold rise in biofilm production withinS. mutans[50]. Ample evidence of cell death after CSP exposure has been documented by several studies, however, biofilm production has not been normalized to the viable bacterial cells that remain[51-53]. Nevertheless, a number of Gram-positive bacterial species show cell viability and subsequent biofilm production correlate with the level of CSP added to the media. Further studies should be done to assess the actual increase in biofilm formation by taking into account the findings that competence is accompanied by massive cellular death.
We also believe that there may be a connection between metabolic dormancy and the BrpR/BrpS TCS. If malate production is interrupted after BrpR binds to the sigma factor binding sites, malate conversion to oxalacetate would be halted. As a consequence of this interruption, any acetyl groups generated by acetyl-CoA would not interact with citrate within the citric-acid cycle. Thus, too much acetyl-CoA would arise within the cell. Because these functional groups would be liberated, it is possible that there would be an epigenetic modification and BrpR would be rapidly released from thesrtAgene enhancer region. By freeing BrpR from thesrtAgene enhancer region, additional BrpR molecules would be available to interact with sigma factors, blocking transcription ofbrpRSthat would lead to even less transcription of themqo2gene. The work by Zhanget al[54] used profiling of lysine acetylomes inS. aureusandE. colito identify a sequence motif, which supports our idea that BrpR may epigenetically block DNA-binding[54]. As part of that study, 412 proteins and 1361 lysine sites were cross-referenced against each other, which led to a conserved motif, N’-RLYELExQLxxxFIRISKxxEIVNC’, being identified. BrpR has this conserved motif, which is very well conserved among a number of bacterial species. By shutting down malate expression, persister cells could form suddenly as a response to late-stage competence or treatment with the SK-03-92 drug. Thus, BrpR repression of malate production could be connected to formation of persister cells that is a feature of late-stage competence.
CONCLUSlON
Our study suggests that BrpR/BrpS is a TCS that may repressS. aureusbiofilm production and be linked to late-stage competence inS. aureus.
ARTlCLE HlGHLlGHTS
Research background
Staphylococcus aureus (S. aureus) is a primary cause of skin/soft tissue infections. Biofilm formation is a key component of S. aureus pathogenesis. Thus, an understanding of what regulates biofilm formation in S. aureus is important.
Research motivation
We were interested in characterizing two open reading frames that we thought were tied to biofilm formation in S. aureus.
Research objectives
Determine if mutations in the brpR and brpS genes affected biofilm formation and what the respective proteins had homologies with.
Research methods
We used biofilm assays and quantitative real-time-polymerase chain reaction (qRT-PCR) analysis to test brpR and brpS mutants compared to the parent strain of S. aureus. Bioinformatic tools were used to determine what roles the BrpR and BrpS proteins may play in S. aureus cells.
Research results
The biofilm and qRT-PCR analyses demonstrated that mutations in the brpR and brpS genes affected biofilm formation in S. aureus and led to transcriptional differences in key biofilm-related genes as compared to the parent strain. Further, the BrpR and BrpS proteins share homologies with proteins involved in late-stage competence in streptococcal species.
Research conclusions
BrpR/BrpS are likely a new two-component system which regulates biofilm formation inS. aureus.
Research perspectives
A better understanding of a new regulator ofS. aureusbiofilm formation has been identified.
ACKNOWLEDGEMENTS
We wish to thank Jean Lee, Ambrose Cheung, Jo Handelsman, and NARSA for bacterial strains and plasmids used in this study as well as Jennifer Klein and the Molecular Biology Laboratory students who ran some of the qRT-PCRs.
FOOTNOTES
Author contributions:Zank A, Wescott A, and Schwan WR designed the research study; Zank A, Schulte L, Brandon X, Carstensen L, Wescott A, and Schwan WR performed the research; Zank A, Brandon X, and Schwan WR contributed new plasmids; Zank A, and Schwan WR analyzed the data and wrote the manuscript; and all authors have read and approved the final manuscript.
Supported byNational Science Foundation Graduate Research Program to Zank A, No. 0002016179620.
lnstitutional review board statement:No humans or samples from human were used in this study.
lnstitutional animal care and use committee statement:No animals were used in this study.
Conflict-of-interest statement:Schwan WR holds a composition of matter and use patent covering the SK-03-92 Lead compound.
Data sharing statement:The authors will share their data with whomever asks.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:United States
ORClD number:Allison Zank 0000-0001-6264-853X; Lillian Schulte 0000-0003-4175-0193; Xavier Brandon 0000-0002-2108-793X; Lauren Carstensen 0000-0002-3976-9023; Amy Wescott 0000-0002-3581-3365; William R Schwan 0000-0003-3076-1815.
S-Editor:Fan JR
L-Editor:A
P-Editor:Cai YX
