Methanotrophy-driven accumulation of organic carbon in four paddy soils of Bangladesh
2022-05-11NasrinSULTANAJunZHAOYuanfengCAIGMustafizurRAHMANMohammadSaifulALAMMohammadFAHEEMAdrianHOandZhongjunJIA
Nasrin SULTANAJun ZHAOYuanfeng CAIG.K.M.Mustafizur RAHMANMohammad Saiful ALAMMohammad FAHEEMAdrian HO and Zhongjun JIA*
1 State Key Laborator y of Soil and Sustainable Agriculture,Institute of Soil Science,Chinese Academy of Sciences,Nanjing 210008(China)
2 University of Chinese Academy of Sciences,Beijing 100049(China)
3 Depar tment of Agroforestr y and Environmental Science,Faculty of Agriculture,Sher-e-Bangla Agricultural University(SAU),Sher-e-Bangla Nagar,Dhak a-1207(Bangladesh)
4 Depar tment of Soil Science,Faculty of Agriculture,Bangabandhu Sheikh Mujibur Rahman Agricultural University(BSMRAU),Gazipur-1706(Bangladesh)5 Institute for Microbiology,Leibniz Universität Hannover,Herrenhäuser Straße 2,Building 4104,Hannover D-30419(Ger many)
ABSTRACT Biological methane oxidation is a crucial process in the global carbon cycle that reduces methane emissions from paddy fields and natural wetlands into the atmosphere.However,soil organic carbon accumulation associated with microbial methane oxidation is poorly understood.Therefore,to investigate methane-derived carbon incorporation into soil organic matter,paddy soils originated from different parent materials(Inceptisol,Entisol,and Alfisol)were collected after rice harvesting from four major rice-producing regions in Bangladesh.Following microcosm incubation with 5%(volume/volume)13CH4,soil 13C-atom abundances significantly increased from background level of 1.08%to 1.88%—2.78%,leading to a net methane-derived accumulation of soil organic carbon ranging from 120 to 307 mg kg-1.Approximately 23.6%—60.0%of the methane consumed was converted to soil organic carbon during microbial methane oxidation.The phylogeny of 13C-labeled pmoA(enconding the alpha subunit of the particulate methane monooxygenase)and 16S rRNA genes further revealed that canonicalα(type II)andγ(type I)Proteobacteria were active methane oxidizers.Members within the Methylobacter-and Methylosarcina-affiliated type Ia lineages dominated active methane-oxidizing communities that were responsible for the majority of methane-derived carbon accumulation in all three paddy soils,while Methylocystis-affiliated type IIa lineage was the key contributor in one paddy soil of Inceptisol origin.These results suggest that methanotroph-mediated synthesis of biomass plays an important role in soil organic matter accumulation.This study thus supports the concept that methanotrophs not only consume the greenhouse gas methane but also serve as a key biotic factor in maintaining soil fertility.
Key Words: 16S rRNA gene,DNA-based stable-isotope probing(DNA-SIP),methane oxidation,methanotroph,pmoA,rice soil,soil organic carbon,soil organic matter
INTRODUCTION
Soil organic carbon(SOC)plays a vital role in soil fertility by shaping soil physical structure and modulating soil biogeochemical processes(Panakouliaet al.,2017;Schjnninget al.,2018).As the key component of soil organic matter(SOM),SOC is originated from three major sources,including shoot litter,root residues/exudates,and microbial biomass(Nguyen,2003).Microbe-derived carbon(C),rather than plant originated C,was arguably considered to be the primary source of SOC(Barret al.,2018),particularly in the deeper soil layers(Fontaineet al.,2007;Schmidtet al.,2011).The microbial residues and exudates occupied up to 80%of C in SOM fractions(Gleixneret al.,2002;Kiem and Kgel-Knabner,2002;Liang and Balser,2011),and microbial biomass accounts for 1%—5%of SOC(Jenkinson and Ladd,1981),which is relatively stable and essential in the biochemical processes of SOC and soil elemental cycling.The microbial biomass serves as an engine for SOC turnover by balancing SOMmineralization and stabilization processes(Srensen,1983;Simpsonet al.,2007;Liang and Balser,2011).
Methane(CH4)is a potent greenhouse gas emitted from both the natural wetlands and anthropogenic rice paddies,and its concentration in the atmosphere has exhibited a remarkable increase relative to its level in the pre-industrial era(Wahlen,1993;Etminanet al.,2016).Inundated paddy fields are one of the vital sources of atmospheric methane(Minami and Neue,1994),broadly distributed in Southeast Asia and other(sub)tropical areas worldwide.Bangladesh is a subtropical low-lying deltaic country in South Asia and the third largest rice-producing country with 36million tons produced every year(BRRI,2019).The tropical monsoon climate,low-lying land,and irrigated rice farming system enable enhanced methane emissions during the rice-growing seasons.It was estimated that the average methane release from the rice paddies in Bangladesh is as high as 1 071 Gg year-1(Khan and Saleh,2015).Therefore,an adequate understanding of methane consumption and C metabolism associated with active methane oxidation is important for sustainable rice production in Bangladesh,as aerobic methanotrophs arguably account for up to 80%of the methane produced in paddy soils.
Methane is continuously produced in inundated rice fields by methanogenic degradation of organic matter under anoxic conditions(Reeburgh,1996).Methanotrophs are the only biological sink of atmospheric methane in terrestrial ecosystems.They can consume methane at high concentrations(Conrad,2009).For example,methane up to 50 000 μL L-1was detected and consumed within the oxic zone of rice fields(Nouchiet al.,1990;Eller and Frenzel,2001).It is estimated that nearly 80%of the methane originated from submerged rice fields is reduced by methanotrophs(Frenzelet al.,1992),and methane emission would likely double if the anaerobically produced methane is not consumed by methanotrophs(Le Mer and Roger,2001).During methane oxidation,organic intermediates,e.g.,formaldehyde,are produced and further assimilated into the methanotrophic cell biomass(Anthony,1982).The synthesized microbial biomass either supports other organisms as C substrate(e.g.,through predation and cross-feeding(Bastvikenet al.,2003)or becomes soil organic components(Kindleret al.,2009),contributing to the stable SOC as recently proposed(Miltneret al.,2012;Kallenbachet al.,2016).
Soil organic Calso contains humic fractions that are heterogeneous high-molecular-weight organic materials,which can serve as electron acceptors for the anaerobic oxidation of organic compounds and hydrogen(Lovleyet al.,1996).It is speculated that humic substances catalyze the oxidation of organic C by mediating the reduction of the oxidized iron(Scottet al.,1998),as oxidized iron indeed acts as an electron acceptor for anaerobic oxidation of methane(AOM(Bealet al.,2009).Subsequent flux measurement showed significantly lower methane emission upon the addition of humic acids in peat soils,implying the reduction of humic substance coupled to AOM(Blodau and Deppe,2012).Stable-isotope probing(SIP)has demonstrated that humic substances could serve as electron shuttles to fuel anaerobic methane oxidation coupled to iron(III)reduction(Heet al.,2019).Recent studies have shown that humic substances facilitate anaerobic methane oxidation in a variety of environments,e.g.,denitrifying reactors(Baiet al.,2019),wetland sediments(Valenzuelaet al.,2019),and paddy soils(Fanet al.,2020).It is also anticipated that humic substancemediated AOMpotentially contributes to the global sink of methane at levels of up to 114 Tg CH4year-1in coastal wetlands(Valenzuelaet al.,2017)and plays an important role in nitrous oxide(N2O)reduction as well(Valenzuelaet al.,2020).Intriguingly,the accumulation of SOC derived from microbial methane oxidation in wetland environments remains poorly understood.
Aerobic methanotrophs that belong toαandγProteobacteria are termed as type II and type I methanotrophs,respectively.They thrive under high methane conditions and regulate the rate of aerobic methane oxidation in wetland and rice fields(Caiet al.,2016;Zhaoet al.,2020).Although more recently discovered,Verrucomicrobia(designated as type III)(Op den Campet al.,2009;Knief,2015)and candidate division NC10(Ettwiget al.,2009)also play important roles in reducing methane emission to the atmosphere from acidic and/or inorganic nitrogen(N)-rich environments.However,conventional methanotrophs contribute to the majority of microbial methane oxidation in rice soil owing to their extraordinarily faster growth rates than those of NC10 and Verrucomicrobia(Caiet al.,2016).Despite the high diversity of methanotrophs in paddy soils,methane oxidation is usually controlled by distinct phylotypes of methanotrophs,the biogeography of which is best explained by soil pH(Zhaoet al.,2020).However,it remains unclear whether soils dominated by distinct active methanotrophic groups have different methane consumption efficiency,since it is challenging to estimate the proportion of methane-derived C converted to SOC during methane oxidation and to assess the contribution of methanotrophs to SOM formation in paddy fields with different soil parent materials.Methanotrophy is an active microbial process in paddy soils.We hypothesized that methane-derived C could be an important source of new SOC in soils,and it is largely determined by active methanotrophic composition in paddy field soils.In this study,we employed a stable isotope-tracing approach with DNA-based molecular tools to assess the conversion efficiency of methane-derived C into SOC,and to identify the active methanotrophs responsible for methane oxidation and assimilation based on high-throughput sequencing of the 16S rRNA gene and thepmoAgene(encoding the alpha subunit of the particulate methane monooxygenase).ThepmoAgene is a widely used functional molecular marker for the detection of active methanotrophs during methane oxidation,and we applied the A189F/mb661R primer set to amplify thepmoAgene fragments(Costello and Lidstrom,1999).
MATERIALS AND METHODS
Paddy soils used
Soil samples were collected from four different regions,including Shirajgong(SG),Kushtia(KT),Mymensingh(MS),and Gazipur(GZ),representing 90%of rice cultivated areas in Bangladesh(Fig.S1,see Supplementary Material for Fig.S1).The geographical and physicochemical properties of the four paddy soils are shown in Table I.
From each field(around 300—400 m2),a composite soil sample was collected by mixing three random cores from 0—20 cm depth after rice harvesting on May 2015.The soil samples were homogenized by passing through a 2-mm sieve and stored at 4°C prior to incubation.A subsample of about 200 g of each sample was stored at-20°C for microbial and molecular analyses.Another subsample was air-dried to measure physicochemical properties,including soil pH,soil total organic C,total N,and cation exchange capacity(CEC),as previously described(Wanget al.,2015).
Microcosm incubation experiment
A microcosm incubation experiment was set up with three replicates.The four paddy soils were incubated under three different atmospheric conditions,including control(ambient air),high12CH4(with 5%12CH4),and high13CH4(with 5%13CH4),in a similar manner to that of a previous study(Sultanaet al.,2019).The fresh soil(equivalent to an air-dried weight of 6.0 g of soil)was placed into a 120-mL serum bottle sealed with a butyl stopper and incubated at 60%maximum water holding capacity at 28°Cin the dark.For the high13CH4and high12CH4treatments,the13C-labeled(>99%13C-atom purity)and unlabeled CH4gas,respectively,was injected into the bottles on the first day of incubation to reach a methane gas mixing ratio of approximately 5%(equivalent to 50 000μL L-1).Methane concentration in the headspace was monitored twice a day by a gas chromatograph equipped with a flame-ionization detector(Shimadzu GC12-A,Shimadzu Corporation,Japan),and the standard curve was generated with methane concentrations of 100,1 000,5 000,10 000,50 000,and 80 000μL L-1.Destructive sampling was conducted when methane concentration decreased to below 5 000μL L-1in the microcosms,i.e.,more than 90%of the methane added had been consumed.Soil samples were collected and frozen at-20°C after incubation for 7,15,18,and 24 d for SG,KT,MS,and GZ,respectively.
Methane oxidation and production rates
Methane oxidation rate was calculated according to the following equation:

whereRoxis the methane oxidation rate(μmol g-1h-1);Vis headspace volume of the bottle(L);ρis the methane density under standard conditions(1.01×105Pa,273.15 K),i.e.,0.72 g L-1;Ciis the CH4initial concentration in headspace(μL L-1);Ceis the CH4concentration at the end of incubation(μL L-1);Mis the molar mass of CH4(g mol-1);mis the dry weight of soil(g)used in the incubation;andtis the incubation time(h).More than 10 data points were used for each sample.
Methane production rate was determined as previously described with slight modification(Wassmannet al.,1998).In brief,fresh soil samples(equivalent to 10 g of air-dried soil)were placed in 120-mL serum bottles,and 40 mL distilled water was added to make soil slurries.The bottles were sealed with a butyl stopper,and the headspace gas of each bottle was replaced with dinitrogen(N2)by vacuum flushing prior to incubation at 28°C in the dark.Methane concentration in headspace was measured after 3,7,11,15,and 19 d of incubation using a gas chromatograph as described above.Methane production rate was calculated as the slope of temporal increase in methane concentration according to the following equation:
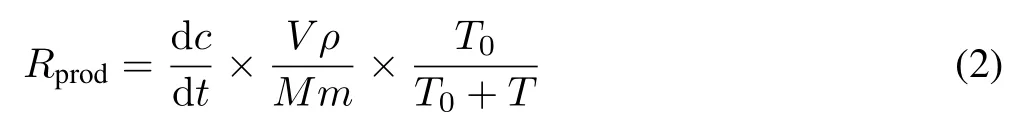
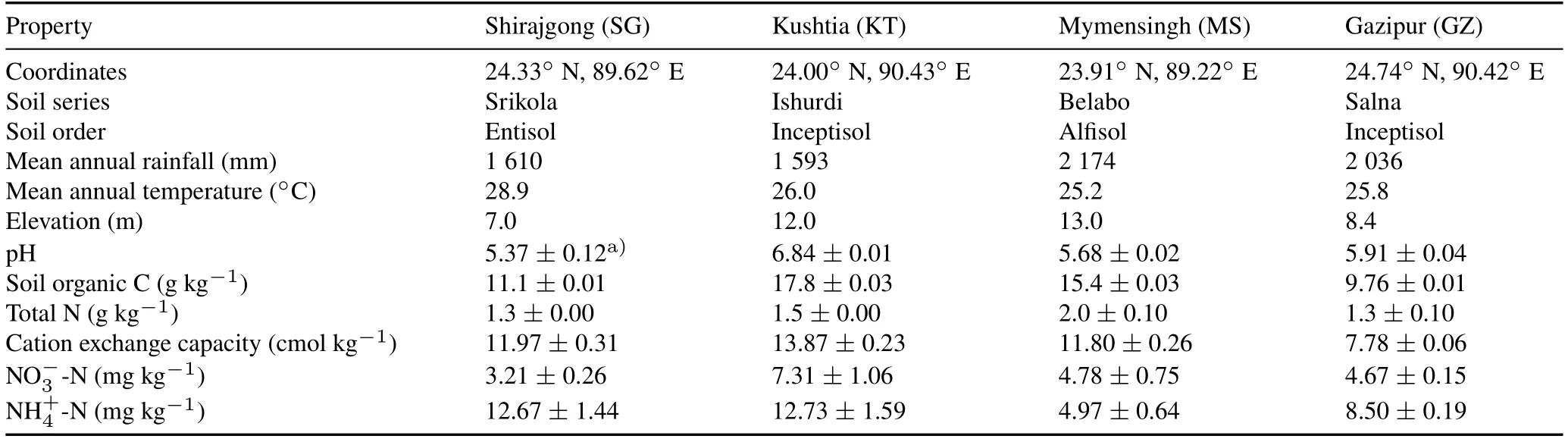
TABLE I Geographical and physicochemical properties of the four paddy soils of Bangladesh used in the microcosm incubation experiment
whereRprodis the methane production rate(μmol g-1d-1);dc/dtis the methane concentration change in headspace(μL L-1d-1);T0is the standard temperature(K);andTis the incubation temperature(K).
Determination of 13 C-abundance and 13 C-SOC conversion rate
Soil samples of about 1.5 g from the control and high13CH4treatments were stored at-80°C for about 6h and then frozen-dried for 48 h to obtain fine soil particles.The relative abundance of soil13C-atom and SOC content were measured by an elemental analyzer/isotope ratio mass spectrometer(Flash-2000 Delta VAdvantage,Thermo Fisher Scientific,USA)as previously described(Liet al.,2019).Soil13C-atom abundance and13C-SOC conversion rate were calculated using the following equations.
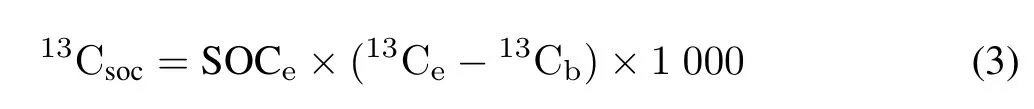
where13Csocis the net increase of13C-SOC in the high13CH4treatment(mg kg-1);SOCeis the SOC content after incubation in the high13CH4treatment(g kg-1);13Ceis the soil13C-atom abundance after incubation in the high13CH4treatment(%);13Cbis the soil13C-atom abundance after incubation in the control treatment(%);and 1 000 is a unit conversion factor.

where13Csocrateis the conversion rate of CH4-C to SOC(%)and CH4consumed is the13CH4-C oxidized(μmol g-1).
DNA extraction and SIP gradient fractionation
Soil DNA was extracted from 0.5 g soil using the FastDNA spin kit(MP Biomedicals,USA)following the manufacturer’s instructions.DNA quantity and quality were determined by a NanoDrop ND-1000 UV-vis spectrophotometer(NanoDrop Technologies,USA).Soil DNA extracts from the high12CH4and high13CH4treatments were subjected to CsCl isopycnic ultracentrifugation and fractionated to 14—15 density gradient fractions to separate the heavy13Clabeled DNA from the light12C-DNA fractions as detailed in previous studies(Xiaet al.,2011;Lu and Jia,2013).A digital refractometer(AR200,Reichert Tech.,Ametek Inc.,USA)was used to determine the density of each gradient fraction.The polyethylene glycol 6000(PEG 6000)was used to precipitate DNA in CsCl solution prior to precipitation by 70%ethanol.The final DNA was dissolved in 30μL of sterile water for downstream analysis.
Real-time quantitative PCR(qPCR)
DNA samples(including the total DNA extracts and DNArecovered from CsCl gradient fractions)were subjected to real-time qPCR using the primer pair A189F/mb661R to quantify the copy number ofpmoAgene on a CFX96Optical Real-Time Detection System(Bio-Rad,Laboratories Inc.,Hercules,USA).The PCR reactions were carried out in a 20-μL mixture containing 10μL of SYBR Premix Ex TaqTM(Takara Bio Inc.,Japan),0.25μL of each primer,8.5μL sterile distilled water,and 1μLof DNAtemplate.The thermal conditions are described in Table SI(see Supplementary Material for Table SI).Plasmid standards containing 102—107copies ofpmoAgene were prepared and used as described in a previous study(Sultanaet al.,2019).The amplification efficiencies ranged from 92%to 105%,withR2values of 0.996—0.999.
MiSeqsequencing of 16S rRNA genes
MiSeq sequencing of the 16S rRNA genes was employed to assess the community structure of methanotrophs in soil total DNA and the active methanotrophs in the13C-DNA heavy fractions from the high13CH4treatment.The DNA of the same buoyant density fractions from the high12CH4treatment was also used for control.The universal primer pair 515F/907R was used to amplify the V4—V5 region of the 16S rRNA gene(Caporasoet al.,2012).The thermal conditions and primer details of the PCR reaction are described in Table SI.Paired-end sequencing(2×300 bp)was conducted using the MiSeq system(Illumina,USA).Raw sequences were subjected to key quality control steps using the Quantitative Insights Into Microbial Ecology(QIIME)software(Caporasoet al.,2010),and low-quality sequences(quality score below 25,containing mismatched primers,ambiguous bases,and chimeras)were removed.A total of 1 131 823 high-quality reads were acquired.The high-quality sequences were applied to the Ribosomal Database Project(RDP)MultiClassifier for taxonomic assignment(Wanget al.,2007).All sequences of known methanotrophs have been deposited in the RDP database so that the sequences in this study could be aligned and phylogenectically classified into the two methanotroph families(MethylococcaceaeandMethylocystaceae).The taxonomic name of target sequences was converted to a txt file to construct an accnos file that is used by the Mothur software.The command“get.seq”in Mothur was then used to pick out the target methanotrophic sequences from the fasta file.These sequences were then clustered into operational taxonomic units(OTUs)at the 97%sequence similarity threshold using the UPARSE algorithm(Edgar,2013).The representative sequences of the major methane-oxidizing bacteria(MOB)OTUs(containing≥2%of total methanotrophic sequences in at least one of the samples)were used to construct a neighbor-joining phylogenetic tree using the MEGA software.It should be noted that the sequencing data is of compositional nature and ecological methods should be carefully evaluated using multiple statistical tests(Glooret al.,2017).The high-throughput raw sequencing reads of both the 16S rRNA andpmoAgenes were deposited in the National Center for Biotechnology Information(NCBI)Sequence Read Archive(SRA)under the project accession number PRJNA643911.
MiSeqsequencing of pmoA gene
ThepmoAgene fragments in the13C-DNA heavy DNA fractions of the high13CH4treatment were amplified using A189F/mb661R primers and applied to paired-end sequencing(2×300 bp)on the MiSeq system(Illumina,Inc.,USA)as previously described(Caiet al.,2016;Sultanaet al.,2019).The cycling profile for the PCR process is detailed in Table SI.The raw paired-end reads were first assembled and quality filtered by Mothur(Schlosset al.,2009).Putative frame-shifting reads were corrected or removed using the FrameBot program(Wanget al.,2013).A total of 59 902 high-quality sequences were obtained.The FunGene Pipeline was used to cluster the reads into OTUs at the genus-level with a distance cutoffof 0.07(Degelmannet al.,2010).The phylogenetic analysis of the representative sequences of majorpmoAgene OTUs(containing≥2%of sequences in at least one sample)was performed by MEGA using the neighbor-joining method.
Statistical analysis
One-way analysis of variance(ANOVA)followed by Tukey’spost-hoctest was conducted using SPSS version 10.0 to determine any significant differences in methane oxidation rate,soil13C-atom abundance,andpmoAgene abundance between different treatments.
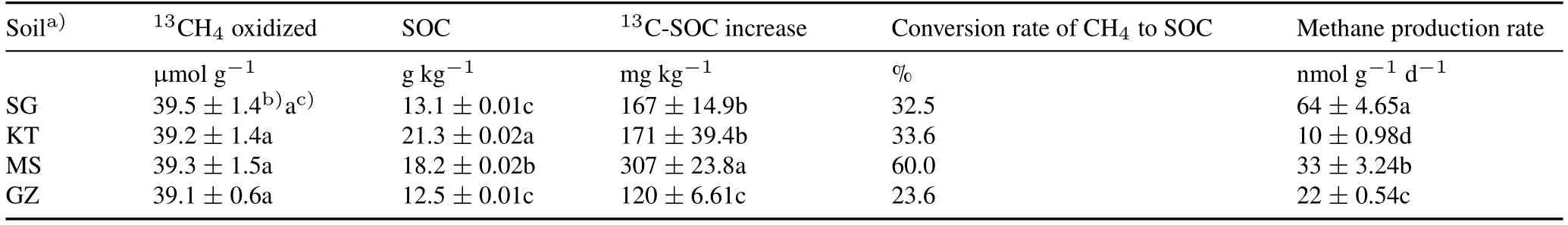
TABLE IIMethane oxidation and soil organic carbon(SOC)accumulation in four paddy soils after microcosm incubation under air enriched with 5%(volume/volume)13C-labeled methane(13CH4)
RESULTS
Accumulation of SOC by methanotrophy
Assuming linear kinetics,following 7—27 d of soil incubation for the same amount of CH4consumption,the highest CH4oxidation rate was observed in SG soil,and the other three soils showed similar but significantly lower rates of CH4oxidation(Fig.1a).The soil13C-atom abundances were significantly increased from the background level of 1.08%to 2.36%(SG),1.88%(KT),2.78%(MS),and 2.04%(GZ),with a mean value of 2.27%(Fig.1b).The calculated net increase of soil13C-SOC in the high13CH4treatment was 167,171,307,and 120 mg kg-1in SG,KT,MS,and GZ soils,respectively(Table II).The conversion rate of CH4-C to SOC was 32.5%,33.6%,60.0%,and 23.6%in SG,KT,MS,and GZ soils,respectively.
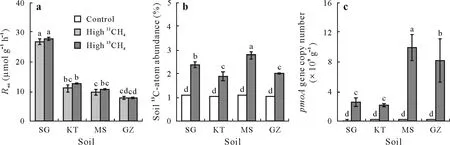
Fig.1 Methane oxidation rate(R ox)(a),soil 13C-atom abundance(b),and pmoA gene copy number(c)in four paddy soils after microcosm incubation under ambient air(control)or air enriched with 5%(volume/volume)13C-labeled(high 13CH4)or unlabeled methane(high 12CH4).The four soils were collected from Shirajgong(SG),Kushtia(KT),Mymensingh(MS),and Gazipur(GZ)regions of Bangladesh.The pmoA gene encodes the alpha subunit of the particulate methane monooxygenase.The error bars represent standard errors(n=3).Different letters above the bars indicate significant differences between soil samples(P<0.05).
Dynamics of methanotrophic communities
Consistent with the incorporation of methane-derived C into SOM,the abundances of methanotrophs,estimated by qPCR of thepmoAgene,exhibited a significant increase in the soils following incubation under a high concentration of methane(Figs.1c and 2).The addition of methane led to about 1.2—4.5-fold increase inpmoAgene abundance in the soils(Fig.1c).Similarly,high-throughput sequencing of the 16S rRNA gene from the total DNA extracts showed 5.9—39.2-fold increase in relative abundance of the methanotrophs following methane-amended incubation,relative to the ambient air control(Fig.2).Particularly,MethylobacterandMethylosarcina-affiliated type Ia methanotrophs were the most abundant lineages of methanotrophs in SG,MS,and GZ soils,while a similar abundance ofMethylocystis-affiliated type IIa andMethylobacter-affiliated type Ia methanotrophs were observed in the KT soil.
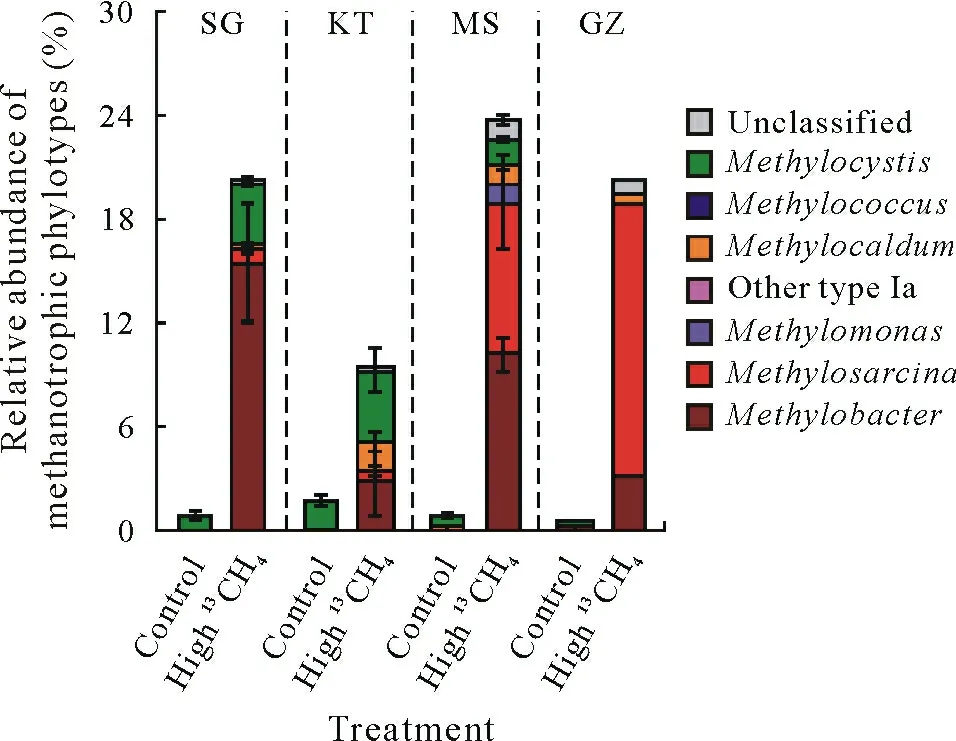
Fig.2 Composition of methanotrophs based on 16S rRNA gene sequences in four paddy soils after microcosm incubation under ambient air(control)or air enriched with 5%(volume/volume)13C-labeled methane(high 13CH4).The four soils were collected from Shirajgong(SG),Kushtia(KT),Mymensingh(MS),and Gazipur(GZ)regions of Bangladesh.The error bars represent standard errors(n=3).
Stable-isotope probing of 13C-methanotrophs
Following the isopycnic centrifugation of the extracted DNA,quantification of thepmoAgene in each fractionated DNA extract demonstrated active cell propagation of methanotrophs(Fig.3a),which is consistent with the13C increase of the SOC(Fig.1b)in all four soils fed with high concentrations of13CH4.ThepmoAgene was mostly detected in the light fractions(with CsCl buoyant density ofca.1.715 g mL-1)in the high12CH4treatment,but the highest abundance ofpmoAgene moved toward heavy DNA fractions(buoyant density ofca.1.735 g mL-1)in the high13CH4treatment(Fig.3a).
The labeling of methanotrophs was further supported by high-throughput 16S rRNA gene sequencing analysis of the whole microbial communities in the heavy DNA fractions(the 5th fraction)from both the high13CH4and12CH4treatments.The relative abundances of methanotrophs-affiliated 16S rRNA gene in the13C-DNA were 49.1%(GZ),47.1%(SG),35.1%(KT),and 16.9%(MS),with low abundances of 6.4%(GZ),0.5%(SG),1.2%(KT),and 2.6%(MS)in the heavy DNA fraction of the high12CH4treatment(Fig.3b).This result thereby suggests that methanotrophs were13C-labeled and spun down to the lower fraction of the ultracentrifugation tube with a heavy buoyant density,leading to a markedly enriched abundance of methanotrophs in the high13CH4treatment compared with the high12CH4treatment.
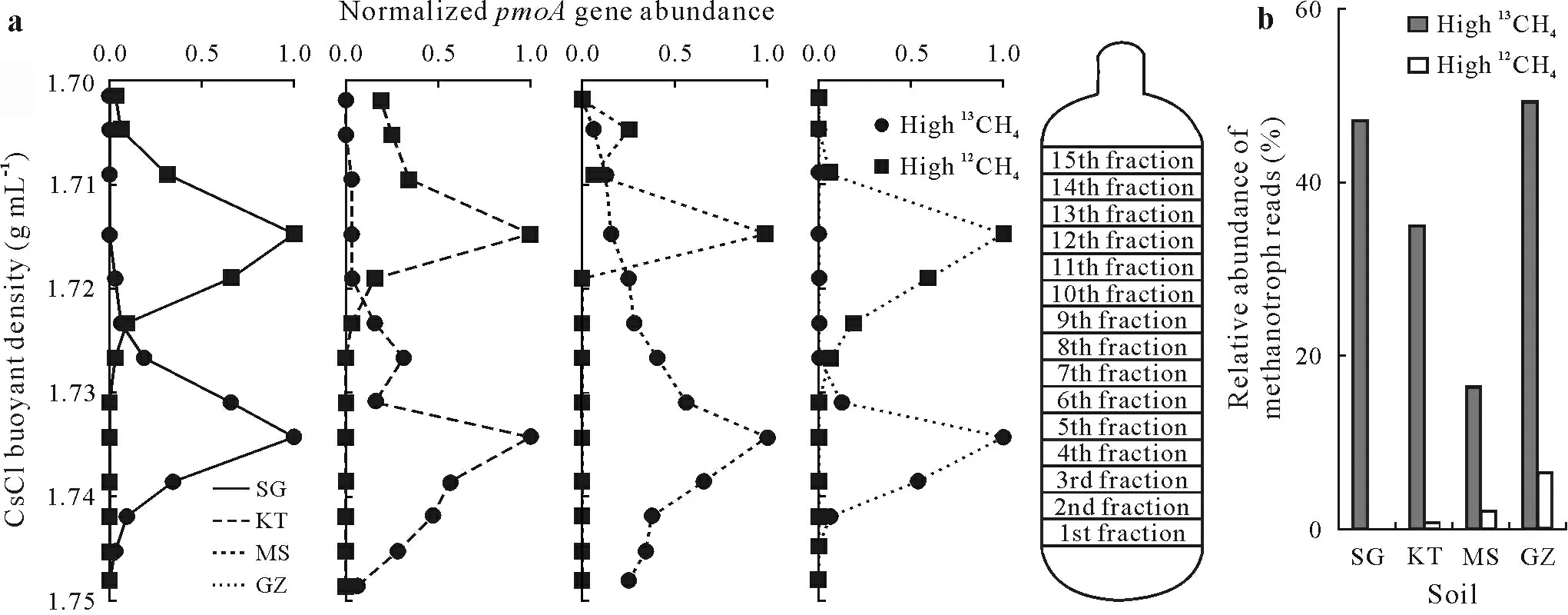
Fig.3 Normalized abundance distribution of pmoA gene across the entire buoyant density gradient of the DNA fractions extracted from four paddy soils after microcosm incubation under air enriched with 5%(volume/volume)13C-labeled(high 13CH4)or unlabeled methane(high 12CH4)(a)and relative abundance of methanotroph-affiliated reads in the total 16S rRNA gene sequenced from the heavy DNA fraction(the 5th fraction)of both the high 13CH4 and 12CH4 treatments(b).The normalized abundances are the ratios of the pmoA gene copy number for each DNA gradient to the maximum copy number of each treatment.The four soils were collected from Shirajgong(SG),Kushtia(KT),Mymensingh(MS),and Gazipur(GZ)regions of Bangladesh.The pmoA gene encodes the alpha subunit of the particulate methane monooxygenase.
Active methanotrophs in soils
To identify the taxonomic identity of active methanotrophs,phylogenetic analyses were performed on both13Clabeled 16S rRNAandpmoAgenes amplified from the heavy DNA fraction of the high13CH4treatment.The taxonomic classification of13C-labeled methanotrophic 16S rRNA genes demonstrated that type Ia methanotrophs dominated active methane oxidizers in SG,MS,and GZ soils,but the compositions of active type Ia-related methanotrophs in these three soils differed(Figs.4a and S2,see Supplementary Material for Fig.S2).Particularly,Methylobacter-like phylotypes were the most abundant phylotypes in soil SG,accounting for about 63.4%of all methanotrophs,whileMethylosarcina-like phylotypes contributed 66.6%of active methanotrophs in soil GZ.In the MS soil,however,
Methylobacter-andMethylosarcina-like phylotypes showed similar relative abundances.Analysis of the13C-labeledpmoAgene revealed a generally higher relative abundance of type Ia methanotrophs in these soils relative to the 16S rRNA gene analysis,i.e.,the predominance of eitherMethylobacterorMethylosarcina(Figs.4b and S3,see Supplementary Material for Fig.S3).Notably,in the KT soil,Methylocystiswas the most abundant active methanotrophs(77.5%)based on the 16S rRNA gene phylogenetic classification(Fig.4a).It should also be noted that the phylogeny of the13C-labeledpmoAgene revealed that the most abundant in this soil was theMethylobacterrelated type Ia methanotrophs(Fig.4b).
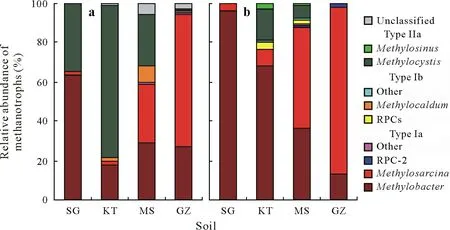
Fig.4 Composition of 13C-labeled methanotrophs based on MiSeq amplicon sequencing of the 16S rRNA(a)and pmoA(b)genes following DNA-based stable isotope probing of four paddy soils subjected to microcosm incubation under air enriched with 5%(volume/volume)13C-labeled methane.The four soils were collected from Shirajgong(SG),Kushtia(KT),Mymensingh(MS),and Gazipur(GZ)regions of Bangladesh.RPC=rice paddy cluster.
DISCUSSION
The present study employed a stable isotope-tracing technique to investigate the contribution of methane oxidizers to SOC accumulation.Following an incubation of four paddy soils with methane concentration(50 000μL L-1)found under field conditions(Shresthaet al.,2008),soil13Ccontent significantly increased in all paddy soils(Fig.1b).The newly accumulated13C in these soils might represent viable soil microbiota and/or dead biotic biomass,but it should derive exclusively from methanotrophic metabolisms(either directly orviathe methane-driven food web),given that methanotrophs are capable of using methane as sole C and energy source for their growth.It is estimated that,on average,37.4%of the methane-C was assimilated by the MOB to synthesize cell organic molecules in the soils studied(Table II),which was at a level comparable to the atmospheric methane assimilation rate of 31%—43%found in forest soils(Roslevet al.,1997).Flooded paddy fields are an important source of atmospheric methane,releasing up to 36Tg CH4year-1globally(Saunoiset al.,2016).However,this might account for a minor part of the methane produced from paddy fields,and the majority of methane(up to 80%)has already been consumed by methanotrophs prior to being released to the atmosphere(Frenzelet al.,1992).Therefore,up to 1.07 Tg CH4year-1were microbially consumed(Khan and Saleh,2015),which could contribute to 0.25—0.64 Tg of methane-derived SOC,assuming that the conversion rate of methane-derived C to SOC varied from 23.6%to 60.0%in this study(Table II).It should be noted that,from an ecosystem level perspective,plantdriven methanogenesis may also play important roles in SOM accumulation by delivering large amount of substrates that can be utilized by specific trophic guilds of Bacteria and/or Archaea.Meanwhile,different levels of inorganic species(particularly ammonium)could have profound impacts on methanotrophic communities and methanogenesis either directly and/or indirectly(Bodelier and Laanbroek,2004).Nonetheless,not only can methanotrophs relieve the emission of the greenhouse gas methane from paddy soils,but can also mitigate the loss of SOC due to soil heterotrophic respiration and stabilize SOM in waterlogged paddies(Stein,2020;Valenzuelaet al.,2020).Multi-omics approaches and singlecell techniques in the future may provide important tools to decipher these complex interactions at the level of the whole microbial community in soil.
Methanotrophs rely on methane-derived C assimilation for growth,oxidizing methane to carbon dioxide(CO2)for energy acquisition.The methane C assimilated by methanotrophs was used for the synthesis of the Cbackbone of organic molecules(Hanson and Hanson,1996),including nucleic acids.Therefore,13C-DNA-based SIP(DNA SIP)can be used as a powerful tool to track C assimilation by a diverse guild of microorganisms that perform methane oxidation and to evaluate their relative contribution to methane uptake and the soil food web(Radajewskiet al.,2003).In the present study,thepmoAgene was greatly enriched in the heavy DNA fractions from soils fed with13CH4(Fig.3a),indicating the active growth of methanotrophsviamethane-C assimilation and successful13C-labeling of these organisms.The proportion of methane oxidizers may account for up to>45%of the total13C-labeled microbes based on the 16S rRNA gene sequencing analysis(Fig.3b),and was considerably lower in samples without supplemented with high concentration of methane.This suggested that MOB can store a considerable amount of methane-derived C through biomass synthesis and cell propagation under high concentrations of methane,such as in inundated wetland ecosystems.In addition,it is likely that the other13C-labeled heterotrophic microorganisms could be associated with methane oxidation(Murase and Frenzel,2007).
Type Ia methanotrophs exhibited a high growth rate in all soils following methane amendment(Fig.2),and they became the dominant community of active methanotrophs in three of the soils(Fig.4a),consistent with the previous observation that high concentration of methane stimulated the growth of type I rather than type II methanotrophs(Krauseet al.,2012).However,it should be noted that the concentration of methane in this study was exceptionally high,up to 5%,approaching the upper limits of methane concentrations observed in the dissolved water of wetlands under field conditions(Zhanget al.,2010).Intriguingly,type II methanotrophs were also detected with potentially high methane oxidizing activity based on13C-labeled 16S rRNA gene analysis in the KT soil(Figs.4a and S2),which was not frequently observed in paddy soils(Sultanaet al.,2019).It was proposed that soil pH may have played an important role in the distribution of active methanotrophs in paddy soils,and dominant type II MOB methanotrophic activity would most likely occur in acidic environments(Zhaoet al.,2020).However,the KT soil with potential high methanotrophic activity in the present study demonstrated the highest pH(6.84)among all four soils tested(Table I),indicating that other environmental factor(s)may also be important in modulating the activity and growth of type II methanotrophs in this soil.It is noteworthy that phylogenetic analysis of13C-pmoAgenes from KT soil revealed more type Ia MOBparticipating in methane oxidation.This discrepancy between different marker genes could be due to primer bias,and it appeared that thepmoAprimers used here resulted in more frequent detection of type I methanotrophic communities(Fig.4b).Nevertheless,both thepmoAand 16S rRNA gene phylogenies confirmed a higher activity of type II methanotroph in methane consumption in the KT soil,compared to the type II methanotroph activities in the other three soils.
The diversity and habitat preferences of aerobic methanotrophs in physicochemically distinct environments have been explicitly summarized(Knief,2015).After long-term adaption to abiotic and/or biotic factors,type I and type II methanotrophs have evolved distinct phenotypes,such as differing cell sizes,morphologies,and specific growth rates.Microbes are considered the engine that drives global elemental cycling processes(Falkowskiet al.,2008),but the contribution of microbial biomass to SOMfrom specialized functional guilds is rarely investigated(Miltneret al.,2012).Methane oxidation is an ecologically and agriculturally important process with significant consequence(Knief,2015),and it appears that the conversion rate of CH4to SOC is affected by the size of methanotrophic communities,as expressed by the relative abundances of methanotrophic phylotypes,although the rate was less affected by the presence of type I methanotrophs(Fig.1).For instance,the influence of type IMethylobacterwas not apparent in the conversion of CH4to SOC in the SG soil(Table II).Intriguingly,the fixed C derived from methane(Table II)accounted for only a small fraction of the total SOC pool(Table SII,see Supplementary Material for Table SII),representing 1.28%,0.80%,1.68%,and 0.96%in the13CH4-amended paddy soils of SG,KT,MS and GZ,respectively.Meanwhile,our results also showed significant labeling of the obligate methanol oxidizersMethylobacteriumin the incubated soils(Table SIII,see Supplementary Material for Table SIII).Cross-feeding of methylotrophs on methanol released during methane oxidation has been demonstrated previously(Zhenget al.,2014),but the specific compound actually exchanged has not yet been fully deciphered.Future research is warranted regarding a more accurate budget of methane-driven accumulation and turnover of SOM in a wide variety of wetland ecosystems.
Due to the limited number of soil samples used in this study,confident correlations cannot be obtained between multiple environmental factors tested and the variations in methanotrophic compositions(Table SIV,see Supplementary Material for Table SIV).However,there are some discernible patterns which may be of importance in explaining the distribution of different methanotrophic phylotypes in the paddy fields.For instance,the methane production rate varied among the four soils(Table II,Fig.S4,see Supplementary Material for Fig.S4).It is also interesting to note that the highest relative abundance ofMethylobacteraffiliated type Ia methanotrophs was in the SG soil with the highest methane production rate,while the KT soil,with the lowest methane production rate,contained the most abundantMethylocystis-affiliated type IIa methanotrophs(Fig.4).Since our incubation was conducted after a flooding procedure and replacing all the headspace gas with N2in the bottle,the emitted methane gas should mostly originate from the methanogenic archaea in the soils without being partially oxidized by methanotrophs.Therefore,the methane production rate observed in the microcosms might well reflect the potential activities of methanogens.Methanogens and methanotrophs are the most important microbial groups in methane cycles and are closely associated with each other in inundated paddy fields(Conrad,2007),and different combinations of dominant methanogenic and methanotrophic phylotypes were indeed observed in paddy fields(Leeet al.,2014;Liuet al.,2016;Vaksmaaet al.,2017).It is thus reasonable to speculate that the selection of different types of methanotrophs might be related to the methane-producing organisms and dynamics in soils.Meanwhile,the KT soil with more abundant active type II methanotrophs also had the highest pH and CEC.Nonetheless,the emission of CO2and likely N2O was not measured in this study,and the production of these greenhouse gases is closely associated with microbial oxidation of CH4by aerobic methanotrophs(Stein,2020).The total global warming potential considering all greenhouse gases would provide a more comprehensive perspective on the sequestration and respiration of SOC in paddy fields.Since abiotic properties greatly influence the assembly of active methanotrophs(Kaupperet al.,2020),future studies need to include adequate samples and/or abiotic factors to decipher the biochemical processes responsible for microbial turnovers of CH4,CO2,and N2O in soils.
It should be noted that SOC is relatively stable,but an apparent increase was observed in the soils after microcosm incubation(Table II).The significant enrichment of13C atom in SOC serves as strong evidence of the net increase of SOC in soils.Meanwhile,methanotrophs-induced SOC may be closely associated with diazotrophs in paddy soils,as previously shown in peatlands(Larmolaet al.,2014).Accumulation of SOC can lead to a high C/N ratio,thus inducing biological N fixation.It is shown by15N2tracing that up to 45 kg N ha-1year-1could be fixed by biological N fixation in the paddy soils that received no fertilizers(Beiet al.,2013).Recent studies provide solid evidence on the taxonomic identities of the active diazotrophs that are linked to N fixation activity in paddy soils by DNA-SIP(Liet al.,2019).Understanding the turnover rate of fresh microbial biomass and its coupling with N fixation would be the key to better understanding of microbially-mediated processes related to soil health.
CONCLUSIONS
In this study,we revealed a significant incorporation of methane-derived C into SOM through microbial biomass buildup associated with microbial methane oxidation under methane-enriched air in four different paddy soils of Bangladesh.The oxidation and assimilation of13C-labeled CH4occurred in association with methanotrophs,providing strong evidence of the accumulation of13C-SOCviathe methane-driven trophic web.The conversion efficiency of methane-derived C into SOC varied greatly among the soils studied,which may be explained by the phylogenetically distinct methanotrophs.Type Ia lineages dominated the methanotrophs that mediated methane-derived C conversion to SOC in three soils,while type IIa methanotrophs were more actively engaged in methane-C turnover in one soil sample.Our study highlights the important roles of methanotrophs not only in reducing greenhouse gas emission but also in sustaining soil organic matter.
ACKNOWLEDGEMENTS
This study was financially supported by the National Natural Science Foundation of China(Nos.91751204,41630862,41701302,41530857,and 41877062).The first author,Ms.Nasrin Sultana,gratefully acknowledges the Organization for Women in Science for the Developing World(OWSD)Ph.D.Fellowship.The authors thank Mr.Zhiying Guo at Soil Sub-center of Chinese Ecological Research Network,Institute of Soil Science,Chinese Academy of Sciences(CAS)for bioinformatic analysis.We also thank the staffof the Analysis Center at the Institute of Soil Science,CAS for technical support,including Ms.Rong Huang and Mr.Zuohao Ma for Illumina MiSeq sequencing,Ms.Deling Sun for13C-atom abundance assay,Ms.Yufang Sun for soil organic carbon and total nitrogen content assay,Mr.Ruhai Wang for ammonia and nitrate nitrogen content assays,and Mr.Guoxing Lu for soil organic matter assay.
SUPPLEMENTARY MATERIAL
Supplementary material for this article can be found in the online version.
杂志排行
Pedosphere的其它文章
- Ultraviolet B radiation-mediated stress ethylene emission from rice plants is regulated by 1-aminocyclopropane-1-carboxylate deaminase-producing bacteria
- Drying-rewetting rather than sieving stimulates soil respiration
- Complementary effect of zoo compost with mineral nitrogen fertilisation increases wheat yield and nutrition in a low-nutrient soil
- Comparison of potential potassium leaching associated with organic and inorganic potassium sources in different arable soils in China
- A conceptual framework and an empirical test of complementarity and facilitation with respect to phosphorous uptake by plant species mixtures
- Evaluation of immobilizing agents as soil quality conditioners in addition to their metal(loid)immobilizing effect
