Pathogen Identification and Phylogenetic Analysis of Sugarcane
2022-03-07XiaoyanWANGWenfengLIJiarongPURongyueZHANGJinanPUHongliSHANYinhuLIZhaoguiYANGYingkunHUANG
Xiaoyan WANG Wenfeng LI Jiarong PU Rongyue ZHANG Jinan PU Hongli SHAN Yinhu LI Zhaogui YANG Yingkun HUANG
Rust in Plateau Areas at Low Latitudes
Abstract [Objectives] This study was conducted to clarify the species, occurrence and distribution characteristics of sugarcane rust pathogens in low-latitude plateau sugarcane rust areas of Yunnan Province and the phylogenetic relationship between the pathogens and that between them and other rust fungi of the genus Puccinia.
[Methods]Fifty seven sugarcane rust samples collected from the sugarcane areas of Baoshan, Lincang, Menghai, Menglian, Ximeng, Lancang and Wenshan in Yunnan were subjected to molecular identification.
[Results] The four sugarcane rust samples from Haiyin1hao in Menghai, Yunnan Province belonged to orange rust of sugarcane, and the pathogen was Puccinia kuehnii Butler.; and other 53 samples of sugarcane rust belonged to brown rust of sugarcane, and the pathogen was Puccinia melanocephala Syd. et P. Syd. The rDNA sequences of other rust fungi from the genus Puccinia were downloaded from GenBank and used for the construction of an NJ tree for systematic evolution analysis together with the sequences obtained in this study. The results showed that the four P. kuehnii sequences obtained in this study were clustered with Puccinia polysora (GenBank accession number: GU058024) into one group, and they were close genetically; the 53 P. melanocephala sequences were clustered with P. nakanishikii (GenBank accession number: GU058002), P. rufipes (GenBank accession number:AJ296545), Aecidium deutziae (GenBank accession number:KU309317) and P. coronata (GenBank accession number: DQ354526) into one group, and they had a close genetic relationship; and the genetic relationship of the two species of sugarcane rust fungi identified in this study was relatively distant.
[Conclusions]This study provides a scientific basis for sugarcane rust epidemic prediction, disease resistance breeding, and precise prevention and control.
Key words Sugarcane rust; Pathogen identification; Puccinia melanocephala; Macroropyxis fulva; Phylogenetic evolution
Received: September 19, 2021 Accepted: November 15, 2021
Supported by China Agriculture Research System of MOF and MARA(CARS-170303); Joint Special Project of Basic Agricultural Research in Yunnan Province (2018FG001-029); "Yunling Industrial Technology Leading Talents" Training Project (2018LJRC56); Special Fund for the Construction of Modern Agricultural Industry Technology System in Yunnan Province (YNGZTX-4-92).
Xiaoyan WANG (1981-), female, P. R. China, associate researcher, devoted to research about sugarcane diseases.
*Corresponding author. E-mail: huangyk64@163.com.
Sugarcane rust is one of the most important diseases of sugarcane worldwide, which often causes huge economic losses[1-2]. According to the symptoms and pathogen characteristics, sugarcane rust is divided into three types, namely, sugarcane brown rust caused by Puccinia melanocephala Syd. et P. Syd., sugarcane orange rust caused by Puccinia kuehnii Butler., and sugarcane orange-brown rust caused by Macroropyxis fulva sp. nov.[6]. Since sugarcane brown rust was first discovered in Java in 1890, it has rapidly spread to most sugarcane-growing countries and regions in the world, and has been epidemic multiple times, causing some high-yield high-sugar varieties such as Co475, T34362 and CP78-1247 to be eliminated[7-12]. The distribution range of sugarcane orange rust is relatively narrow, mainly in countries and regions such as India, Australia, the United States, and Latin America[13-17]. Sugarcane orange-brown rust is a new type of sugarcane rust that was discovered in Swaziland sugarcane variety N25 in 2008. The disease is currently common in South Africa and Swaziland sugarcane regions[6]. In mainland China, Ruan et al.[18] first reported the occurrence of P. melanocephala in Yunnan sugarcane areas in 1982, followed by other sugarcane areas such as Fujian, Guangdong, Sichuan, Jiangxi, Guangxi and Hainan[19-23].
Yunnan is the second largest sugarcane producing area in China. Sugarcane rust is common in this producing area, and has a tendency to spread year by year and increase the damage. However, its pathogen types, occurrence and distribution characteristics and the phylogenetic relationship between the pathogens and that between them with other rust fungi of the genus Puccinia are still unclear, and there is no scientific basis for effective prevention and control. In this study, the occurrence and distribution of sugarcane rust in different sugarcane areas of Yunnan at low latitudes were systematically investigated by molecular biology methods through the identification of the species of 57 samples of sugarcane rust fungi and the analysis of the phylogenetic relationship between the sugarcane rust fungi and that between them and other rust fungi of the genus Puccinia based on the cloning and sequencing of the rDNA sequences by molecular biology methods, so as to clarify the species of sugarcane rust pathogens in Yunnan sugarcane regions at low latitudes and reveal the molecular evolution of sugarcane rust isolates. This study provides a scientific basis for the epidemic prediction, disease resistance breeding and precise prevention and control of sugarcane rust.
Materials and Methods
Sample collection
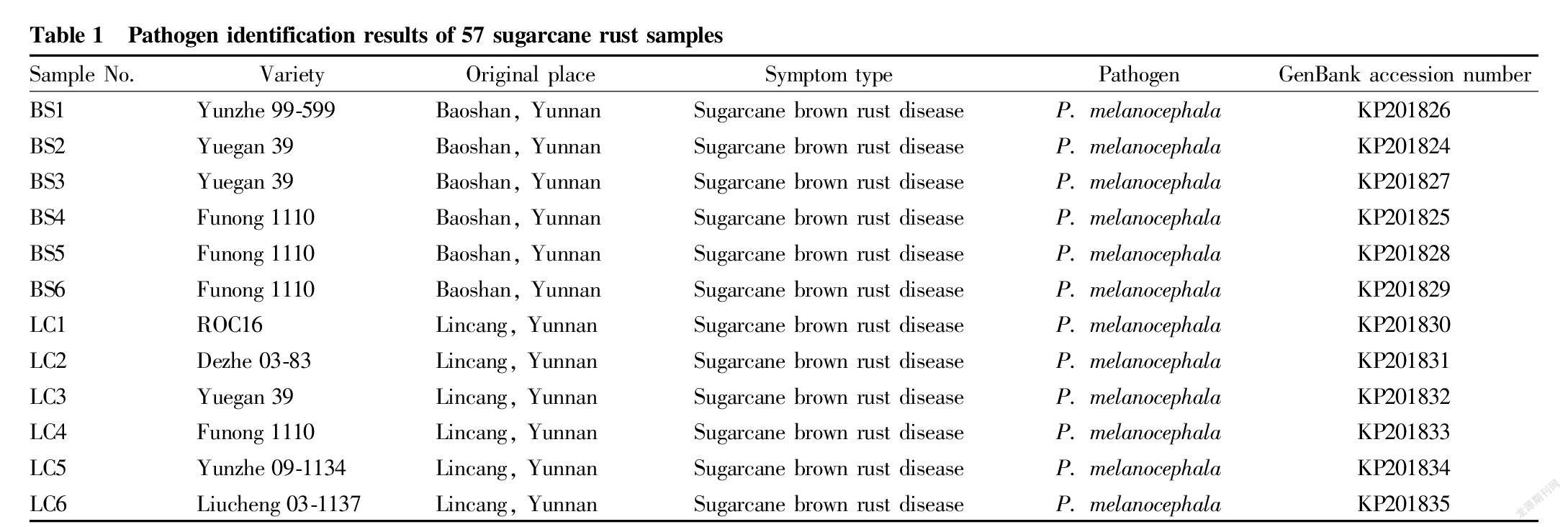
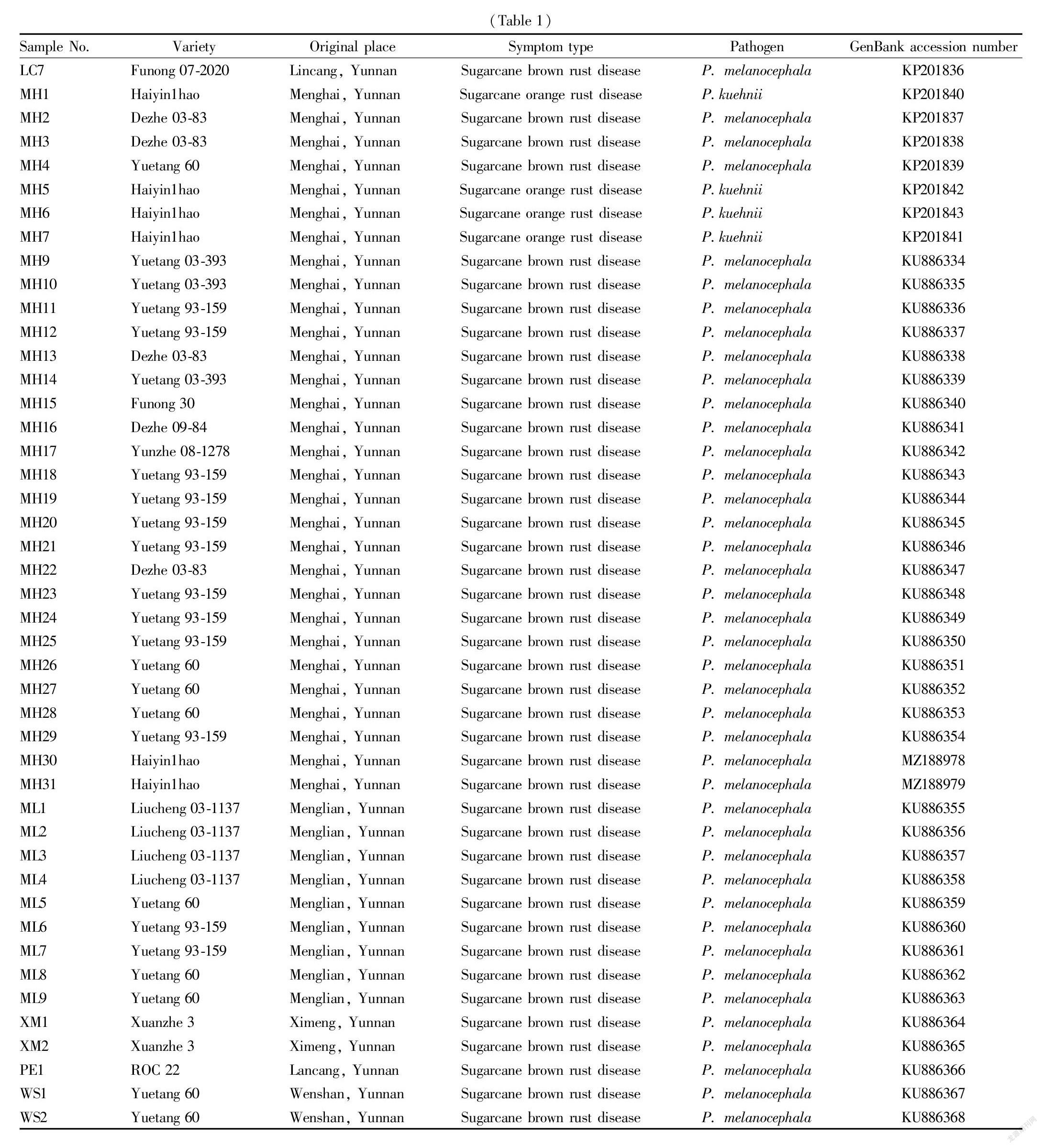
From 2014 to 2018, we successively investigated the occurrence and distribution of sugarcane rust in sugarcane areas such as Baoshan, Lincang, Menghai, Menglian, Ximeng, Lancang, Xinping and Wenshan in Yunnan province, and collected 57 parts of diseased leaves with different rust symptoms and typical characteristics (Table 1).
Molecular identification of pathogens
Primer design and synthesis
Referring to rust specific primers reported in the literature[24], we commissioned Sangon Biotech (Shanghai) Co., Ltd. to synthesize following sequences: Rust2inv: 5’-GATGAAGAACACAGTGAAA-3’; LR6: 5’-CGCCAGTTCTGCTTACC-3’. The expected length of the amplified product was 1 500 bp.
Extraction of plant total DNA
First, 0.2 g of diseased leaves was weighed from each sugarcane rust sample, and extracted with Easy Pure plant Genomic DNA Kit of TransGen Biotech for the total DNA of leaves. The specific steps followed the instructions. After extraction, the extraction quality was identified with an Eppendorf AG 22331 protein/nucleic acid analyzer.
PCR and sequence analysis
The PCR amplification system was 25 μl in volume and contained 10 μl of ddH2O, 12.5μl of 2×PCR Taq mixture, 0.5 μl of DNA template, and 1.0 μl each of the upstream and downstream primers (20 μg/μl). The PCR reaction program was started with pre-denaturation at 94 ℃ for 2 min, followed by 40 cycles of denaturation at 94 ℃ for 30 s, annealing at 57 ℃ for 1 min and extension at 72 ℃ for 1.5 min, and finally completed with extension at 72 ℃ for 7 min. The amplified products were detected by 1.5% agarose gel electrophoresis.
Cloning and sequencing of PCR products
The PCR products were purified using a fast agarose DNA recovery kit [Tiangen Biotech (Beijing) Co., Ltd.]. The purified PCR product was ligated with the pMD 18-T vector (TaKaRa company) overnight at 16 ℃, and the ligation product was transformed into Escherichia coli DH5ɑ competent cells, which were then coated on LB solid medium containing antibiotic Amp and placed in a 37 ℃ incubator for inverted culture. The monoclonal colonies on the plate were picked and cultured in LB liquid medium with shaking, and an appropriate amount of the culture product was taken for PCR detection. For each isolate, six clones that were identified as positive by PCR were selected and sent to Beijing Liuhe BGI Co., Ltd. for sequencing. The sequencing results were aligned with the published sequences on the GenBank using BLAST search on the NCBI website to determine the taxonomic status of pathogens. DNAMAN was used for multiple sequence alignment analysis.
Phylogenetic analysis
According to the alignment results, the rDNA sequences of other rust fungi of the genus Puccinia were obtained from the GenBank database, and a phylogenetic tree was constructed by the Neighbor-Joining method of the MEGA 6.0[25] software in the mode of Kimura 2-parameter with Coleosporium asterum (HQ31750) and Melampsora sp. (GU058014) as outgroups under 1 000 times of repetition in bootstrap test.
Results and Analysis
Molecular identification results of pathogens
With the extracted total DNA of sugarcane rust diseased leaves as a template, the 57 sugarcane rust samples were amplified by PCR using Rust2inv/LR6 specific primers of rust fungi, and a target band of about 1 500 bp in size was obtained from the amplification. The amplified PCR products were sequenced, and the obtained sequences were submitted to GenBank, as shown in Table 1. Homologous sequences were searched in GenBank using BLAST. The results showed that the four nucleotide sequences of sugarcane rust pathogens (GenBank accession number: KP201840-KP201843) from Haiyin1hao in Menghai, Yunnan Province and corresponding nucleotide sequence of P. kuehnii registered in GenBank (GenBank accession number: GU058021) ) shared more than 99.9% of homology, indicating that the pathogens are P. kuehnii (Table 1); and 53 nucleotide sequences of sugarcane rust pathogens from other sugarcane varieties in Baoshan, Lincang, Menghai, Menglian, Ximeng, Lancang and Wenshan in Yunnan (GenBank accession numbers: KP201824-KP201839, KU886334-KU886368, MZ188978-MZ188979) shared more than 99.8% of homology with corresponding nucleotide sequences of P. melanocephala registered in GenBank (GenBank accession numbers: GU058001, JX036025 and MG564638), indicating that the pathogens are P. melanocephala (Table 1).
Agricultural Biotechnology2022
Phylogenetic analysis
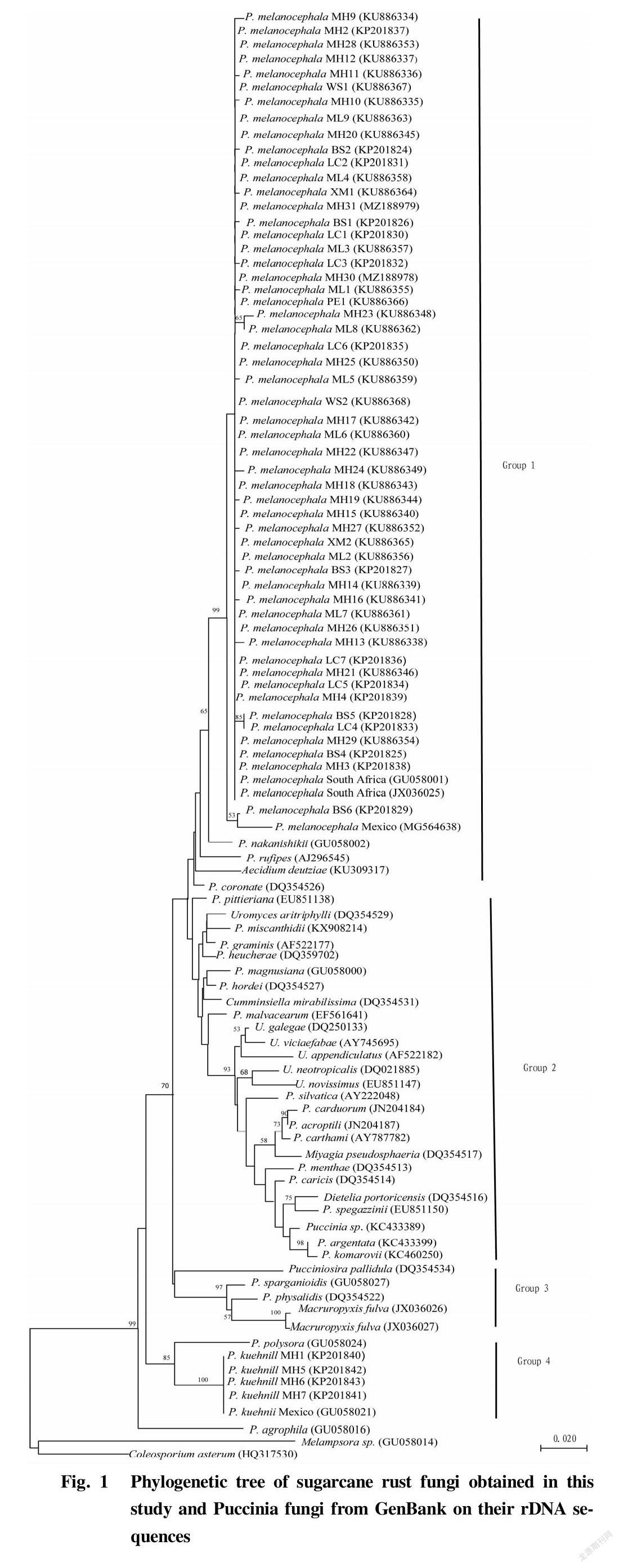
According to the alignment results, the rDNA sequences of sugarcane rust pathogens and other Puccinia fungi were selected and downloaded from the GenBank database, and used to build a tree by the Neighbor-Joining method together with the 57 sugarcane rust sequences obtained in this study. From the phylogenetic tree (Fig. 1), it can be seen that the 57 sugarcane rust rDNA sequences obtained in this study and other Puccinia fungi were divided into 4 groups in the entire phylogenetic tree. Among them, 53 P. melanocephala sequences were clustered with corresponding nucleotide sequences of P. melanocephala reported in GenBank (GenBank accession numbers: GU058001, JX036025 and MG564638), and P. nakanishikii (GenBank accession number: GU058002), P. rufipes (GenBank accession number: AJ296545),
Aecidium deutziae (GenBank accession number: KU309317) and P. coronata (GenBank accession number: DQ354526) into group 1. Four P. kuehnii sequences were clustered with corresponding nucleotide sequence of P. kuehnii reported in GenBank (GenBank accession number: GU0580021) and P. polysora (GenBank accession number: GU058024) into group 4. Sugarcane orange-brown rust pathogen M. fulva sp. nov. (GenBank accession number: JX036026 and JX036027) was clustered with P. physalidis (GenBank accession number: DQ354522), P. sparganiodis (GenBank accession number: GU058027) and Pucciniosira pallidula (GenBank accession number: DQ354534) into group 3. The rest of rust fungi of the genus Puccinia that had been reported in GenBank were clustered into group 2.
Discussion
In this study, morphological observation and molecular identification methods were adopted for detection and sequence analysis of 57 sugarcane rust pathogen samples collected from sugarcane areas such as Baoshan, Lincang, Menghai, Menglian, Ximeng, Lancang and Wenshan in Yunnan Province. For the first time, P. kuehnii, which causes yellow rust of sugarcane, was found in the sugarcane area of Menghai, Yunnan Province, and it was confirmed that P. melanocephala is the main pathogen causing brown rust. It lays a foundation and provides a basis for the in-depth research and scientific prevention and control of sugarcane rust in China.
The phylogenetic analysis of the rDNA sequences of sugarcane rust fungi and other rust fungi of the genus Puccinia showed that the 53 isolates of P. melanocephala in the low-latitude plateau sugarcane areas of Yunnan Province had no obvious variety and geographic groupings in the phylogenetic tree, indicating that there is no molecular variation or small variation among these 53 isolates of P. melanocephala. All the isolates of P. melanocephala used for analysis in this study had the closest genetic relationship with P. nakanishikii, but were relatively farther from another pathogen of sugarcane rust, i.e., P. kuehnii[26], which is consistent with the phylogenetic analysis results of Dixon et al.[26] on the phylogenetic relationship between sugarcane rust isolates from 164 sugarcane areas in 23 countries and other rust fungi.
P. kuehnii, which causes orange rust of sugarcane, is a sudden pathogen. Once the disease breaks out, it often causes huge economic losses. In Australia and Florida, the annual economic loss caused by sugarcane orange rust is more than 1.77 billion U.S. dollars and 400 million U.S. dollars[27-28], which greatly affects the sustained and stable development of local sugar industry. The occurrence and prevalence of sugarcane rust is closely related to the disease resistance of varieties. Large-scale planting of susceptible varieties is an important reason for the epidemic. Breeding and planting disease-resistant varieties is the most economical and effective measure to prevent and control sugarcane rust. Yunnan is an important distribution center of wild sugarcane resources in China and one of the origin centers of wild sugarcane in the world. Its complex and diverse geographical and climatic conditions have formed rich sugarcane germplasm resources, and it is one of the natural and precious gene banks for sugarcane genetic improvement in China and the world[28]. So far, the identification and evaluation of rust resistance in sugarcane germplasm resources and variety materials in China has been only for sugarcane brown rust[29-31], and the identification and evaluation of sugarcane orange rust resistance has not been carried out. In the future, it is also necessary to adjust the breeding strategies for sugarcane rust resistance and strengthen the evaluation and selection of sugarcane orange rust-resistant materials and disease-resistant varieties while further carrying out the evaluation and screening of sugarcane brown rust-resistant materials and disease-resistant varieties, so as to provide a scientific basis and excellent source materials for the selection and utilization of disease-resistant varieties to effectively prevent and control sugarcane rust.
Sugarcane orange-brown rust caused by M. fulva sp. nov. is a new type of sugarcane rust discovered in Swaziland sugarcane variety N25 in 2008. Its lesions are linear, parallel to the midrib, and are about 2 to 10 mm long. The uredinia are bright orange when fresh, then turn dark reddish brown, and reach 3 mm in length. The uredospores are elliptical, obovate or spherical, and bright orange to orange-red; the walls are evenly thickened, with a thickness of 1.5-4.9 μm; and the size is (25-32) μm×(20-26) μm[6]. Therefore, it is necessary to strengthen the quarantine of varieties introduced from abroad and the monitoring of rust diseases in sugarcane and Miscanthus or other related weeds that can serve as alternative hosts to prevent sugarcane orange-brown rust from being introduced into China from the source and effectively reduce the potential threat of the disease to the cane sugar industry, so as to promote the high-quality development of China’s cane sugar industry.
References
[1] ROTT P, BAILEY RA, COMSTOCK JC, et al. A guide to sugarcane diseases[M]. Montpellier: CIRAD and ISSCT, 2000.
[2] Hoy JW, HOLLIER CA. Effect of brown rust on yield of sugarcane in Louisiana[J]. Plant Disease, 2009(93): 1171-1174.
[3] CUMMINS GB, HIRATSUKA Y. Illustrated genera of rust fungi[M]. Minnesota: American Phytopathology Society, 1983.
[4] RYAN CC, EGAN BT. Rust, Chap. XIII. In: RICAUD C, EGAN B T, GILLASPIE A G, et al. Diseases of sugarcane: Major diseases[M]. Amsterdam: Elsevier, 1989.
[5] SIVANESAN A, WALLER JM. Sugarcane diseases [M]. Oxon: CAB International, 1986.
[6] MARTIN LA, LLOYD EVANS D, CASTLEBURY LA, et al. Macruropyxis fulva sp. nov., a new rust (Pucciniales) infecting sugarcane in southern Africa[J]. Australasian Plant Pathology, 2017, 46(1): 63-74.
[7] HUGHES CG, ABBOTT EV, WISMER CA, et al. Sugarcane diseases of the world[M]. CHEN QL trans. Beijing: Agriculture Press, 1982. (in Chinese).
[8] CSIRO. Unlocking success through change and innovation: Options to improve the profitability and environmental performance of the Australian sugar industry[M]. Submission to Sugarcane Industry Assessment, 2005.
[9] RAID RH. Florida sugarcane handbook[M]. Florida: University of Florida, 2006.
[10] COMSTOOK JC. Effect of rust on sugarcane growth and biomass[J]. Plant Disease, 1992(76): 172-177.
[11] COMSTOOK JC, SHINE JM, RAID RN. Effect of early rust infection on subsequent sugarcane growth [J]. Sugar Cane, 1992(4): 7-9.
[12] MA L. Current status of sugarcane production and scientific research in Thailand[J]. World Agriculture, 1995(9): 23. (in Chinese).
[13] EGAN BT. A review of the world distribution of Puccinia spp. attacking sugarcane[J]. Plant Pathology, 1980(17): 1373-1381.
[14] COMSTOCK JC, SOOD SG, GLYNN NC, et al. First report of Puccinia kuehnii, causal agent of orange rust of sugarcane, in the United States and in the Western Hemisphere[J]. Plant Disease, 2008(92): 175.
[15] OVALLE W, COMSTOCK JC, GLYNN NC, et al. First report of Puccinia kuehnii, causal agent of orange rust of sugarcane, in Guatemala[J]. Plant Disese, 2008(92): 973.
[16] CHAVARRIA E, SUBIROS F, VEGA J, et al. First report of orange rust of sugarcane caused by Puccinia kuehnii in Costa Rica and Nicaragua [J]. Plant Disese, 2009(93): 425.
[17] FLORES RC, LOYO JR, OJEDA RA, et al. First report of orange rust of sugarcane caused by Puccinia kuehnii in Mexico, El Salvador and Panama[J]. Plant Disese, 2009(93): 1347.
[18] RUAN XY, YANG W, SUN CJ. Puccinia erianthi on sugarcane found in Yunnan Province[J]. Mycosystema, 1983, 2: 260-261. (in Chinese).
[19] National Collaborative Group for Research on Important Diseases of Sugarcane. The preliminary report of sugarcane diseases investigation in the sugarcane planting provinces (partly), mainland China[J]. Sugarcane and Canesugar, 1991(1): 1-8. (in Chinese).
[20] LI XM, LIU WB, SHI HH. Pathogen identification and biological characteristics of sugarcane rust in Danzhou[J]. Sugar Crops of China, 2008(2): 30-32. (in Chinese).
[21] WEI JJ, DENG ZY, HUANG WH, et al. Control methods and pathogen biological characteristics of sugarcane rust in Beihai[J]. Journal of Anhui Agricultural Sciences, 2010(38): 14997-14999. (in Chinese).
[22] HUANG YK, LI WF. Xiandai Ganzhe Bingchongcaohai Yuanse Tupu[M]. Beijing: China Agriculture Press, 2011. (in Chinese).
[23] HUANG YK, LI WF. The epidemic causes of sugarcane rust in Yunnan sugarcane regions and countermeasures[J]. Plant Protection Technology and Extension, 1998(18): 22-23. (in Chinese).
[24] AIME MC. Toward resolving family-level relationships in rust fungi (Uredinales)[J]. Mycoscience, 2006(47): 112-122.
[25] TAMURA K, Stecher G, Peterson D, et al. MEGA6: Molecular evolutionary genetics analysis version 6.0 [J]. Molecular Biology and Evolution, 2013(30): 2725-2729.
[26] DIXON LJ, CASTLEBURY LA, AIME MC, et al. Phylogenetic relationships of sugarcane rust fungi[J]. Mycological Progress, 2010, 9(4): 459-468.
[27] BRAITHWAITE KS, CROFT BJ, MAGAREY RC, et al. Phylogenetic placement of the sugarcane orange rust pathogen Puccinia kuehnii in a historical and regional context[J]. Australasian Plant Pathology, 2009(38): 380-388.
[28] CHEN H, FAN YH, SHI XW, et al. Research on genetic diversity and systemic evolution in Saccharum spontaneum L.[J]. Acta Agronomica Sinica, 2001(27): 645-652. (in Chinese).
[29] LI WF, CAI Q, HUANG YK, et al. Identification of sugarcane wild germplasm resources resistant to Puccinia erianthi[J]. Plant Protection, 2005, 31(2): 51-53. (in Chinese).
[30] LI WF, CAI Q, HUANG YK, et al. Identification of fine sugarcane parent and innovative germplasm to resistant Puccinia erianthi[J]. Sugar Crops of China, 2007(4): 10-12. (in Chinese).
[31] LI WF, CAI Q, HUANG YK, et al. Identification of sugarcane cultivated original species resistance to Puccinia erianthi[J]. Journal of Yunnan Agricultural Unversity, 2008, 23(1): 25-28. (in Chinese).
Editor: Yingzhi GUANG Proofreader: Xinxiu ZHU
杂志排行
农业生物技术(英文版)的其它文章
- Expression Analysis of Heat Shock Protein 70 Gene in Rice (Oryza sativa L.)
- Changes in Physiological and Biochemical Characteristics of Floral Organ
- Research Progress on Lonicera japonica Thunb. Affected by Environmental Stress
- Research Progress on Genetic Diversity of Snap Bean
- Allelopathic Effects of Cedrus deodara Needle Extracts on Seed
- Development Status and Countermeasures of Passiflora spp. Seedling Industry in Qinzhou, Guangxi
