Expression Analysis of Heat Shock Protein 70 Gene in Rice (Oryza sativa L.)
2022-03-07FeiTONGJiweiLIUYuannaCHENGTingYEZhaodongLIUNiZHANHuiYELiquanWUJuanLI
Fei TONG Jiwei LIU Yuanna CHENG Ting YE Zhaodong LIU Ni ZHAN Hui YE Liquan WU Juan LI

Abstract [Objectives] This study was conducted to construct a pColdI-HSP70 recombinant prokaryotic expression vector. [Methods] With rice ‘N22’ as the test variety, hydroponic experiments were set up for rice seedlings with normal growth (CK), high temperature treatment (H), drought treatment (D) and drought-high temperature cross treatment (DH). The recombinant prokaryotic expression vector was constructed by the method of prokaryotic expression, and its induction expression time, IPTG concentration and temperature were optimized. [Results] The pColdI-HSP70 expression vector was successfully constructed, and the fusion protein was highly expressed in host strain BL21, and the expressed proteins were all in a soluble form. By optimizing the induction expression conditions, it was found that the optimal expression conditions were the IPTG concentration of 0.5 mmol/L and induction at 20 ℃ for 36 h. The expression analysis of the rice HSP70 gene under different stress treatments was carried out by qRT-PCR technology, and it was found that H, D and DH stresses all could induce its expression, and its expression levels were 4.65, 1.40 and 17.66 times higher than that of the CK group, respectively. [Conclusions] This study lays a solid foundation for the isolation, purification and functional study of rice HSP70 proteins.
Key words Rice; HSP70; qRT-PCR; Prokaryotic expression
Received: October 28, 2021 Accepted: December 29, 2021
Supported by Anhui Provincial Education Department Project (KJ2019A0213); National Natural Science Foundation of China (31501245); Undergraduate Innovation and Enterpreneurship Training Program of Anhui Province (S202010364185; XJDC2020541; 202110364788).
Fei TONG (1996-), male, P. R. China, master, devoted to research about crop cultivation and farming sciences.
Jiwei LIU (2000-), male, P. R. China, devoted to research about plant stress physiology.
#These authors contributed equally to this work.
*Corresponding author. E-mail: lj_welcome_ok@126.com.
Heat shock proteins (HSP), also known as heat stress proteins or heat shock proteins, are a class of protective proteins that organisms respond rapidly to adversity stress[1-2]. As a molecular chaperones, on the one hand, they are involved in the folding of nascent peptide chains, the assembly of oligomeric proteins, and the transmembrane transport of proteins, and on the other hand, they stabilize protein structures, prevent protein aggregation and deformation, and repair damaged proteins under stress conditions[3-5]. According to different molecular weights, plant heat shock proteins are divided into six families: HSP100, HSP90, HSP70, HSP60, HSP40 and sHSP[6-7], of which the HSP70 family with a molecular weight of 70 kDa is the most common and the most conserved, and the most abundant in cells. It is not only indispensable in the process of plant development, but also plays an important role in plant responses to various stresses. Studies have shown that under various stress conditions such as high temperature, low temperature, drought, high salt, and heavy metals, the Hps70 genes of plants are induced to express, which greatly enhances the stress resistance of plants[8-21]. Thirty-two rice HSP70 genes have been found in rice, and most of the genes are expressed to varying degrees in different stages of rice tissues and organs[22]. Rice ER HSP70 (BiP) is involved in the entry of nascent polypeptides into the endoplasmic reticulum cavity and promotes the folding and assembly of peptide chains during gliadin transport[23], and the overexpression of BIP can also alleviate "endoplasmic reticulum stress"[24]. Mitochondrial HSP70 is thought to inhibit hyperthermia or hydrogen peroxide-induced programmed cell death[25]. Heat shock on the radicles of rice seedlings before and after low temperature treatment can significantly improve the cold tolerance of seedling radicles[26]. Rice gene Os03g16920 is an important member of the HSP70 gene family, and its expression is induced to varying degrees under high temperature, drought and high salt stress[22]. In this study, the induced expression of this gene under different forms of stress was detected by qRT-PCR, and on the basis of the existing sequence, a pColdI-HSP70 recombinant prokaryotic expression vector was constructed and successfully induced in the host bacteria BL21, laying a solid foundation for the isolation, purification and functional study of rice HSP70 protein.
Materials and Methods
Materials
Indica rice variety Nagina 22 (Oryza sativa L.) was selected as the test material. Uniform and plump rice seeds were selected, surface sterilized and soaked, and then placed in a petri dish for germination. The seeds with uniform germination were selected and sown in plastic pots with grids and cultured in Hoagland nutrient solution (Hoagland formula) in an artificial climate box (day/night temperature 30 ℃/25 ℃, day/night photoperiod 12 h/12 h, relative humidity 75%, light intensity 3 000 Lx)[27]. The developmental process of young panicles was diagnosed by the leaf age remainder method[28], and the stress treatment was started when the auriculae of the flag leaves were just exposed.
Drought treatment (DT): The plants was subjected to drought stress simulated with a 10% PEG-6000 Hoagland nutrient solution for 3 d; high temperature treatment (HT): the plants were stressed at a high temperature of 38 ℃ for 3 d; drought-high temperature cross stress (DH): the plants were stressed with 10% PEG-6000 for 3 d, restored for 3 d and re-stressed at 38 ℃ for 3 d; and rice growing normally was used as control (CK). Four groups of young rice panicles were cut respectively, frozen with liquid nitrogen, and then stored in a -80 ℃ refrigerator for later use.
Methods
Primer design and synthesis
According to the rice HSP70 gene sequence (Accession No. XM_015775442 in GenBank), primers P1 (5’-CGCGGATCCATGGGCGGGCAACAAGGGAG-3’) and P2 ( 5’-CCCAAGCTTGTCCACTTCCTCGATCTTG-3’) containing BamH I and Hind III restriction sites at both ends of the open reading frame, respectively, and qRT-PCR primers P3 (5’-AGAGGAAGCACAAGAAGGA-3’) and P4 (5’-GTGATGGTGGCGTAGAAG-3’), were designed. Rice reference gene primers were 18SrRNA-F (5’-CAACTTTCGATGGTAGGATAGGG-3’) and 18SrRNA-R (CCAATTACCAGACACTAAAGCGC).
Extraction of total RNA and synthesis of the first strand of cDNA and RT-PCR
Rice total RNA was extracted using RNAiso Plus kit (TaKaRa), and the specific operation was carried out according to the instructions. The first strand of cDNA was synthesized using the Quant Reverse Transcriptase Kit (Tiangen), and the operation was performed according to the instructions. The first strand of the synthesized cDNA was used as a template for PCR with a high-fidelity Taq enzyme Prime-STAR Max DNA Polymerase (TaKaRa). The PCR conditions were 94 ℃ for 5 min, followed by 35 cycles of 94 ℃ for 30 s, 55 ℃ for 30 s and 72 ℃ for 2 min, and 1 cycles of 72 ℃ for 10 min. The amplification products were detected by 1.2% agarose electrophoresis, and the PCR products were recovered and purified by gel DNA recovery kit (Tiangen).
Construction of prokaryotic expression vector
The HSP70 gene and the expression vector pColdI (TaKaRa) were subjected to double-enzyme digestion with Bam HI and Hind III, respectively. After purifying the digested fragments, they were ligated and transformed into E. coli Top10 competent cells. Positive colonies were selected for suspension culture, and plasmids were extracted. The transformants inserted into the target gene were identified by double-enzyme digestion, and sequenced to ensure the correctness of the recombinant expression vector.
Induced expression of pColdI-HSP70 fusion protein
The correctly sequenced pCold I-HSP70 recombinant plasmids were transformed into BL21 competent cells, and single colonies were selected and inoculated into 5 ml of LB medium (containing 50 μg/ml Amp and 20 μg/ml chloramphenicol), cultured at 37 ℃ overnight, and then inoculated to 500 ml of fresh LB medium (containing 50 μg/ml Amp, 20 μg/ml chloramphenicol and 5 ng/ml tetracycline) at a ratio of 1∶100 (V∶V). Culture was performed at 37 ℃ until the bacterial liquid OD600=0.4-0.6, and the bacterial liquid was placed in a refrigerator at 4 ℃ for 20 min, and then stood on a shaker at 15 ℃ for 20 min to cool it down. The induced expression of the bacterial liquid was carried out according to the orthogonal experimental design (Table 1). Meanwhile, the engineered bacteria without IPTG-induced pColdI-HSP70 expression and the engineered bacteria with empty vector pColdI induced by IPTG were designed as controls.
SDS-PAGE analysis of expression products
First, 1 ml of each bacterial liquid was centrifuged at 12 000 r/min for 10 min at 4 ℃, and the supernatant was discarded. Then, 1 ml of 1×PBS was added to re-suspend the precipitate, which was then lysed ultrasonically (power 300 w) for 20 min, and then centrifuged at 12 000 r/min at 4 ℃ for 30 min. The supernatant and the precipitate were respectively taken for 12% SDS-PAGE electrophoresis detection and Coomassie brilliant blue staining, and photographed after destaining with a destaining solution.
qRT-PCR analysis of the HSP70 gene
qPCR reactions were performed using Cham QTM Universal SYBR qPCR Master Mix Kit (Vazyme) on a ViiATM 7 real-time fluorescence quantitation system (Thermo Fisher). The reaction system was 20 μl, containing 2×ChamQTM Universal SYBR qPCR Master Mix 10.0 μl, diluted cDNA (40 ng/μl) 2 μl, primer P3 (10 μmol/L) 0.4 μl, primer P4 (10 μmol/L) 0.4 μl and RNAase free ddH2O 7.7 μl. The qPCR conditions were 95 ℃ for 30 s, followed by 40 cycles of 95 ℃for 5 s and 60 ℃ for 30 s. Three technical replicates were set for each sample, and rice 18S rRNA gene was selected as the reference gene of the experiment. The experimental data were statistically analyzed by the 2-ΔΔCt method.
Results and Analysis
Extraction of total RNA and PCR amplification of the HSP70 gene
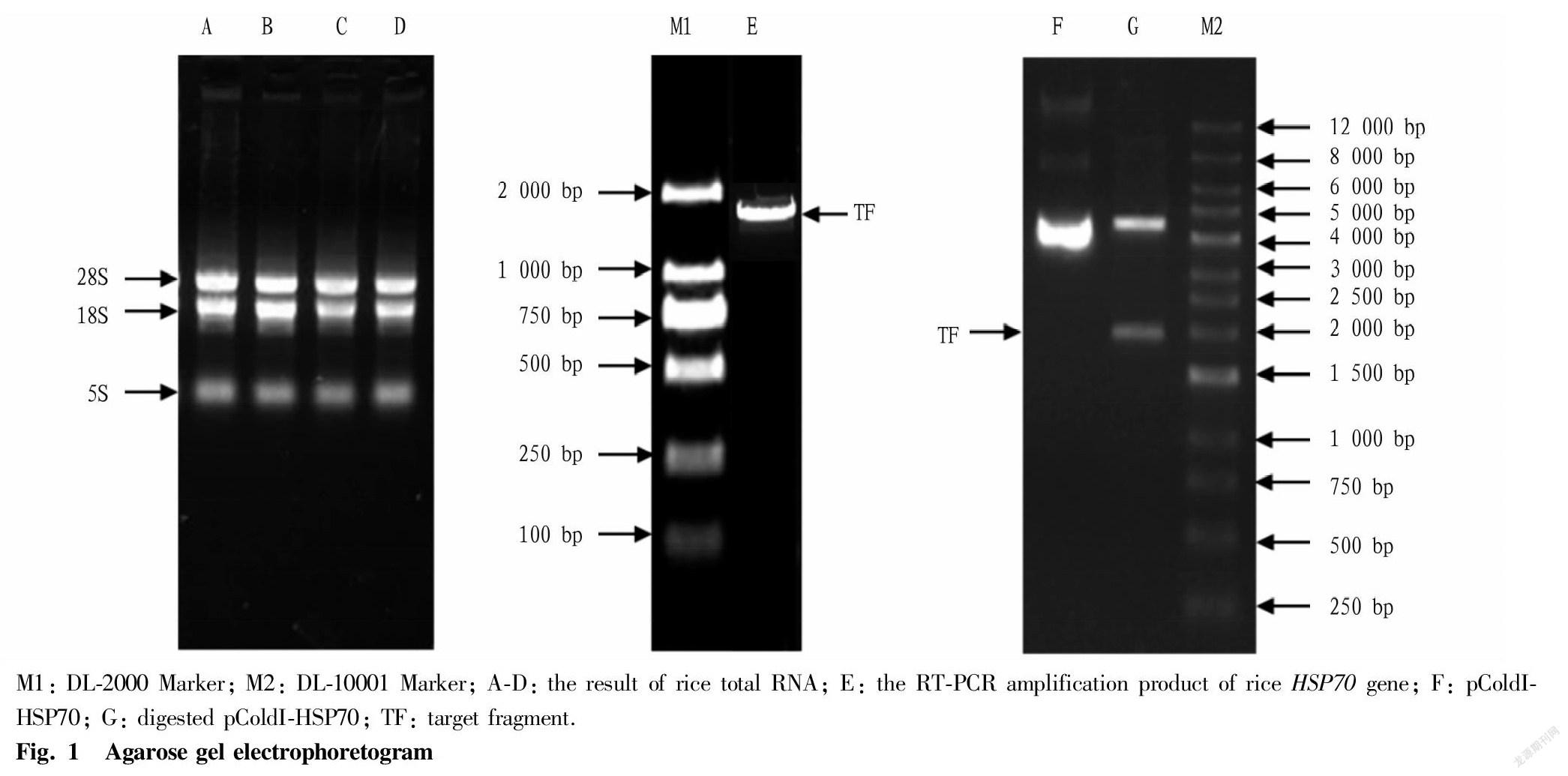
Total rice RNA was extracted according to the instructions of RNAiso Plus kit, and detected by agarose gel electrophoresis. The results showed that the RNA samples showed three bands, of which the 28S and 18S bands were clear and bright (Fig. 1, lanes A, B, C, and D). The OD260/OD280 detected by a nucleic acid analyzer was between 1.8 and 2.2, and the OD260/OD230 was greater than 1.8, indicating that the RNA extracted in this study had good integrity and quality, and could be used for the synthesis of the first strand of cDNA. The primers P1 and P2 were used for RT-PCR, and the products were analyzed by agarose gel electrophoresis. A specific band appeared at about 1.9 kb (Fig. 1, lane E), which was consistent with the expected size of the HSP70 gene.
Construction of prokaryotic expression vector pColdI-HSP70
In Fig. 1, G lane was the electrophoresis result of recombinant plasmid pColdI-HSP70 after double digestion with Bam HI and Hind III, and it can be seen that there is a clear band at about 4.5 and 1.9 kb, respectively, which was consistent with the expected result. Meanwhile, the sequencing result of the inserted DNA fragment of the recombinant vector was exactly the same as that of the rice HSP70 gene, indicating that the prokaryotic expression vector of the rice HSP70 gene was successfully constructed.
Fei TONG et al. Expression Analysis of Heat Shock Protein 70 Gene in Rice (Oryza sativa L.)
SDS-PAGE analysis of HSP70 gene recombinant protein
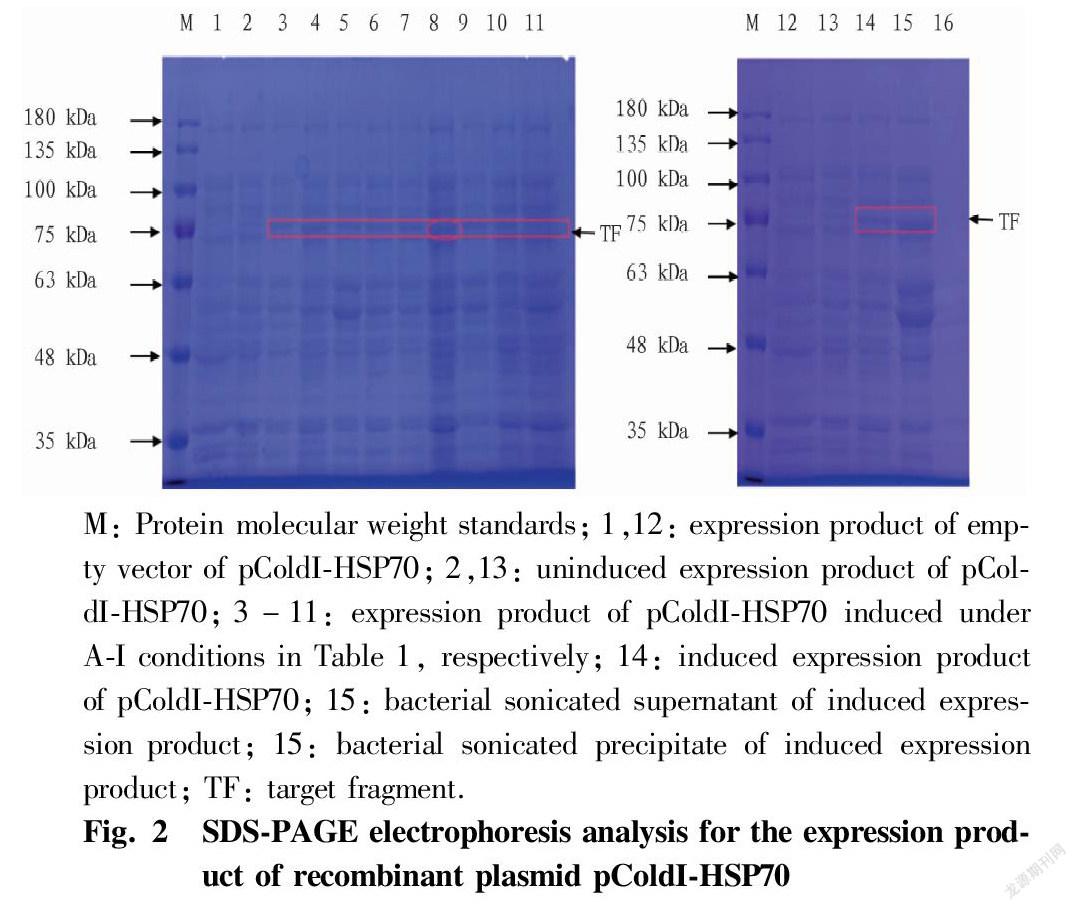
After the recombinant plasmid containing HSP70 was transformed into the expression strain, nine orthogonally-designed induced experimental groups were obtained by concentration (0.1, 0.5 and 1.0 mmol/L), induction temperature (10, 15 and 20℃) and induction time (12, 24 and 36 h). Fig. 2 (lane 1-11) shows the SDS-PAGE results of various induced expression experimental groups, as well as the recombinant bacteria uninduced by IPTG and the control bacteria with empty vector induced by IPTG. The results showed that a nascent protein band appeared at 75 kDa in the samples of the nine experimental groups induced by IPTG, and excluding the 3.5 kDa protein expressed by the pColdI vector itself, the results were consistent with the expected target protein (71.6 kDa). Among them, the abundance of the expressed protein in the F experimental group was the highest, so the optimal induction conditions of the fusion protein pColdI-HSP70 were 0.5 mmol/L IPTG and induction at 20 ℃ for 36 h. The uninduced recombinant bacteria and the empty vector did not express the protein. From the protein electrophoresis images of the supernatant and precipitate after sonication (Fig. 2, lanes 15-16), it can be seen that the HSP70 gene expression product mainly existed in the supernatant of the bacterial lysate, and there was almost no in the precipitate, so most of the rice HSP70 protein existed in the cells in a soluble form.
Expression of the rice HSP70 gene under different stresses
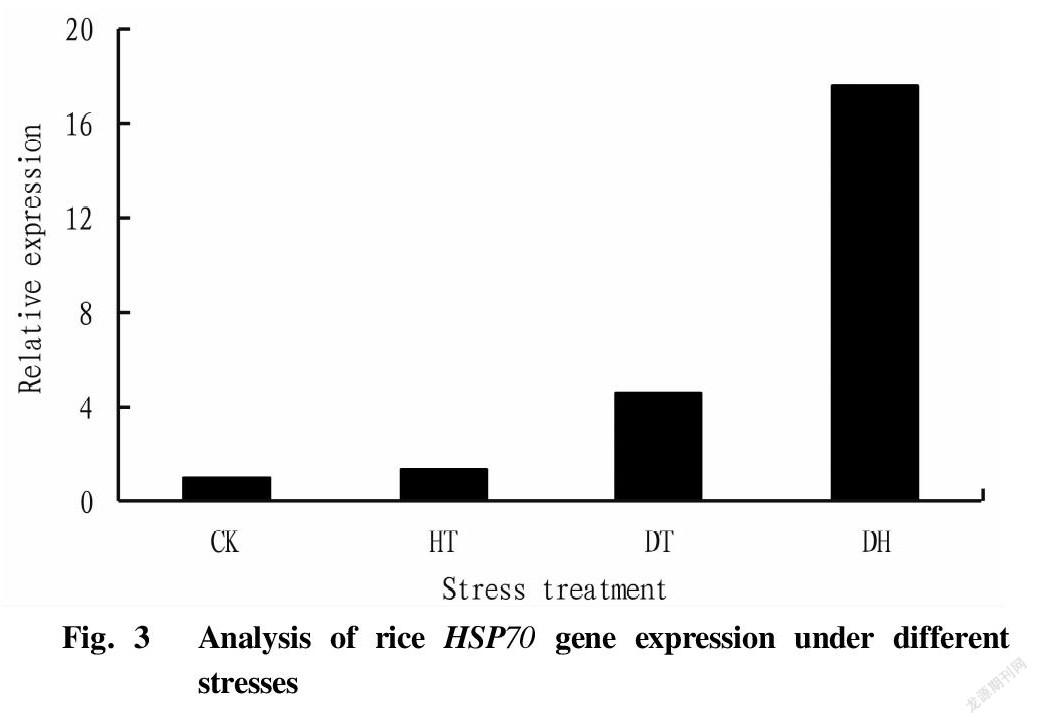
The expression of rice HSP70 gene after different forms of stress is shown in Fig. 3. As can be seen from the figure, compared with the control (CK), the expression of the HSP70 gene was induced in rice under stress. The expression level of the HSP70 gene in the high temperature stress treatment (HT) group was 4.65 times that of the CK group; the drought stress treatment (DT) was 1.40 times that of the CK group; and the expression level of the drought-high temperature cross stress treatment group was the highest, 17.66 times higher than that of the CK group. The results showed that the cross stress of drought and high temperature significantly induced the expression of the HSP70 gene, which might be because compared with the single high temperature treatment, the pretreatment of drought stress stimulated the expression of the HSP70 gene in rice, thereby enhancing the resistance of rice to high temperature, that is, cross adaptability was generated.
Conclusions and Discussion
Plant HSP70s are a family of molecular chaperone proteins with important functions that maintain homeostasis in plants. It is widespread in plants and plays an important role in enhancing plant stress tolerance[2,4]. In this study, a gene (Os03g16920) in the rice HSP70 gene family was analyzed by qRT-PCR and prokaryotic expression. According to subcellular localization, plant HSP70 is mainly divided into four categories, namely HSP70 located in cytoplasm, HSP70 in endoplasmic reticula, HSP70 in mitochondria and HSP70 in chloroplasts, and the Os03g16920 gene is located in the cytoplasm. Jung et al.[22] used gene chips to detect differential expression patterns of 32 rice HSP70 genes under different stresses (heat, drought, light and salt), and the results showed that the responses of rice cytoplasmic HSP70 genes to drought, salt and heat were all up-regulated, and Os05g38530, Os01g62290 and Os03g16920 among them had the greatest responses. In this study, HSP70 was also up-regulated under different forms of stress, and the expression levels of the HT, DT and DH groups were 4.65, 1.40 and 17.66 times higher than that of the CK group, respectively. The expression of HSP70 was significantly induced under DH stress, indicating that the rice HSP70 (Os03g16920) gene plays an important role in rice drought and high temperature cross adversity, and may be an important gene related to rice cross adaptation, which provides a theoretical basis for the screening of rice stress resistance genes.
Few studies have been reported on the prokaryotic expression of rice HSP70 genes. Prokaryotic expression can obtain a large number of recombinant proteins with biological activity, has the advantages of simple operation, good stability, and economic benefits, and thus lays a foundation for further research on protein functions. In the process of prokaryotic protein expression, expression vectors, host strains and expression induction conditions are the keys to the success of experiments. In this study, the vector pColdI with the cold shock gene cspA promoter was used, which effectively overcame the problems of difficult gene expression and insolubility of expressed proteins. In the selection of host strains, E. coli BL21 was selected in this study, and a kind of molecular chaperone protein plasmid (pG-Tf2) was transformed in its competent cells and co-expressed with the HSP70 protein to increase the ratio of soluble proteins. The induced expression conditions were optimized by designing a three-factor and three-level orthogonal experiment. The results showed that the optimal induced expression conditions of the fusion protein pColdI-HSP70 were 0.5 mmol/L IPTG and induction at 20 ℃ for 36 h. Prokaryotic expressed proteins are generally divided into two parts, soluble proteins in supernatants after sonication and inclusion bodies in precipitates. The soluble proteins in supernatants generally have the same molecular structure as natural proteins, and often have the biological activity of natural proteins, which is convenient for the next step of protein separation and purification. The protein expressed in this study mainly existed in the supernatant, and purification and analysis will be further carried out.
References
[1] HAN SM, LIU Y, CHANG A. Cytoplasmic HSP70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1[J]. The Journal of biological chemistry, 2007, 282(36): 26140-26149.
[2] WANG MQ, ZHANG DY. Research advance of heat shock protein 70 gene family and its biological functions in plant[J]. Genomics and Applied Biology, 2015, 34(2): 421-428. (in Chinese).
[3] QI MD, XIE X, WEI FJ. Research progress of HSP70s in Poaceae[J]. Plant Physiology Communications, 2019, 55(8): 1054-1062. (in Chinese).
[4] GEORGOPOULOS C, WELCH WJ. Role of the major heat shock proteins as molecular chaperones[J]. Annual Review of Cell Biology, 1993, 9(1): 601-634.
[5] LINDQUIST S, CRAIG EA. The heat-shock proteins[J]. Annual review of genetics, 1988, 22(1): 631-677
[6] DING YD, SHU HY, GAO CL, et al. Analysis of heat shock protein 70 gene family in Capsicum chinense Jacq.[J]. Plant Science Journal, 2021, 39(2): 152-162. (in Chinese).
[7] GONG XG, YU H. Functions of HSP60[J]. Progress in Biochemistry and Biophysics, 2003(4): 649. (in Chinese).
[8] ZHENG HX, GAO YL, ZHANG FM, et al. Cloning of heat shock protein gene Ld-HSP70 in Leptinotarsa decemlineata and its expression characteristics under temperature stress[J]. Scientia Agricultura Sinica, 2021, 54(6): 1163-1175. (in Chinese).
[9] ZHAO X, ZHANG TT, XING WT, et al. Genome-wide identification and expression analysis under temperature stress of HSP70 gene family in Dendrobium catenatum[J]. Acta Horticulturae Sinica, 2021, 48(9): 1743-1754. (in Chinese).
[10] SUNG DY, VIERLING E, GUY CL. Comprehensive expression profile analysis of the Arabidopsis HSP70 gene family[J]. Plant physiology, 2001, 126(2): 789-800
[11] SWINDELL WR, HUEBNER M, WEBER AP. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways[J]. BMC genomics, 2007, 8(1): 1-15
[12] FENG BY, LI R, LAI ZX, et al. Cloning and expression analysis of HSP70 in Oncidium hybridum[J]. Chinese Journal of Tropical Crops, 2020, 41(4): 745. (in Chinese).
[13] LUO L, XU XH, YANG K, et al. Senescence and heat shock protein in plants in response to abiotic stress[J]. Pratacultural Science, 2020, 37(11): 2320-2333. (in Chinese).
[14] CHO EK, CHOI YJ. A nuclear-localized HSP70 confers thermos protective activity and drought-stress tolerance on plants[J]. Biotechnology letters, 2009, 31(4): 597-606.
[15] LOPEZ-MATAS MA, NUNEZ P, SOTO A, et al. Protein cryoprotective activity of a cytosolic small heat shock protein that accumulates constitutively in chestnut stems and is up-regulated by low and high temperatures[J]. Plant Physiology, 2004, 134(4): 1708-1717.
[16] LI YB, LI T, HAN YY, et al. Cloning and function analysis of heat-shock-protein LsHsp70-2711 gene under high temperature stress in leaf lettuce (Lactuca sativa L.)[J]. Scientia Agricultura Sinica, 2017, 50(8): 1486-1494. (in Chinese).
[17] SABEHAT A, LURIE S, WEISS D. Expression of small heat-shock proteins at low temperatures: A possible role in protecting against chilling injuries[J]. Plant Physiology, 1998, 117(2): 651-658.
[18] ZHANG XY, CHEN FH, WU GB. Heat-shock proteins (HSP) and relationship of HSP with chilling tolerance of fruits and vegetables[J]. Food Science, 2008, 29(12): 726-730. (in Chinese).
[20] ZOU J, LIU C, LIU A, et al. Overexpression of OsHsp17.0 and OsHsp23. 7 enhances drought and salt tolerance in rice[J]. Journal of plant physiology, 2012, 169(6): 628-635.
[21] KIM BM, RHEE JS, JEONG CB, et al. Heavy metals induce oxidative stress and trigger oxidative stress-mediated heat shock protein (hsp) modulation in the intertidal copepod Tigriopus japonicus[J]. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology, 2014(166): 65-74.
[22] KI-HONG JUNG, HYUN-JUNG GHO, MINH XUAN NGUYEN, et al. Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress[J]. Funct Integr Genomics, 2013(13): 391-402.
[23] MUENCH DG, WU Y, ZHANG YS, et al. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue[J]. Plant and Cell Physiology, 1997, 38(4): 404-412.
[24] LEBORGNE-CASTEL N, JELITTO-VAN DOOREN EP, CROFTS AJ, et al. Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress[J]. 1999, Plant Cell, 11(3): 459-469.
[25] QI YC, WANG HJ, ZOU Y, et al. Overexpression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice[J]. 2001, FEBS Letters, 585(1): 231-239.
[26] SALTVEIT ME. Chilling injury is reduced in cucumber and rice seedlings and in tomato pericarp discs by heat-shocks applied after chilling[J]. Postharvest Biology and Technology, 2001, 21(2): 169-177.
[27] MA WZ, WANG K, ZHAO K, et al. Study on phosphorus distribution in growth and development of greenhouse cucumber under different nutrient solution concentrations[J]. Northern Horticulture, 2015(1): 42-44. (in Chinese).
[28] HUANG DS. The leaf age remainer at the beginning of the differentiation of young panicles of hybrid rice is 3[J]. Hybrid Rice, 1995(6): 33-35. (in Chinese).
Editor: Yingzhi GUANG Proofreader: Xinxiu ZHU
杂志排行
农业生物技术(英文版)的其它文章
- Changes in Physiological and Biochemical Characteristics of Floral Organ
- Research Progress on Lonicera japonica Thunb. Affected by Environmental Stress
- Research Progress on Genetic Diversity of Snap Bean
- Allelopathic Effects of Cedrus deodara Needle Extracts on Seed
- Development Status and Countermeasures of Passiflora spp. Seedling Industry in Qinzhou, Guangxi
- Pathogen Identification and Phylogenetic Analysis of Sugarcane
