Difficult-to-culture bacteria in the rhizosphere:The underexplored signature microbial groups
2022-03-02SadafKALAMAnirbanBASUandAppaRaoPODILE
Sadaf KALAM,Anirban BASU and Appa Rao PODILE
Department of Plant Sciences,School of Life Sciences,Universityof Hyderabad,Hyderabad 500046(India)
ABSTRACT Microorganisms represent a substantial portion of the earth’s biodiversity and biomass,and the plant rhizosphere is an innate reservoir teeming with heterogeneous microbes predominated by bacterial communities.Rhizospheric microbial diversity(genetic,phenotypic,and metabolic)has been extensively studied to understand the key ecological roles played by the microbial members,including plant growth promotion.The application of 16S rRNA gene sequencing and next-generation sequencing(NGS)technologies has revolutionized the discovery of novel bacterial groups that have remained undetected by traditional cultivation-based approaches.Such technological advancements have opened new vistas in our current understanding of predominant but concealed and missed bacterial diversity referred to as difficult-to-culture bacterial lineages,especially the predominant phyla Acidobacteria,Verrucomicrobia,Planctomycetes,and Gemmatimonadetes.Regardless of their ubiquity and prevalence,little is known about their ecophysiology because of the non-availability of culturable members.More recently,there has been increased interest in understanding the cosmopolitan distribution and diversity of the difficult-to-culture bacteria,focusing on their role in driving complex plant-microbial interactions and mobilizing nutrients in soil and their potential as sources of novel bioactive metabolites.As an initial step,we review the distribution and significance of such bacterial phyla in soil,their ecophysiological roles,and their hidden plant growth promoting potential.The ability to select and deploy plant probiotic bacteria from the difficult-to-culture fraction of the bacterial community might open new avenues for improving crop health.
KeyWords:Acidobacteria,ecophysiological roles,Gemmatimonadetes,microbial diversity,Planctomycetes,plant growth promotion,Verrucomicrobia
INTRODUCTION
Microbial biodiversity in terrestrial ecosystems potentially influences important ecophysiological processes because microbes play crucial roles in driving biogeochemical cycles.Soil harbors a complex microbial consortium with an abundant bacterial population.The complex below-ground plant-bacterial interactions,especially in the rhizosphere,are major determining factors of plant health and soil fertility.Rhizosphere bacteria that benefit plants through nitrogen fixation,phosphate solubilization,or by several other indirect and direct mechanisms,broadly termed plant growth promoting rhizobacteria(PGPR)(Dutta and Podile,2010),have been studied extensively.Excellent reviews have discussed the role of such rhizobacteria that are easily obtained as pure cultures on media plates.However,a vast disparity between the bacterial numbers visible on plates and those observed through microscopy ultimately led to the discovery of difficult-to-culture microbes(Stewart,2012).So far,a significant number of microbes have remained cryptic in different ecosystems because of their recalcitrance to cultivation;hence they are referred to as difficult-to-culture microbes.Difficult-to-culture bacteria have been a major challenge for microbiologists seeking to understand their biodiversity,cultivation,and ecological roles.To find ways to explore difficult-to-culture bacteria,a comprehensive understanding of their physical,chemical,and biological processes is required.
Culture-dependent and-independent approaches have together provided new insights about the rhizosphere microbial ecophysiology of difficult-to-culture phyla.The 16S rRNA gene sequencing approach has been a useful and powerful tool in determining the presence,diversity,and abundance of the bacterial members of the difficult-to-culture phyla in soil and rhizosphere bacterial communities(Pisaet al.,2011;da Rochaet al.,2013;Rosselliet al.,2016).Direct isolation and cultivation strategies have been employed to confirm whether these bacteria are really rhizosphere-relevant(Sanguinet al.,2006;Zulet al.,2007).Genetic analyses of these bacterial phyla shed light on their metabolic functions and environmental physiology(da Rochaet al.,2009).The involvement of difficult-to-culture bacteria in fundamental biogeochemical cycles,such as the degradation of organic compounds and mobilization of essential nutrients,has been confirmed.Studies on the rhizosphere microbiome of diverse plant species have reconfirmed the abundance of difficult-toculture bacterial groups in the rhizosphere but have provided very little information on their ecological roles.Such bacteria or bacterial groups may influence plant growth promotion(PGP).Ascertaining PGP activities and understanding the relationships between the triad of the difficult-to-culture bacteria,the rhizosphere,and the plant root system becomes imperative for deciphering their ecological roles.This review presents the most recent advances in understanding the ecophysiology of the underexplored difficult-to-culture rhizosphere bacteria with a particular focus on their role in PGP.
RHIZOSPHERE MICROBIAL COMMUNITIES
Soil is a complex,dynamic,and diverse environment populated with a plethora of diverse microbes that keep it healthy and productive.Rhizosphere soil is under the direct influence of plant roots and differs considerably from the bulk soil in terms of its physico-biochemical properties(Hinsinger,1998)and associated microbial communities.The rhizosphere is home to a wide range of prokaryotic and eukaryotic taxa,of which bacteria and fungi comprise the most abundant groups(Buéeet al.,2009),exhibiting various significant ecological functions.Rhizosphere bacterial communities and their diversity in various soils are primarily determined by soil and climatic parameters across different ecological niches(Lladóet al.,2018),as well as by plant root architecture(Kotani-Tanoiet al.,2007),exudates,biochemistry,and development(Hinsingeret al.,2005).Roots release carbonaceous root exudates into the rhizosphere,which influence the root-associated microbial populations(Ankatiet al.,2019).
Soil bacteria can generally be ecologically classified as copiotrophs or oligotrophs.Copiotrophs prefer nutrient-rich soils and exhibit high growth rates under favorable conditions.On the contrary,oligotrophs prefer low nutrient soils and exhibit slow growth rates(Fiereret al.,2007).Oligotrophs are difficult to isolate from a mixed bacterial community since fast growers dominate.Although researchers have highlighted the crucial roles played by copiotrophs,the roles of difficult-to-culture oligotrophs and facultatively oligotrophic bacteria remain obscure.
A large disparity exists between the total number of bacterial cells inherently present in a given soil sample and that recovered on a solid medium using the same soil as inoculum.This inconsistency has restricted the estimation of actual soil bacterial diversity.Molecular ecological technologies have aided in overcoming this limitation.In this regard,the 16S rRNA genes derived directly from environmental samples have proved crucial in revealing the presence of previously uncultured and undetected difficult-to-culture groups of bacteria(Tringe and Hugenholtz,2008).
Based on the available 16S rRNA gene sequences,nine phyla,Proteobacteria,Actinobacteria,Firmicutes,Bacteroidetes,Planctomycetes,Verrucomicrobia,Acidobacteria,Gemmatimonadetes,and Chloroflexi,dominate soil bacterial communities(Janssen,2006).Of these,the first seven phyla predominate in plant rhizospheres(da Rochaet al.,2009).The relative distribution of cultured and uncultured members of the nine dominant phyla,expressed as a percentage of the total 16S rDNA sequences obtained from the latest accession(release 11.5)of the Ribosomal Database Project(Coleet al.,2014),is shown in Fig.1.Not surprisingly,the percentage of uncultured sequences significantly outnumbers the percentage of cultured sequences for each phylum.Thus,there is a need for an in-depth study of the uncultured members from the rhizosphere to understand their ecological significance.
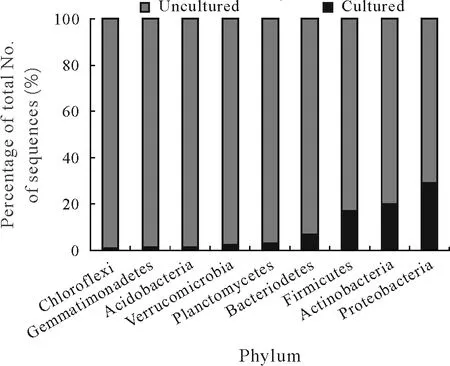
Fig.1 Relative distribution of the nine dominant soil bacterial phyla,expressed as a percentage of the total number of 16S rRNA gene(rDNA)sequences,obtained from cultured and uncultured sources(i.e.,from culturedependent and culture-independent analyses).The figure is a reanalyzed and updated representation of the data from da Rocha et al.(2009).All sequences of near full length(≥1 200 bases)derived from different ecosystems were obtained from the latest accession of the Ribosomal Database Project(release 11.5).
MISSED DIVERSITY:THE RARE BIOSPHERE AND MICROBIAL DARK MATTER
Novel or unrecognized bacterial taxa are present in low abundance within a microbial community and constitute the largely unexplored rare biosphere(Soginet al.,2006;Lynch and Neufled,2015;Bull and Goodfellow,2019).Unprecedented access to this rare biosphere has been accomplished with the aid of amplicon-based 16S rRNA gene biodiversity studies,including pyrosequencing(Janssen,2006;Joneset al.,2009;Shadeet al.,2012)and Illumina sequencing(Bartramet al.,2011).Despite its rarity,the rare biosphere represents a genomic reservoir of microbial diversity influencing ecosystem functioning(Joussetet al.,2017;Pascoalet al.,2020).The microbial dark matter represents the abundant uncultured microbial majority in diverse ecosystems(Rinkeet al.,2013;Ramondet al.,2015;Soldenet al.,2016).It can be detected through molecular studies targeting 16S rRNA gene sequences but is as yet uncultivated or difficultto-culture(Hedlundet al.,2014;Bull and Goodfellow,2019).It is represented by candidate phyla(CP)solely composed of hitherto-uncultured representatives that cannot be ascribed to validly published taxa(Soldenet al.,2016).Investigation of the CP members offers further opportunities and avenues to explore their hidden biotechnological potential(Hedlundet al.,2014).A major interest in sequencing the genomes of CP to understand their genetic makeup and study their metabolic pathways has resulted in new information.The difficult-to-culture bacterial phyla such as Acidobacteria,Verrucomicrobia,Planctomycetes,and Gemmatimonadetes represent a vital component of the microbial dark matter(Rinkeet al.,2013;Soldenet al.,2016)and might play significant ecological roles in the rhizosphere.
The concepts of microbial dark matter and the rare biosphere are often correlated due to their shared lack of cultivable members and representation of missed and as yet undetected microbial diversity(Lynch and Neufled,2015;Ramondet al.,2015;Pascoalet al.,2020).Molecular studies employing culture-independent methods such as metagenomics and single-cell genomics have enabled significant advances in exploring the undetected microbial diversity and have revealed novel candidate taxa of dark matter and rare biosphere microorganisms at or above the suprageneric rank(Bull and Goodfellow,2019).These studies have also led to a better and more detailed understanding of the ecological significance of these largely inaccessible,unknown,and underexplored microbial lineages from a fundamental perspective.
OVERVIEW OF CULTURE-DEPENDENT AND INDEPENDENT APPROACHES TO STUDY DIFFICULTTO-CULTURE BACTERIA
Overmannet al.(2017)reviewed the techniques available for culturing difficult-to-culture bacterial phyla and predicted that such underexplored phyla could represent new and innovative research opportunities.Culture-dependent studies revealed that conventional cultivation media favor the growth of fast-growing bacteria,masking the growth of slow growers,including oligotrophs(Koch,1997;Connon and Giovannoni,2002);however,the use of dilute nutrient media has been successful in isolating some difficult-to-culture bacteria from soil and rhizosphere samples(Watveet al.,2000).Other methods used to isolate bacteria from mixed samples by physical reduction of the bacterial load were reviewed by Pham and Kim(2012)and Stewart(2012).Isolation of the slow-growing oligotrophic bacteria requires extended incubation periods(Stevensonet al.,2004;Daviset al.,2005).Co-cultivation of difficult-to-culture bacteria with helper species has also proved fruitful(D’Onofrioet al.,2010).Certain soil bacteria produce extracellular polymeric substances which could be dissolved with surfactants like pyrophosphate and Tween 80(Böckelmannet al.,2003)to aid their recovery during isolation.
Other techniques for improving recovery of difficult-toculture bacteria,including replacement of agar with gellan gum(Otsukaet al.,2013;Tanakaet al.,2017),the inclusion of polymeric growth substances like xylan(Daviset al.,2005;Sangwanet al.,2005;Saitet al.,2006),and culturing with CO2-enriched air(Navarreteet al.,2013),have been extensively used.Additional approaches include modified media formulations and simulated natural microbial environments,including diffusion bioreactors(Kaeberleinet al.,2002;Chaudharyet al.,2019)and agarose gel microdroplet encapsulation(Zengleret al.,2002).The addition of selected components of root exudates to cultivation media enhanced the recovery of difficult-to-culture bacteria.A summary of the different cultivation strategies adopted to culture the difficult-to-culture phyla and the phyla subsequently recovered is provided in Table I.
The role of difficult-to-culture microbes in the critical rhizosphere processes has been assessed using cultureindependent techniques such as genomics,transcriptomics,proteomics,and metabolomics(Jerez,2008;Chaparroet al.,2014).Multiple genomic and metagenomic rhizosphere studies have revealed the existence of several novel microbial genes involved in many vital pathways(Mireteet al.,2007),including biogeochemical cycles of nutrients,and rhizosphere proteomics has brought to light several proteins and enzymes typically involved in plant-microbe interactions(Kielyet al.,2006).The metabolomics approach has revealed that certain uncommon nutrients found in root exudates are efficiently metabolized by rhizospheric microbes(Micallefet al.,2009).The application of integrated multi-omics technologies to decode the rhizosphere microbial communities has been comprehensively reviewed by Whiteet al.(2017a,b).Multi-omics-based information from environmental metadata can be harnessed to devise novel strategies for culturing hitherto-uncultivable bacteria from various environments(Gutlebenet al.,2018);however,there are still some technological constraints affecting complete multi-omics analyses of environmental samples.
Culture-independent techniques like metagenomics have bypassed traditional culture-dependent methods for assessing microbial diversity in natural ecosystems(Fiereret al.,2012).These DNA-based techniques allow the simultaneous screening of samples for an array of microorganisms.High-throughput sequencing of metagenomic DNA with next-generation sequencing(NGS)platforms has enabledthe discovery of novel difficult-to-culture microbial taxa.While culture-independent techniques like dot-blot and fluorescentin situhybridization(FISH)enable the detection of novel taxa,radioisotope-based techniques like FISHmicroautoradiography,isotope array,and stable isotope probing of nucleic acids provide insights into the functional aspects as well(Dumont and Murrell,2005).The stable isotope probing method,in combination with genomics and transcriptomics,can provide information on the structure,function(including gene expression),and diversity of the active rhizosphere microbial community(Achouak and Haichar,2019).
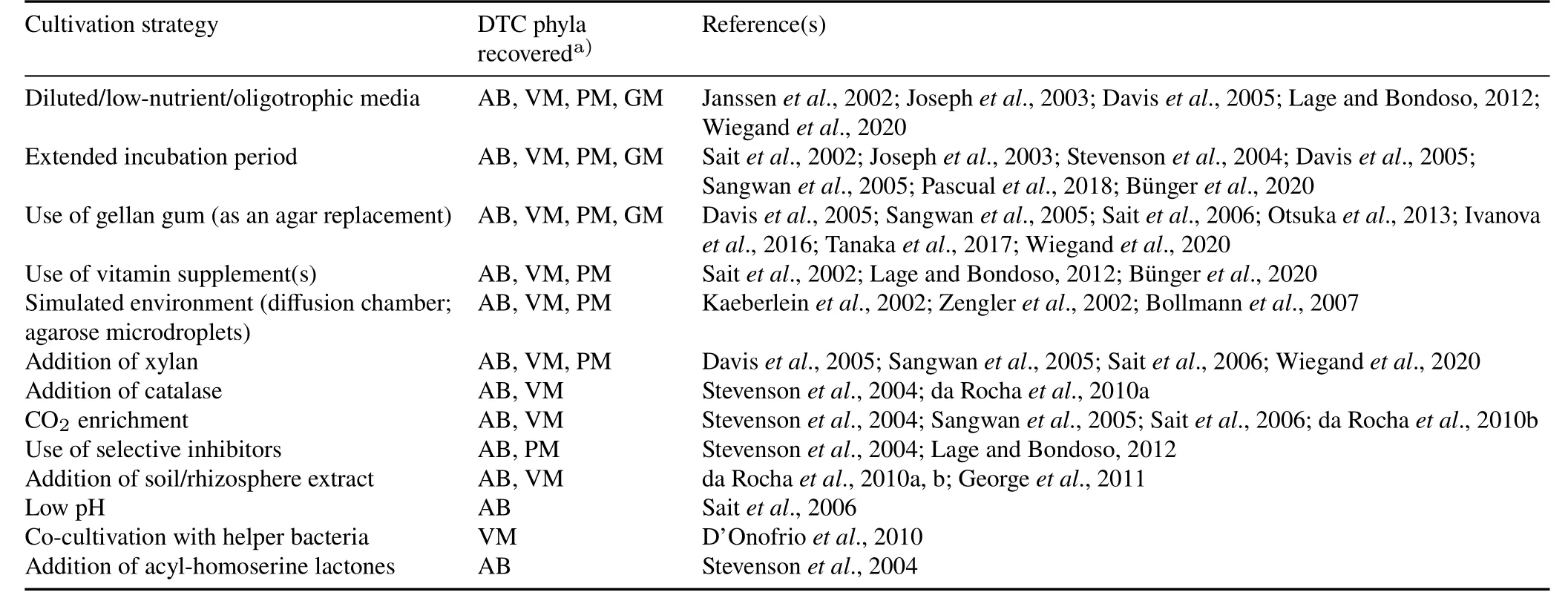
TABLE I Cultivation strategies adopted to culture difficult-to-culture(DTC)phyla
During the past few decades,the use of phylogenetically relevant markers like 16S rRNA gene sequences has been a significant breakthrough in the study of microbial ecology and diversity(Tringe and Hugenholtz,2008;Rosselliet al.,2016).Before the advent of NGS technologies,cultureindependent analysis of microbial community structure,diversity,and dynamics was mostly dependent on 16S rDNAbased molecular fingerprinting techniques such as denaturing gradient gel electrophoresis(DGGE),single-strand conformation polymorphism(SSCP),and terminal restriction fragment length polymorphism(T-RFLP)(Kowalchuket al.,1997;Bäckmanet al.,2003;Fourattet al.,2003;Smallaet al.,2007).Multiple studies have employed culturedependent and culture-independent approaches to target the low-abundance species of the rare biosphere and bacteria belonging to the uncultured dark matter clades within the rhizosphere soil-associated microbial community(Haoet al.,2008;Shadeet al.,2012;Rinkeet al.,2013;Ramondet al.,2015;Pascualet al.,2016).Thus,with the amalgamation of culture-dependent and-independent approaches,identification of several novel and unexplored microbial lineages has been achieved.
INSIGHTS INTO THE ECOLOGICAL ROLES OF PREDOMINANT DIFFICULT-TO-CULTURE BACTERIAL PHYLA
Physiologic,genomic,and metagenomic studies provide evidence of the crucial roles played by difficult-to-culture bacterial phyla inhabiting the soil environment.Nutrient cycling is one of the most essential ecological processes in which microorganisms play a crucial role.Determining rhizospheric difficult-to-culture bacterial interactions and their dynamics within soil ecosystems is of high priority from an ecological perspective.The predominant difficultto-culture bacterial phyla Acidobacteria,Verrucomicrobia,Planctomycetes,and Gemmatimonadetes are genetically and geographically diverse and are also present in extreme environments ranging from plant-soil ecosystems to acid mines.This signifies their possible involvement in several physicochemical and metabolic processes,interactions with plants,and participation in biogeochemical cycles in different ecosystems(Fig.2).This review explores the diversity,distribution,genetic makeup,and ecological significance of these four difficult-to-culture bacterial phyla in the soil.Their salient ecophysiological features are summarized in Table II.
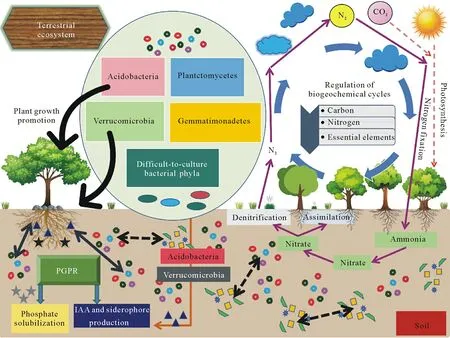
Fig.2 Insights into the ecological roles of difficult-to-culture bacterial phyla in soil designed using resources from freepik.com and pinclipart.com.Intricate plant-microbe interactions(solid bidirectional arrows)and microbe-microbe interactions(dashed bidirectional arrows)operating in soil environment contribute to the regulation of the fundamental processes of nutrient cycling,ecosystem functioning,and plant growth promotion.Predominant difficult-to-culture bacterial phyla in soil,Acidobacteria,Verrucomicrobia,Planctomycetes,and Gemmatimonadetes,are involved in regulating vital biogeochemical pathways,including the cycling of carbon,nitrogen,and other elements.Among them,the phyla Acidobacteria and Verrucomicrobia are known to actively interact with plants as plant growth promoting rhizobacteria(PGPR)through their ability to produce indole-3-acetic acid(IAA)and siderophores and to solubilize phosphates.
Acidobacteria
Based on culture-dependent and-independent studies,the phylum Acidobacteria constitutes one of the most abundant and cultivation-recalcitrant taxa(Georgeet al.,2011).The members of the Acidobacteria are ubiquitous but rarely cultured and are poorly understood(Hugenholtzet al.,1998;Eichorstet al.,2018).The phylum Acidobacteria exhibit oligotrophic characteristics and require low nutrient concentrations for growth,along with long incubation periods(Fiereret al.,2007;Daviset al.,2011).Barnset al.(2007)have extensively analyzed over 400 acidobacterial 16S rRNA gene sequences and classified Acidobacteria into 26 subdivisions.The phylum Acidobacteria and its subdivisions exhibit high genetic diversity and probably are more divergent in their function and physiology than other difficult-to-culture bacterial phyla.
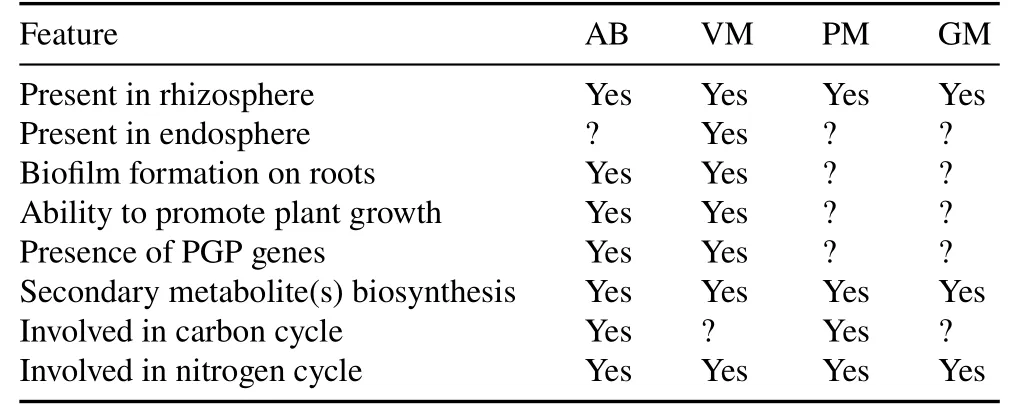
TABLE II Salient ecophysiological features of predominat difficult-to-culture(DTC)bacterial phylaa)in soil
Cultivating Acidobacteria members.The number of cultivable Acidobacteria members is very low compared to the 16S rRNA gene sequence abundance data(Barnset al.,2007).Tanakaet al.(2017)have worked towards improving and modifying cultivation strategies for recovering difficult-to-culture Acidobacteria members.In addition,several approaches mimicking environmental conditions have met with success in improving Acidobacteria recovery(Janssenet al.,2002;Kaeberleinet al.,2002;da Rochaet al.,2009).The approaches include the use of diluted growth media(for creating an oligotrophic environment),growth media supplemented with environmental extracts,humic acid inclusions,antioxidants like catalase(to protect cells from peroxide effects),signaling molecules,and quorum sensing molecules like acyl-homoserine lactones(Saitet al.,2002;Stevensonet al.,2004;Daviset al.,2005).Simple modifications of cultivation techniques,including extending incubation periods and using selective inhibitors to prevent the growth of non-desirable bacteria,were also successful.Organic nutrients,as supplements,hindered Acidobacteria recovery(Stevensonet al.,2004),while lowered pH with elevated CO2levels facilitated the cultivation of Acidobacteria members(Saitet al.,2006).Employing such culture-dependent strategies has led to the recovery of several acidobacterial lineages from environmental samples,rendering this once underrepresented phylum now documented as one of the most widespread phyla in soil.
Distribution of Acidobacteria in the soil environment.The phylum Acidobacteria is a dominant taxonomic group in forest soils involved in potentially important ecological activities(Mushinskiet al.,2018).Represented mainly by subdivisions 1,4,and 6,Acidobacteria constitutes up to approximately 20%of the bacterial 16S rRNA genes recovered from soil(Janssen,2006).Subdivisions 1,2,and 3 are dominant in soil with very low organic carbon and nutrient content.Leeet al.(2008)reported the dominance of Acidobacteria-like sequences in both DNA-and RNA-based clone libraries after analyzing the 16S rRNA gene sequences of chestnut(Castanea crenata)rhizosphere soil.The greater diversity and abundance of acidobacterial populations in bulk soil,in comparison to rhizosphere soil(Sanguinet al.,2006;Fiereret al.,2007;Kielaket al.,2008),could be correlated with carbon availability(de Chaveset al.,2019).This assertion has been supported in studies that employed a similar carbon concentration in cultivation strategies for isolating Acidobacteria(Eichorstet al.,2007;Foeselet al.,2016).da Rochaet al.(2010b)used several culture-dependent and-independent approaches and recovered six distinct acidobacterial groups in the clone library,indicating ubiquity of the phylum in the potato(Solanum tuberosum)rhizosphere.Acidobacteria subdivision 6 members have been reported in plant rhizospheres of grain legumes,maize(Zea mays),Taxussp.,Phragmites australis,Scirpus mariqueter,andSpartina alterniflora(Schmalenberger and Tebbe,2003;Haoet al.,2008;Kielaket al.,2009).
Physicochemical factors affecting acidobacterial abundance
Soil pH is the most important factor determining the abundance of Acidobacteria in the bulk soil and rhizosphere(Eichorstet al.,2007;Naetheret al.,2012;Wanget al.,2019).Evidence in support of this is provided in the phylogenetic clustering data,which becomes more robust when moving from neutral to lower pH(Männistöet al.,2007;Joneset al.,2009).Other physicochemical factors influencing and regulating the abundance of Acidobacteria include the availability of phosphate,potassium,and total carbon in soil(Joneset al.,2009;Zhanget al.,2018).Oligotrophic environments exhibited a selective effect on Acidobacteria abundance and growth(Dunbaret al.,1999;Leeet al.,2008).Among the most typical representatives of soil-inhabiting Acidobacteria,it appears that subdivisions 1 and 3 prefer acidic soils,while subdivisions 4 and 6 show affinity for arid soils(Dedysh and Damsté,2018).The presence of great diversity provides compelling evidence for the existence of highly contrasting metabolic strategies within this phylum.
Insights into the Acidobacteria genome.Understanding the genetic makeup has helped to explain the role of Acidobacteria members in terrestrial biogeochemical cycles and environmental physiological processes(Eichorstet al.,2018).Genomes of three acidobacterial strains(two from subdivision 1(Acidobacterium capsulatumandKoribacter versatilisstrain Ellin 345)and one from subdivision 3(Ellin 6076Solibacter usitatus))have been analyzed by Wardet al.(2009).These strains possess genes encoding enzymes for degradation of complex carbohydrates,xylan,hemicelluloses,pectin,starch,and chitin(Wardet al.,2009;Kielaket al.,2016a,b).Acidobacteria might reduce nitrate,nitrite,and possibly nitric oxide content due to the presence of homolog candidate genes for nitrate reductase(nirA)in Ellin 345,while genes for nitrate transport(nrtABCD)are present in certain other acidobacterial strains.Acidobacterial genomes are also equipped with several secondary metabolite biosynthetic genes,including those for diverse polyketides and nonribosomal peptides(Crits-Christophet al.,2018).Several iron-redox reactions are catalyzed in acidobacterial systems.Acidobacterial genomes containfeoABgenes encoding a high affinity ferrous(Fe2+)iron transporter(Blötheet al.,2008).TheA.capsulatumgenome contains a complete operon for cellulose biosynthesis(Udeet al.,2006;Whiteet al.,2006).Acidobacterial genomes are equipped with lower-specificity cellular translocation machinery systems for sugar transport and high-affinity ABC transporters required for survival in oligotrophic environments(Paulsenet al.,1998).Acidobacteria genomes also contain multiple copies oftonB,exbB,andexbDgenes required for siderophore transport(Llamaset al.,2006).Certain Acidobacteria are known to contain putative addiction modules(toxin-antitoxin systems)encoding genes that constitute an essential mechanism for plasmid maintenance.These genes may enable Acidobacteria to survive oligotrophic environmental conditions.The three sequenced acidobacterial strains,A.capsulatum,Ellin 345,and Ellin 6076,are moderately acidophilic,with candidate genes in the AR3 system suggesting the existence of an acid tolerance mechanism(Wardet al.,2009).
Ecological significance of Acidobacteria.Acidobacteria are endowed with the potential to degrade polymeric carbonaceous complexes.Consequently,they act as decomposers in soil(Wardet al.,2009;Naetheret al.,2012;Rawatet al.,2012)and actively participate in the cycling of organic matter from plants,fungi and/or insects.The acidobacterial genomes are flexible and novel in their carbon metabolizing activity.Thus,a significant contribution is made by acidobacterial enzyme machinery in regulating the carbon biogeochemical cycle(King and Weber,2007).Acidobacteria members also play vital roles in nitrogen transformation cascades in terrestrial ecosystems(Liuet al.,2017).
Verrucomicrobia
Verrucomicrobia is a phylogenetically divergent bacterial phylum(Leeet al.,2009;Bergmannet al.,2011).The members of Verrucomicrobia are cosmopolitan in distribution,occurring in a wide range of terrestrial and aquatic habitats(Haukkaet al.,2006;Dedyshet al.,2006;Breweret al.,2017),suggesting their importance in terrestrial ecological processes.The phylum largely remains defined through the 16S rRNA gene inventories deposited in public databases from diverse environmental samples.The phylum Verrucomicrobia has been divided into seven monophyletic subdivisions(Schlesneret al.,2006).Recently,a putative novel subdivision 8 was proposed(Büngeret al.,2020).Since Verrucomicrobia members are recalcitrant to cultivation,relatively few species have been characterized,and the phylum classification remains informal.
Cultivating Verrucomicrobia members.Despite their abundance and diversity in soil ecosystems,very few members have been recovered in pure culture(Otsukaet al.,2013;Tanakaet al.,2017);however,novel methods devised to overcome their recalcitrant nature have led to the detection of more than 40 new Verrucomicrobia members(Scheuermayeret al.,2006;Yoonet al.,2007).Although Verrucomicrobia members have been isolated and reported from marine environments,their isolation from plant-soil ecosystems is still in early development.To date,Verrucomicrobia abundance in natural ecosystems was identified by employing primers or probes covering multiple subdivisions of this phylum.Thus,minority groups within this phylum might have been ignored,probably leading to underestimating Verrucomicrobia distribution in plant-soil communities.
Distribution of Verrucomicrobia in the soil environment.Verrucomicrobia representatives are highly abundant in soils,constituting more than one-fifth of the total soil bacterial community(Springet al.,2016).However,their biology and ecology remain largely unexplored(Buckley and Schmidt,2002;Breweret al.,2017).Chinet al.(2001)isolated three obligately anaerobic strains of Verrucomicrobia belonging to the genusOpitutusfrom paddy(Oryza sativa)soil microcosms.Sangwanet al.(2004)isolated and characterized the first known pure culture representative of subdivision 2,an aerobic heterotrophic soil bacterium—Chthoniobacter flavus.Cultivable strains of Verrucomicrobia subdivision 1 have been isolated from potato(Solanum tuberosum)and leek(Allium porrum)rhizospheres(da Rochaet al.,2010a,b).According to molecular ecological surveys,the members of subdivisions 2 and 3 occur more abundantly in soil bacterial communities(Chowet al.,2002;Lileset al.,2003;da Rochaet al.,2010a).Conflicting reports on the distribution of Verrucomicrobia members indicate both rhizosphere and bulk soil inhabitation(Buckley and Schmidt,2001;Sanguinet al.,2006;Kielaket al.,2008).Understanding the physiology,functions,and genetics of Verrucomicrobia will help to determine their ecological roles and soil abundance(Sangwanet al.,2004).One approach to gaining more insight into Verrucomicrobia ecology involved isolating and analyzing the 16S rRNA gene sequences from different ecological niches.Based on the 16S rRNA gene libraries and real time quantitative PCR analyses,a dominant presence of verrucomicrobial numbers in the rhizosphere was identified and shown to vary between plant types(Kielaket al.,2008).
Physicochemical factors affecting verrucomicrobial abundance.Verrucomicrobia have been recovered from a variety of soil conditions.The community structure of Verrucomicrobia is affected mainly by soil types and nutritional regimes(Buckley and Schmidt,2001;Kuramaeet al.,2012),plant type(Berg and Smalla,2009;Hartmannet al.,2009),and soil pH,moisture,and carbon/nitrogen(C:N)ratio(Shenet al.,2017).The other factors influencing verrucomicrobial abundance and distribution include temporal and seasonal variations,soil management history,and soil depth(Buckley and Schmidt,2001).Although reports indicate the importance of Verrucomicrobia as rhizosphere colonizers(da Rochaet al.,2009,2010b),detailed information regarding their genotypic and/or phenotypic characteristics is yet to be determined.
Insights into the genome of phylum Verrucomicrobia.Wertzet al.(2012)analyzed the draft genome of Verrucomicrobia strain TAV2(Diplosphaera colitermitum)and identified the presence ofnifAandanfAgenes along with the transcriptional activators atnifandanfpromoters.TAV2 also containsglnB,which is an activator ofnifA.In TAV2,the deduced amino acid sequence of the NifH and AnfH proteins revealed the presence of the Walker-A motif and Fe4S4binding sites,which are conserved in all nitrogenase iron-proteins.In addition,TAV2 exhibits growth on a nitrogen-free medium.All these data strongly suggest the functionality of these enzymes and signify that nitrogen fixation could be an essential attribute of the phylum Verrucomicrobia.With the aid of high throughput sequencing technologies,the distribution of nitrogenase genes within the phylum would be easier to assess.Additionally,increased cultivation strategies and nitrogenase assays would be useful in evaluating the importance of nitrogen fixation genes present in the Verrucomicrobia genome(Wertzet al.,2012).Büngeret al.(2020)reported the presence of nitrogen cycling genes likenirA,nirBD,nirK,nosZ,andnasABinvolved in denitrification and transport of nitrite/nitrate in verrucomicrobial genomes.Complete or draft genome sequences available for a few Verrucomicrobia strains may provide new perspectives into the genetic makeup of culturable members of this phylum(Kielaket al.,2009),leading to exploration of their putative biotechnological potential.Single-cell sequencing and micro-cultivation(Inghamet al.,2007;Podaret al.,2007)have provided new windows into their hidden genomic diversities.Analyses of 19 draft verrucomicrobial genomes,through time-series metagenomics,by Heet al.(2017)identified the presence of a substantial number of glycoside hydrolases and carbohydrate transport genes,indicating Verrucomicrobia members to be potential polysaccharide degraders.Additionally,TonB-dependent receptor genes involved in transmembrane carbohydrate and iron(siderophore)transport,sulfatase genes for hydrolysis of sulfate esters in sulfated polysaccharides,and phosphate metabolism-related gene clusters,phoRB,phoA,phnA,andpstABC,are also present in verrucomicrobial genomes(Heet al.,2017;Büngeret al.,2020).Reconstruction of 135 verrucomicrobial genomes has indicated the presence of multiple biosynthetic gene loci for secondary metabolites such as polyketides and nonribosomal peptides(Crits-Christophet al.,2018).
Planctomycetes
The phylum Planctomycetes largely consists of an unexplored group of microorganisms that are ubiquitously distributed in aquatic(freshwater and marine)and soil habitats,including the rhizosphere(Wanget al.,2002;Aghnatioset al.,2015),as well as in extreme habitats(Papineauet al.,2005;Ivanovaet al.,2016).Culture-independent molecular studies have revealed a vast diversity of Planctomycetes(Rosselliet al.,2016).Until 2002,the phylum Planctomycetes had only four described and cultured genera—Planctomyces,
Pirellula,Gemmata,andIsosphaera,along with twoCandidatusgenera(Wanget al.,2002).During the following ten years,the phylum was reported to contain 3 orders,11 described genera,and 14 species along with 5Candidatusgenera with 14Candidatusspecies(Lage and Bondoso,2012).The count of validly described planctomycete species had risen to 35 by 2017(Wiegandet al.,2018).Recently,Wiegandet al.(2020)reported the isolation and functional characterization of 79 Planctomycetes strains from diverse environmental samples ranging from aquatic to soils and sediments and provided new insights into planctomycete biology.
The phylum Planctomycetes primarily contains autotrophic bacteria,capable of unique anammox metabolism,which has revolutionized the perspective on microbial nitrogen cycling in these bacteria(Bengtsson andØvreås,2010).Members of this phylum are comparatively slow-growing with low nutritional carbon and nitrogen requirements,and fast-growing bacteria easily outgrow them on conventional media(Lage and Bondoso,2012).Out of the few cultured Planctomycetes representatives,the majority have been isolated from aquatic sources.Previously,soil Planctomycetes members were identified only by employing 16S rRNA gene sequencing andin situhybridization procedures;however,Wanget al.(2002)demonstrated that Planctomycetes could also be isolated from soil samples.
Genome mining studies have indicated that Planctomycetes possess unique and characteristic features useful for biotechnological applications(Graçaet al.,2016).Planctomycete genomes encode multiple(approximately 2—15)biosynthetic gene clusters involved in the production of polyketides and nonribosomal peptides(Wiegandet al.,2020).Many planctomycete genomes are equipped with gene families encoding various sulfatases,glycosyl hydrolases,polysaccharide lyases,transporters,response regulators,and protein kinases(Kimet al.,2016).Extensive genome studies ofPaludisphaera borealisPX4Tby Ivanovaet al.(2017)have revealed the presence of genes encoding an array of carbohydrate-active enzymes as well as chemotaxis genes,cheA,cheB,cheR,andcheW.Metagenome-assembled genomes from marine ecosystems(TARA Oceans Project)have been characterized(Delmontet al.,2018),providing the first genomic evidence of nitrogen fixing Planctomycetes in the surface ocean.In addition,PCR assays have confirmed the presence ofnifDandnifHgenes in novel diazotrophic Planctomycetes genomes(Delmontet al.,2018).Planctomycetes can utilize N-acetylglucosamine as the sole source of carbon and nitrogen,andFimbriiglobus ruberSP5Tgenome analyses have revealed the presence of genes encoding several chitinolytic enzymes(Ravinet al.,2018).Genome downsizing might be a significant result of adaptation to freshwater environments in the few Planctomycetes families possessing sediment or soil ancestors(Andreiet al.,2019).
Based on comparative analyses of gene sequences,the phylum Planctomycetes has been grouped together with the phyla Verrucomicrobia,Chlamydiae,Lentisphaera,Candidatus Poribacteria,and the candidate division OP3,to form the Planctomycetes-Verrucomicrobia-Chlamydiae(PVC)superphylum(Wagner and Horn,2006).The PVC superphylum possesses environmental relevance and putative ecological roles(van Niftrik and Devos,2017)that need to be explored to reveal its hidden biotechnological potential.
Gemmatimonadetes
Gemmatimonadetes is another phylum whose members are frequently found in diverse environments and exhibit a ubiquitous distribution pattern in terrestrial ecosystems(De-Bruynet al.,2011).The relative abundance and prevalence of Gemmatimonadetes(comprising 2%of soil bacterial communities)indicate their key position as one of the top nine soil bacterial phyla(Janssen,2006).The highest proportion of Gemmatimonadetes phylotypes has been recorded in the following soil types:grassland soil(26.4%of sequences),agricultural soil(13.1%),forest soil(11.1%),and contaminated soil(20.6%)(Janssen,2006).Although members of this phylum are adapted to low soil moisture conditions,mainly thriving in alpine,grassland,prairie,pasture,and other such soils,the 16S rRNA sequences related to Gemmatimonadetes have also been found in marine as well as eutrophic lake sediments(DeBruynet al.,2011).
There are at least six cultured representatives of this rare and understudied phylum.Among them,only three strains are characterized:Gemmatimonas aurantiacaT-27(Zhanget al.,2003),Gemmatirosa kalamazoonensisKBS708(DeBruynet al.,2013),andGemmatimonas phototrophicaAP64(Zenget al.,2015).The latter represents the sole phototrophic strain described in the phylum(Zenget al.,2016).A novel Gemmatimonadetes species,Roseisolibacter agri,has been isolated from an agricultural floodplain soil(Pascualet al.,2018).
Gemmatimonadetes was formerly referred to as a candidate phylum BD or KS-B(Zhanget al.,2003).Information is not available on the physiology and genetic makeup of the phylum owing to the lack of a substantial number of cultured representatives.However,its phylogenetic breadth has been evidenced by environmental sequence data.An analysis of reconstructed genomes from 49 Gemmatimonadetes species revealed that the genomes possess a few secondary metabolite biosynthetic gene clusters for polyketides and nonribosomal peptides(Crits-Christophet al.,2018).Genome analysis ofG.aurantiacaT-27 revealed the presence of nitrite and nitrous oxide reductase genes(nirKandnosZ),indicating its potential involvement in the biogeochemical cycling of nitrogen(Chee-Sanfordet al.,2019).However,it lacks nitrate and nitric oxide reductase genes(nar,nap,nas,andnor)required for complete denitrification.Certain members of this phylum contain fully functional pheophytinquinone(type 2)photosynthetic reaction centers,exhibiting bacteriochlorophyll-based phototrophy(Zenget al.,2014).The members can utilize an array of organic substrates,including sugars,amino acids,and polymers(Daviset al.,2005).
DIFFICULT-TO-CULTURE BACTERIA AS POTENTIAL PGPR
The difficult-to-culture bacterial taxa have been widely reported from rhizosphere soils,suggesting that they may have PGP abilities(da Rochaet al.,2010a,b).As already stated,free-living soil bacteria that inhabit the rhizosphere and improve overall plant health,leading to augmented plant growth,are referred to as PGPR(Dutta and Podile,2010;Goswamiet al.,2016).The PGPR are considered to be important plant probiotics(Flores-Félixet al.,2015)as they increase crop yields,act as biocontrol agents,enhance resistance to phytopathogens,promote nodulation in legumes,and enhance the emergence of seedlings(Parrayet al.,2016).The most common genera exhibiting PGP includeAcinetobacter,Agrobacterium,Arthrobacter,Azotobacter,Azospirillum,Burkholderia,Bradyrhizobium,Rhizobium,Frankia,Serratia,Thiobacillus,Paenibacillus,Pseudomonas,andBacillus(Podile and Kishore,2006;Ahmadet al.,2008).After the rhizosphere was defined as a second genome to the plant(Berendsenet al.,2012),attempts were made to provide an in-depth understanding of PGPR mechanics in the rhizosphere.
The PGPR increase plant growth through a variety of direct mechanisms that involve the plant or indirect mechanisms that occur outside the plant.However,in both cases,there is a direct influence on plant metabolism(Parrayet al.,2016;Egamberdievaet al.,2017).Indirect stimulation of plant growth could be due to the suppression of deleterious microorganisms(that inhibit plant growth and development),inhibition or killing of phytopathogens(by producing siderophores,cyanides,lytic enzymes,chitinases,and/or antibiotics),competition for space and nutrients within the vicinity of plant roots,and/or induction of systemic resistance in the host plants against a wide range of phytopathogens.Direct stimulation includes providing plants with compounds such as phytohormones(e.g.,indole-3-acetic acid,indole butyric acid,cytokinins,and gibberellins),fixed nitrogen,or solubilized mineral nutrients(e.g.,zinc,iron,phosphorus,and potassium)from the soil(Podile and Kishore,2006;Ahmadet al.,2008;Dutta and Podile,2010;Vaikuntapuet al.,2014;Kalamet al.,2017).The PGPR also produce ACC deaminase that promotes plant growth by lowering plant ethylene levels(Glick,2014).
Rhizosphere colonization bydifficult-to-culture bacteria
Extensive root and rhizosphere colonization by PGPR is crucial for the successful establishment of beneficial interactions with plants(Dutta and Podile,2010;Kalamet al.,2020).Rhizosphere colonization broadly refers to the ability of bacteria to migrate towards plant roots to utilize associated nutrients(Lugtenberg and Kamilova,2009).The migration step of bacteria towards plant roots has been pertinently considered the most crucial step in rhizosphere competence(Rudrappaet al.,2008).Rhizosphere competence or rhizocompetence is regarded as the ability of microbes to compete with other rhizosphere dwellers for nutrition from root secretions and for occupying sites on the root surface or in the rhizosphere(Lugtenberg and Kamilova,2009;da Rochaet al.,2013).Rhizosphere competence,which differentiates the rhizosphere colonizing strains from the non-rhizospheric ones,can vary within a phylum or subdivision(da Rochaet al.,2011).Inoculation of seeds and seedlings with microorganisms has proved to be an efficient way to control diseases,and has also enhanced associative nitrogen fixation and the production of plant growth stimulating compounds.However,this approach is unsuccessful when the inoculated species fail to establish in large numbers due to the rhizocompetence of native microbiota(Schroth and Weinhold,1986).
A particular bacterial species might fail to successfully colonize plant roots as a consequence of repelling mechanisms due to the presence of strong microbial competitors in the vicinity(da Rochaet al.,2011).da Rochaet al.(2011)examined the colonization pattern of two soil indigenous Verrucomicrobia subdivision 1 strains,LuteolibacterandCandidatusgenusRhizospheria,in the leek rhizosphere.The latter exhibited preference for rhizosphere compartments,while the former favored bulk soil.Thus,the two strains,although belonging to the same subdivision,may display differences in rhizosphere colonization due to differential responses in the chemotactic migration of cells towards roots along the root exudation gradient.Similarly,Acidobacteria subdivision 8 members(Holophagae)were found proximal to leek roots but not to roots of grass(Lolium perenne)and potato.These findings suggest that differential rhizosphere colonization and competence of soil bacteria are strongly influenced by the plant roots(da Rochaet al.,2013).Such observations indicate the need to investigate novel approaches for exploring the difficult-to-culture groups to understand their rhizosphere competence in plant-soil environments.
Predominat phyla.Members of the phylum Acidobacteria are metabolically active in rhizosphere soil(Leeet al.,2008).The presence of genes regulating carbon,nitrogen,and iron metabolism in soil acidobacterial strains suggests their role in PGP(Wardet al.,2009).Kielaket al.(2016a)discussed the genomic and metagenomic analyses of Acidobacteria members highlighting their innate capacity to produce exopolysaccharides.Further,Kielaket al.(2016b)confirmed active interactions of Acidobacteria members belonging to subdivision 1 with plants and their potential as PGPR.Results ofin vitrodetermination of PGP traits indicated that the test acidobacterial strains of the generaGranulicellaandAcidicapsacould produce siderophores and auxins such as indole-3-acetic acid(IAA).The strains also exhibited biofilm formation,which enables them to grow successfully along root surfaces.TreatedArabidopsis thaliana(Col 0)plantlets showed a significant increase in plant growth.Kalamet al.(2017)investigated the effect of three PGPR,Sphingobacteriumsp.,Variovoraxsp.,andRoseomonassp.,on the population densities of acidobacterial members in the rhizospheres of two crop plants:tomato(Solanum lycopersicum)and black gram(Vigna mungo).There was a gradual increase in equivalent cell numbers of Acidobacteria members with a concurrent increase in PGP by the test PGPR.The potential of two acidobacterial strains of the genusGranulicellato secrete extracellular polymeric substances,including exopolysaccharides(EPSs),was assessed by Kielaket al.(2017).The EPSs might be actively involved in the formation of the soil matrix and soil aggregates,and could be explored as a novel source of biodegradable and non-toxic biopolymers.More information on the specific ecophysiological roles of rhizospheric Acidobacteria members with regard to plant growth and development is yet to come.The results from such studies are expected to elucidate the PGP potential of acidobacterial members.
Verrucomicrobia.The preponderance of Verrucomicrobia members in soils suggests they play a metabolically active role in their respective niches(Felske and Akkermans,1998;Hacklet al.,2004).Certain Verrucomicrobia members possess the genetic machinery to catalyze nitrogen fixation(Wertzet al.,2012).A recent study by Büngeret al.(2020)reported the isolation and characterization of four novel Verrucomicrobia strains associated with paddy(Oryzasp.)roots,two of which were root endophytes(Opitutussp.andAlbicoccussp.,both belonging to subdivision 4).The study established root endosphere colonization by Verrucomicrobia members for the first time.All strains exhibited multiplein vitroPGP traits,including synthesis of IAA and its precursors,siderophore production,and solubilization of organic and mineral phosphates.Under gnotobiotic conditions,two isolates were able to promote the growth of paddy seedlings,as evidenced by an increase in the fresh weight of the roots.Both strains colonized the root surfaces,generally forming large biofilms.Büngeret al.(2020)also analyzed the draft genomes of the novel strains and identified several genes potentially involved in the plant-associated lifestyle of Verrucomicrobia.The genes required for phosphate solubilization and transport,iron transport,siderophore recognition,nitrate/nitrite transport and assimilation,denitrification,and nitrogen fixation were identified.
Planctomycetes and Gemmatimonadetes.Very few reports are available on the environmental physiology of the phyla Planctomycetes and Gemmatimonadetes.Members of Planctomycetes are key players in global carbon and nitrogen biogeochemical cycles and are thus of ecological and biotechnological significance(Wiegandet al.,2018);for example,they act as decomposers of organic matter derived from plants(Ivanovaet al.,2017).Members of the phylum Gemmatimonadetes are reported to prevail in the rhizosphere but not in the endosphere(Mitteret al.,2017).Genomic studies reveal their possible involvement in nitrogen cycling(Chee-Sanfordet al.,2019).However,the ecological significance,including PGP activities of both these phyla,is yet to be evaluated through meticulous culture-dependent studies in plant-soil systems.Further genetic studies are required to decipher whether members of these phyla encode any genes that may be related to PGP.
CONCLUSIONS AND FUTURE CHALLENGES
The varied metabolic and ecophysiological functions of the difficult-to-culture bacterial phyla in soil open the scope to finding the bright side of microbial dark matter.Culture-dependent and-independent approaches together provide a holistic approach to the study of the soil microbial community,especially the difficult-to-culture phyla.As rhizosphere soils are a major reservoir of microbial consortia,more attentionsensu strictoshould be paid to the rhizospheric difficult-to-culture bacteria.The difficult-to-culture bacteria may be potential PGPR and could be efficiently employed as plant probiotics,providing what may prove to be eco-friendly alternatives to hazardous agrochemicals.Thus,fine-tuning studies related to the diversity,distribution,and environmental physiology of the difficult-to-culture rhizobacterial communities would provide valuable information for understanding the underlying microbe-microbe and plant-microbe interactions and would enable harnessing of their beneficial interactions with host plants for sustainable agriculture.
CONTRIBUTION OF AUTHORS
Sadaf KALAM and Anirban BASU contributed equally to this work and shared the first authorship.
ACKNOWLEDGEMENTS
This work was supported by the Department of Science and Technology(DST),Government of India(GoI),in the form of DST-WOS-A Women Scientist Fellowship for Sadaf KALAM(Grant no.SR/WOS-A/LS-294/2012(G))and in the form of J.C.Bose Fellowship for Appa Rao PODILE(Grant no.JCB/2017/000053).We gratefully acknowledge the DST-FIST level II and University Grants Commission Special Assistance Programme(UGC-SAP)support for the Department of Plant Sciences,School of Life Sciences,University of Hyderabad,India.We thank UGC,India,for financial support to Anirban BASU in the form of a Senior Research Fellowship.The funding bodies had no role in the study design;collection,analysis,and interpretation of data;writing of the review;and the decision to submit the article for publication.
杂志排行
Pedosphere的其它文章
- Elevated carbon dioxide stimulates nitrous oxide emission in agricultural soils:A global meta-analysis
- Hydrogen cyanide production by soil bacteria:Biological control of pests and promotion of plant growth in sustainable agriculture
- Effects of different continuous fertilizer managements on soil total nitrogen stocks in China:A meta-analysis
- Microplastics in soil:Impacts and microbial diversity and degradation
- Rhizosphere microbiomes can regulate plant drought tolerance
- Effects of plant growth-promoting rhizobacteria on the molecular responses of maize under drought and heat stresses:A review
