EXPRESSION PROFILES AND SIGNAL TRANSDUCTION OF TRAF6 IN NILE TILAPIA
2022-01-20HANXueQingGAOFengYingLIUZhiGangCAOJianMengKEXiaoLiWANGMiaoYIMengMengGAOYanXiaandLUMaiXin
HAN Xue-Qing , GAO Feng-Ying LIU Zhi-Gang CAO Jian-Meng KE Xiao-Li WANG Miao YI Meng-Meng GAO Yan-Xia and LU Mai-Xin
(1. Pearl River Fisheries Research Institute, Chinese Academy of Fishery Science, Guangzhou 510380, China; 2. Key Laboratory of Tropical & Subtropical Fishery Resource Application & Cultivation, Ministry of Agriculture, Guangzhou 510380, China; 3. National Demonstration Centre for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai 201306, China)
Abstract: Tumour necrosis factor receptor-associated factor 6 (TRAF6) is a key adaptor protein that plays important role in signalling pathways triggered by the Toll-like receptor/interleukin-1 receptor (TLR/IL-1R)superfamily, which is highly involved in innate immunity. In this study, we investigated expression patterns and preliminary functional analyses of Nile tilapia traf6. In healthy fish, traf6 transcripts were broadly expressed in all examined tissues with the highest expression level in the blood and the lowest in the liver. traf6 was also detected at various embryonic developmental stages. After challenge with S. agalactiae in vivo, upregulated mRNA expression of traf6 was observed in most examined tissues. Moreover, traf6 expression could be significantly induced in Nile tilapia macrophages by treatment with LPS, Poly I: C and S. agalactiae. In addition, overexpression in HEK293T cells showed that TRAF6 was distributed in the cytoplasm and could significantly increase NF-κB activation. Co-immunoprecipitation (Co-IP) assays showed that TRAF6 could interact with IRAK1 (interleukin-1 receptor associated kinase 1), which also plays a vital role in the TLR/IL-1R signalling pathway. In vivo, TLR2, TLR21 and TLR13b overexpression could upregulate traf6 expression levels, which indicates that TRAF6 is involved in the signal transduction of TLR2, TLR21 and TLR13b. These findings suggest that TRAF6 plays important roles in the immune response to pathogen invasion.
Key words: Nile tilapia (Oreochromis niloticus); traf6; Expression profile; Immune response;NF-κB activity; Signal transduction
Nile tilapia is one of the most economically important fish and is widely cultured throughout the world[1]. However, in recent years, this species has suffered from increasingly serious diseases, particularly infection byStreptococcusagalactiae, which has threatened the development of Nile tilapia aquaculture and caused significant economic loss[2]. It is therefore of great significance to understand anti-disease immune mechanisms of tilapia. Tumour necrosis factor receptor (TNFR)-associated factors(TRAFs) are key signalling molecules that function in various cellular signalling events including immune responses, cell death and survival, development, and thrombosis[3,4]. To date, seven members (TRAF1-7)have been identified in mammals, which show evolutionarily conserved phylogeny and similar structure,including a RING-finger, several zinc fingers and a TRAF domain[5,6]. Among the TRAF members,TRAF6 has an unique mode of interaction with receptors (available structures of complexes) and mediates a wide range of interactions via its TRAF domain and RING-finger domain[7,8].
Using CD40 as bait, TRAF6 was originally isolated via yeast two-hybrid screening[9]and independently identified by screening of an expressed sequence tag (EST) library[10]. It is shown that TRAF6 participates in signalling pathways mediated by the IL-1R family, TLRs, the NLR family and RIGI/RLRs, all of which have TIR domains[11—14]. Researchers found TRAF6 is involved in signalling pathways mediated by various cytokines and intracellular mediators, participating in both innate and adaptive immune responses[7]. It is also required for the activation of both classical and non-classical NF-кB pathways as well as activation of MAPKs and IRFs[7,15].Among the above-mentioned receptors families that mediate signalling pathways, TLRs are the first pattern recognition receptor (PRRs) family to be discovered, capable of recognizing a variety of pathogen-associated molecular patterns (PAMPs)[16,17]. The recognition and binding of PAMPs by specific TLRs can activate cell signalling cascades through Myd88-dependent and Myd88-independent pathways[18], in which TRAF6 participates in Myd88-dependent pathway. In the Myd88-dependent TLR pathway, IRAK1(interleukin-1 receptor associated kinase 1) dissociates from the Myd88-IRAK4 complex and interacts with TRAF6. Activation of TRAF6 accelerates selfubiquitination and induces activation of TAK1 (transforming growth factor-b-activated kinase 1). Then,TAK1 activates downstream NF-кB and MAPK pathways[11,16,17].
To date,traf6has been identified in many teleost fishes, such as zebrafish (Daniorerio)[19], common carp (Cyprinuscarpio)[20], grass carp (Ctenopharyngodonidella)[21], orange-spotted grouper (Epinepheluscoioides)[22], blunt snout bream (Megalobrama amblycephala)[23], large yellow croaker (Larimichthys crocea)[24], and Nile tilapia (Oreochromisniloticus)[25].Li,et al.[22]demonstrated thattraf6transcripts were broadly expressed in all tissues tested in the orangespotted grouper and increased after infection withCryptocaryonirritans. Furthermore, overexpression strongly activated NF-кB in HEK293T cells. In general, these studies have robustly confirmed that TRAF6 plays important regulatory roles in immune responses to invasion by pathogens. However, studies of TRAF6 in tilapia and its functional properties have been limited. In this study, we cloned the Nile tilapiatraf6gene, and investigated its tissue distribution and expression profile during embryonic development. Thein vivoexpression profiles oftraf6after bacterial infection withS.agalactiaeandin vitroexpression profiles treatment with LPS, Poly I: C, Δcps(capsule polysaccharide gene cluster deleted strains) andS.agalactiaewere also explored. In addition, recombinant plasmids were constructed and overexpressed in HEK293T cells to determine the intracellular localization and signal transduction functionality.
1 Materials and methods
1.1 Ethics statement
The care and use of experimental animals complied with Committee on Animal Care and Use and the Committee on the Ethics of Animal Experiments of Chinese Academy of Fishery Sciences.
1.2 Fish, challenge and sampling
Healthy tilapia [mean±SD: body mass=(70±10) g;total length=(18±2) cm] were obtained from the Gaoyao fish farm of the Pearl River Fisheries Research Institute (Guangzhou, China). The fish were then cultured in a 500 L tank at (30±1)℃ for 2 weeks before all experiments and fed twice a day with a commercial tilapia diet.
For tissue expression analysis, eleven tissue samples (brain, gill, liver, spleen, intestine, heart,trunk kidney, stomach, skin, muscle and blood) were isolated from six healthy individuals. For gene expression analysis during embryonic development,tilapia embryos were sampled at 2d, 3d, 4d, 5d, 6d, 7d and 8d post-fertilization (dpf). Three RNA samples(approximately 6 embryos per sample) were extracted at each time point. We further explored expression profiles induced byS.agalactiaeWC1535 treatment in accordance with the method described by Gao,et al.[26]. Fish were randomly divided into a treatment group (120 individuals) and control group (40 individuals), with 200 μL of bacterial suspension(3×107CFU/mL) and the same volume of phosphatebuffered saline (PBS, pH 7.2) injected intraperitoneally, respectively. At 8h post infection (hpi) and at 1d, 2d, 3d, 6d and 9d post infection (dpi), intestine,gill, spleen, trunk kidney, and blood samples from four fish were collected for total RNA extraction. In addition, 40 fish in the treatment groups died over the course of the challenge experiment, whereas no fish died in the control group.
1.3 Preparation and sampling of head kidney macrophages
Head kidney leukocytes were isolated as previously reported[27], with minor modifications. In brief, the head kidney was removed aseptically and then passed through a 100 μm mesh in Leibovitz medium (L-15)containing antibiotics and heparin. A cell suspension was layered onto a discontinuous (34%/51%) Percoll density gradient and centrifuged at 400×gfor 30min at 4℃. Cells at the interface were transferred into clean tubes and washed twice by centrifugation. The isolated leukocytes consisted of lymphocytes, monocytes, neutrophils and macrophages as previously reported[28,29]. Cells were seeded in cell culture dishes and cultivated in L-15 supplemented with 10% foetal bovine serum (FBS) for 4h. Then many of the leukocytes had adhered to the surface of dishes and the cells exhibited a characteristic irregular morphology.These cells were stained with Giemsa staining and identified as macrophages (data not shown)[27,28]. In this way, highly pure macrophages populations were obtained.
The harvested macrophages were seeded in 12-well plates (1×106cells per well) and treated with LPS (25 μg/mL, Sigma L2630), Poly I: C (50 μg/mL,Sigma P1530),S.agalactiaeWC1535 (2×106CFU/mL), capsule polysaccharide gene cluster-deleted strains (Δcps) (2×106CFU/mL) and PBS (as a control). Each group was examined in triplicate. Cell samples of each treatment group were collected at 6h and 12h after treatment, respectively.
1.4 RNA extraction and cDNA synthesis
Total RNA was extracted from tissues, embryos and cells using TRIzol reagent (Life, USA) according to the manufacturer’s instructions. The purity and concentration of extracted RNA were determined with agarose gel electrophoresis and spectrophotometry(A260/280). The samples were incubated with DNase I(TaKaRa, Japan) to remove genomic DNA contamination. Then reverse transcription of the total RNA (1 μg)was performed using a PrimeScript Ⅱ 1st strand cDNA Synthesis Kit (TaKaRa, Dalian, China) to synthesize first strand cDNA for cDNA cloning and gene expression analysis.
1.5 Sequence amplification and plasmid construction
We cloned the cDNA sequence oftraf6(Gen-Bank accession No. MK227433) using the primers listed in Tab. 1. The open reading frame (ORF) was identified with ORF Finder program (www.ncbi.nlm.nih.gov/gorf/gorf.html). Using Primer Premier 5.0,Primerstraf6-bf/brcontaining Kozak sequence (GC CACC) before the ATG codon were designed to amplify a full-length ORF oftraf6. Reaction conditions were as follows: an initial denaturation at 94℃ for 3min; followed by 30 cycles of denaturation for 30s at 94℃, annealing for 30s at 57℃, extension for 1.5min at 72℃; and a final extension for 10min at 72℃. PCR products were purified using a gel extraction kit(Magen), and then ligated into a pcDNA3.1/CT-GFP plasmid vector (Catalogue No. K4820-01; Invitrogen)to obtainpcDNA3.1-traf6-GFP. Using the same method, the ORFs ofTLR2(XM_003450307),TLR21(KJ010824),TLR22(KJ010825),TLR13a(MG92 4492) andTLR13b(MK420449) were amplified andpcDNA3.1-TLR2-GFP,pcDNA3.1-TLR21-GFP,pcDNA3.1-TLR22-GFP,pcDNA3.1-TLR13a-GFPandpcDNA3.1-TLR13b-GFPplasmids were constructed.
In addition,pCMV-C-traf6-MycandpCMV-Cirak1-Flag[30]recombinant plasmids were constructed by inserting the ORF into a pCMV-C-Myc vector(Beyotime Biotech, Shanghai, China) withEcoR I/XbaIsites (primers:traf6-F1/R1). Next, we transformed recombinant plasmids into competentEscherichia coliDH5α cells (TaKaRa), and recovered plasmids were sequenced by Guangzhou Sangon Biotech Co., Ltd. All recombinant plasmids were constructed successfully and then extracted with an E.Z.N.A Endo-free Plasmid Mini Kit I (Promega;www.promega.com) according to the manufacturer’s instructions.
1.6 Quantitative analysis of traf6 mRNA expression
A quantitative real-time (rt)-PCR assay was performed with a StepOnePlus™ Real-Time PCR System (Life Technologies, Waltham, MA, USA) with PowerUp SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA). In this study, theef-1αgene was used as a reference gene for tissue expression analysis, temporal expression profiling following bacterial challenge and activated tilapia macrophages. During the evaluation of gene expression during embryonic development, we used theβ-actingene as a reference gene based on the most stable expression levels ofef-1α,β-actinand 18S[31]. Specific fragments oftraf6,ef-1αandβ-actinwere amplified and ligated into the pMD19-T vector in order to construct the various plasmids (Tab. 1). Plasmids were then diluted by a 10-fold (10-1—10-8pmol/L) serial dilution as templates for standard curves of target genes and reference genes[32]. All of the standard curves exhibited correlation coefficients higher than 0.99, and the PCR efficiency of each primer was 1.97—2.03.
PCR was performed in a total volume of 20 μL containing 1 μL of cDNA template, 0.3 μL of each primer (20 μmol/L), 10 μL of Power SYBR Green Master Mix, and 8.4 μL of ddH2O. The conditions were as follows: 50℃ for 2min, 95℃ for 2min, followed by 40 cycles of 95℃ for 15s, 60℃ for 15s and 72℃ for 1min. A melting curve analysis was performed at the end of each PCR. Each sample was amplified in triplicate. The cDNA concentration in each sample was predicted according to the threshold cycle number (CT) and gene-specific standard curve.The relative expression oftraf6was calculated using the following formula:F=(a/b)/(c/d), where a is the concentration of the target gene from the treatment group, b is the concentration of the reference gene from the treatment group, c is the concentration of the target gene from the control group, and d is the concentration of the reference gene from the control group[33].
1.7 Dual-luciferase reporter assay
HEK293T cells (Cell Bank, Chinese Academy of Sciences, Shanghai) were cultured in Dulbecco’s modified eagle medium (DMEM, Gibco, Carlsbad,CA, USA) containing 10% foetal bovine serum in an incubator at 5% CO2at 37℃ and passaged every other day. Twenty-four hours before transfection, the cells were seeded in a 96-well plate and transfected with Lipofectamine™ 2000 Reagent (Invitrogen) according to the manufacturer’s instructions. In brief, after the cells were washed with serum-free DMEM, they were transfected with 420 ng of plasmids containing 200 ng ofpcDNA3.1-traf6-GFPorpcDNA3.1/CTGFPplasmid, 200 ng ofNF-κBreporter plasmid(pGL4.32 [luc2P/NF-κB-RE/Hygro] Vector, Catalogue No. E8491, Promega), and 20 ng of thepRL-TKreference plasmid (Promoter-Driven Control Renilla Luciferase Vectors, pGL4.74 [hRluc/TK] Vector,Catalogue No. E6921, Promega). Each condition was tested in triplicate. Six hours later, the medium was replaced with complete medium. Twenty-four hours after transfection, the cells were washed with PBS.Firefly andRenillaluciferase activities were determined with a Dual-Luciferase Reporter Assay System (Promega). The relative luciferase activity of each trial was calculated as the firefly luciferase activity relative to theRenillaluciferase activity.
1.8 Subcellular localization of TRAF6
HEK293T cells were seeded onto coverslips in a 24-well plate and cultured. Twenty-four hours after transfection withpcDNA3.1-traf6-GFPandpcDNA3.1/CT-GFPplasmids (1 μg), HEK293T cells were washed with PBS and then fixed with Immunol Staining Fix Solution (Beyotime, Beijing, China).After washing, the cells were treated with PBST (8 mmol/L Na2HPO4, 2 mmol/L KH2PO4, 10 mmol/L KCl,140 mmol/L NaCl, and 0.05% Tween-20 [v/v]) and stained with l μg/mL 4′,6-diamidino-2-phenylindole(DAPI). The coverslips were then washed and transferred to glass slides. The subcellular localization of TRAF6 was observed using a Leica TCS SP5 laser confocal fluorescence microscope.
1.9 Co-immunoprecipitation assays
Co-IP assays were performed to measure the potential interaction between TRAF6 and IRAK1.Briefly, HEK293T cells were seeded onto 150 mm dishes and cultured. After 24h, 20 μg plasmids(pCMV-C-irak1-Flag:pCMV-C-traf6-Myc/pCMV-CMyc=1∶1) were co-transfected into the cells. The transfected cells were harvested at 48h post-transfection, then transferred to 1.5 mL centrifuge tube and lysed on ice with 1 mL of lysis buffer. 50 μL cell lysates were used to assess as input products. The remaining cell lysates were incubated overnight at 4℃with anti-FLAG®M2 magnetic beads (Sigma-Aldrich, Shanghai, China). The beads were then washed three times with wash buffer and eluted with elution buffer to obtain IP products[34—36]. The input and IP products were both examined using Western blotting analysis with Flag-HRP (Sigma) and anti-Myc antibodies (Santa Cruz, USA).
1.10 Overexpression of TLR2, TLR21, TLR22,TLR13a and TLR13b in Nile tilapia
pcDNA3.1-TLR2-GFP,pcDNA3.1-TLR21-GFP,pcDNA3.1-TLR22-GFP,pcDNA3.1-TLR13a-GFP,pcDNA3.1-TLR13b-GFPandpCDNA3.1-GFPplasmids were diluted in PBS to 200 μg/mL. Healthy Nile tilapia were divided randomly into seven groups and administered intramuscularly (i.m.) with 100 μL of above plasmids or PBS as a control[37]. Muscle, trunk kidney, liver, and spleen samples were collected from the fish at 2d post-plasmid administration. To examine the presence of plasmids in the tissues, DNA was extracted from the tissues with a TIANamp DNA Kit (Tiangen, Beijing, China) and submitted for PCR analysis with the pCDNA3.1-GFP-specific primers T7 (5´-TAATACGACTCACTATAGGG-3´) and TLR2-br, TLR21-br, TLR22-br, TLR13a-br,TLR13b-br, as well as the pcDNA3.1-specific primers T7 and GFP Reverse primers (5´-GGGTAAGCTTTCC GTATGTAGC-3´) (Fig. S1A). To examine the expression of plasmid-derivedTLR2,TLR21,TLR22,TLR13aandTLR13bin the tissues, total RNA was extracted from the liver tissues and submitted to qRTPCR, analysis with primers T7 and TLR2-br, TLR21-br, TLR22-br, TLR13a-br and TLR13b-br which amplify theTLR2,TLR21,TLR22,TLR13aandTLR13bsequences inpcDNA3.1-TLR2-GFP,pcDNA3.1-TLR21-GFP,pcDNA3.1-TLR22-GFP,pcDNA3.1-TLR13a-GFPandpcDNA3.1-TLR13b-GFP.
1.11 Effects of TLR2, TLR21, TLR22, TLR13a and TLR13b overexpression on expression of the traf6 gene in Nile tilapia
Tilapia were injected i.m. withpcDNA3.1-TLR2-GFP,pcDNA3.1-TLR21-GFP,pcDNA3.1-TLR22-GFP,pcDNA-TLR13a-GFP,pcDNA-TLR13b-GFPorpcDNA3.1-GFPas described above. At 1d, 3d and 5d post-plasmid injection, liver tissues from the fish(three fish/time point) were collected, and the expression levels oftraf6in the liver tissues were determined by qRT-PCR as described above.
1.12 Statistical analysis
Data are expressed as the means ± standard errors. The statistical significance of observed differences was determined by One-way ANOVA followed by Duncan’s testing using SPSS statistics 22.0 software (IBM; www.ibm.com). The level of statistical significance was set atP<0.05.
2 Results
2.1 Expression profiles of traf6 in Nile tilapia
Quantitative expression analysis was performed in eleven tissues from healthy fish. The results showed thattraf6was broadly expressed in all examined tissues in healthy Nile tilapia. The highest expression level oftraf6mRNA was detected in the blood, followed by various expression levels in the spleen, gill, brain; the lowest expression level was found in the liver (Fig. 1a). The highest expression level oftraf6in the blood was approximately 32-fold greater than that in the liver tissue (control group). In addition,traf6expression profiles during embryonic development were also determined (Fig. 1b). We found thetraf6transcript was stably expressed from 2 to 7dpf, and significantly increased at 8dpf. The highest expression level oftraf6at 8dpf was 1.5-fold higher than that at 2dpf (control group).
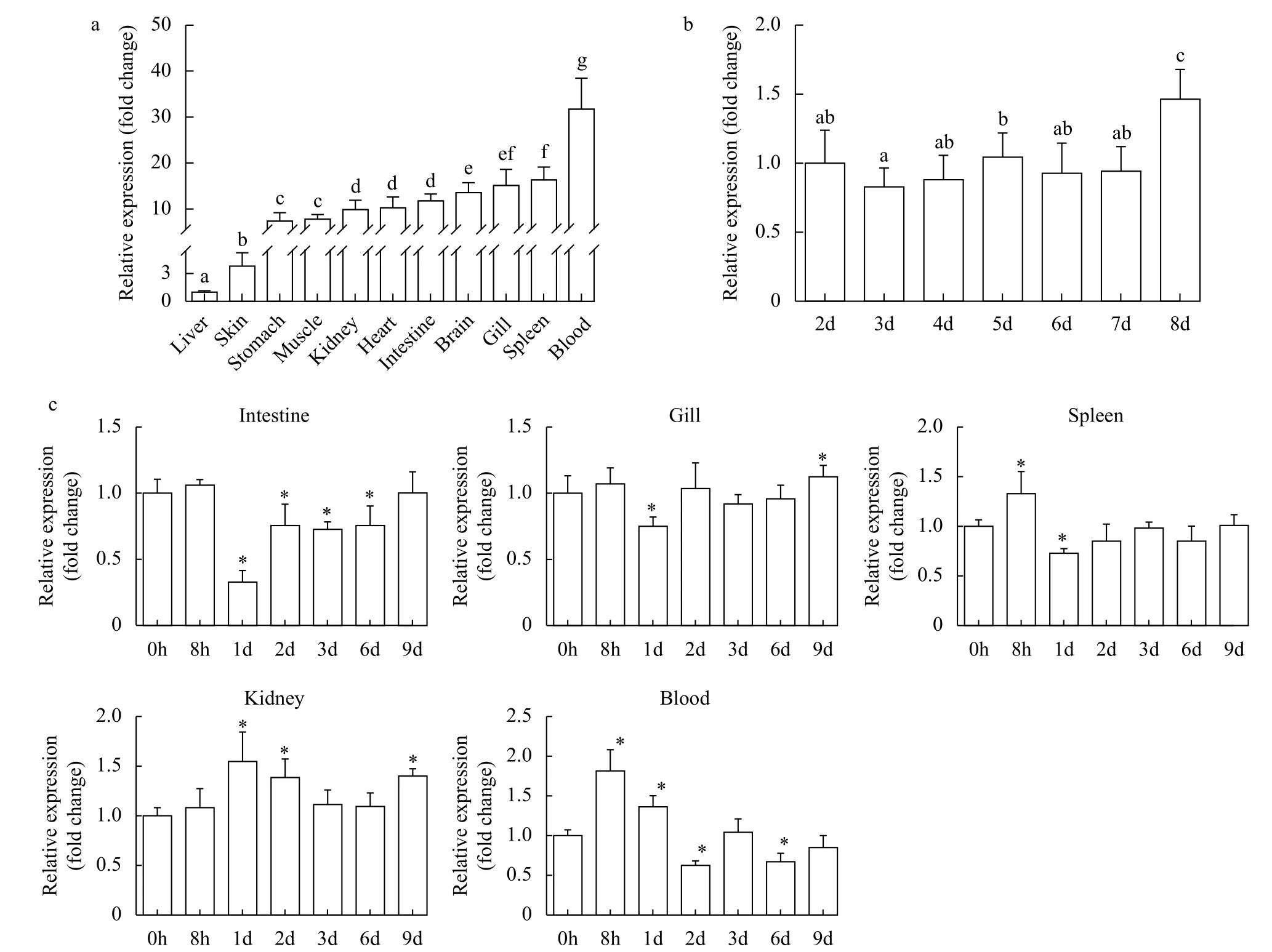
Fig. 1 The expression pattern of Nile tilapia traf6 in tissues (a, c) and during embryonic development (b)
We had studied its tissue distribution in healthy fish. In order to explore whether it participates in the immune response after pathogen stimulation, we next evaluated inducedtraf6expression profiles afterS.agalactiaechallenge in five representative tissues (intestine, gill, spleen, trunk kidney, blood). These five tissues are all involved in (mucosal) immunity, andtraf6had high expression levels in the above-mentioned tissues of healthy tilapia. Thetraf6showed alternating increases and decreases in all five tissues(Fig. 1c). In the intestine and gill,traf6transcript levels first decreased and then rose, showing the lowed expression at 1dpi, 0.3-fold and 0.7-fold compared to that of their control group (at 0h). While the opposite pattern was observed in the trunk kidney,with the highest upregulation at 1dpi (1.5-fold). A similar expression pattern withtraf6in the kidney was detected in the spleen, which peaked at 8hpi (1.3-fold that of 0h) and then recovered to normal. In the blood,traf6transcript had the highest expression at 8hpi (1.8-fold), decreased from 8hpi to 2dpi, and then showed alternating fluctuations of rise and fall. These results suggest thattraf6participates in the antibacterial response and play a role in protecting Nile tilapia fromS.agalactiaeinfection.
2.2 Expression analysis of traf6 in macrophages treated with LPS, Poly I: C, capsule polysaccharide gene cluster deleted strains (Δcps) and S. agalactiae WC1535
The mRNA expression levels oftraf6were quantified in macrophages after treatment with PBS,LPS, Poly I: C, Δcpsand WC1535 at different time points (6h and 12h). We used the expression level of a PBS-treated group at 6h as a blank control. The results showed thattraf6expression decreased after LPS treatment (6h) and was significantly upregulated following Poly I: C challenge (12h), while no change was observed in response to LPS (12h) and Poly I: C(6h) treatment. After treatment with WC1535, the expression level oftraf6first increased (6h) and then significantly decreased (12h). Additionally, the expression levels oftraf6showed no change after Δcpstreatment (Fig. 2).
2.3 traf6 significantly increased NF-κB activation in HEK293T cells
Co-transfection with NF-κB reporter plasmids was performed in HEK293T cells to investigate potential signal transduction roles of TRAF6. Relative luciferase activity was calculated based on the observed firefly luciferase activity relative to theRenillaluciferase activity. The dual-luciferase reporter assay results showed thattraf6overexpression significantly increased NF-κB activation in HEK293T cells, approximately 14-fold greater compared with the control group (Fig. 3a). Activated NF-κB could initiate the expression of target genes. The above showed that TRAF6 does play a role in the NF-κB signalling pathway.
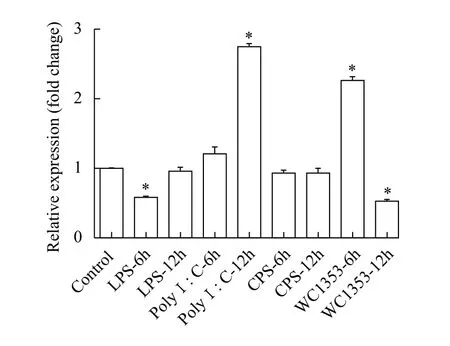
Fig. 2 The expression of Nile tilapia traf6 in macrophages treatment with LPS, Poly I∶C, Δcps and Streptococcus agalactiae WC1535
2.4 Subcellular localization of TRAF6
The subcellular localization of TRAF6 was determined bypcDNA3.1-traf6-GFPfusion protein expression in HEK293T cells. As shown in Fig. 3b, the green fluorescence of the TRAF6-GFP fusion protein was distributed largely in the cytoplasm, where several intense fluorescence signal sites were observed. In pcDNA3.1/CT-GFP-transfected cells, however, green fluorescence was detected in the cytoplasm and in the nucleus, and no sites of intense fluorescence were observed. The results showed that TRAF6 is located in the cytoplasm and plays a role in the cell.
2.5 Interaction between TRAF6 and IRAK1
Our research found that TRAF6 is involved in the intracellular NF-KB signalling pathway. To further clarify the role of TRAF6 in the TLR signalling pathway, we investigated the interaction between TRAF6 and IRAK1 by a Co-IP experiment. In this study, two sets of plasmids were transfected into HEK293T cells (control group:pCMV-C-irak1-FlagandpCMV-C-Myc; experimental group:pCMV-Cirak1-FlagandpCMV-C-traf6-Myc), respectively.Western blotting analysis of input products showed that IRAK1-Flag and TRAF6-Myc were both expressed normally (Fig. 3c), and Co-IP was subsequently performed. The results showed that TRAF6-Myc was co-precipitated with IRAK1-Flag,indicating that TRAF6 interacts with IRAK1 in HEK293T cells and plays a role in the TLR signalling pathway (Fig. 3c).
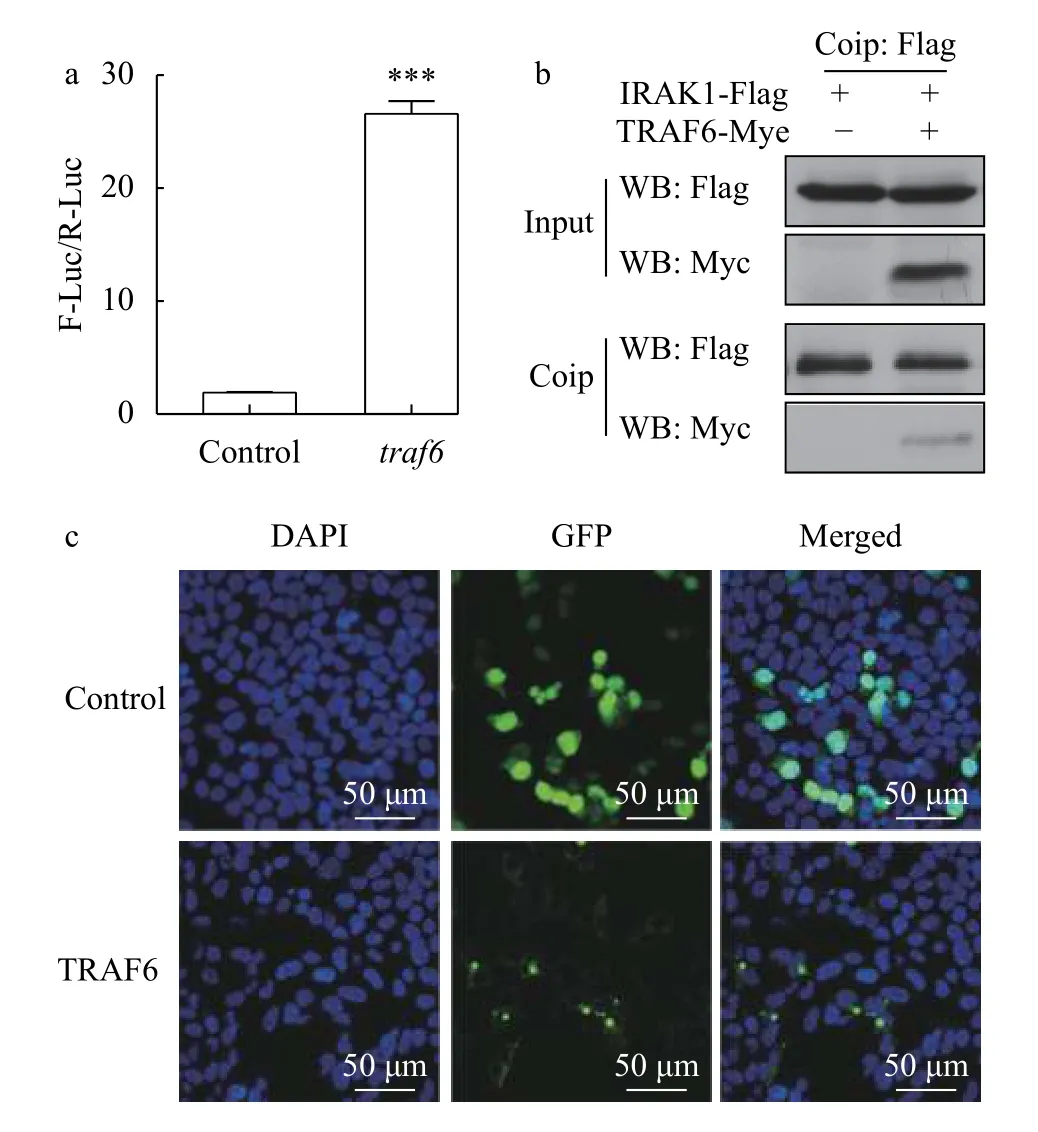
Fig. 3 Overexpression of traf6 in HEK293T cells
2.6 Effect of TLR2, TLR21, TLR22, TLR13a and TLR13b overexpression on the expression of the traf6 gene in Nile tilapia
To further confirm that TRAF6 is involved in TLR-mediated signalling pathways, we determine the mRNA levels oftraf6in individual fish withTLR2,TLR21,TLR22,TLR13aandTLR13boverexpression by qRT-PCR. The results showed that the expression level oftraf6inTLR2,TLR21andTLR13boverexpression group was significantly upregulated compared with the control group (Fig. 4).
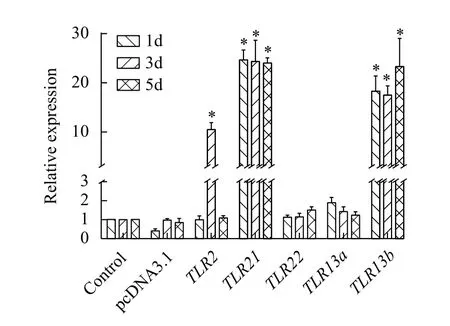
Fig. 4 Effect of TLR2, TLR21, TLR22, TLR13a and TLR13b overexpression on expression of the traf6 gene in Nile tilapia
3 Discussion
3.1 Expression profiles of traf6 in Nile tilapia
In this study, expression patterns and preliminary functional analyses oftraf6were performed in Nile tilapia. In this fish,traf6was widely distributed in all examined tissues (Fig. 1a). The same ubiquitous expression oftraf6has also been reported in other teleost fish[20—22,25]. However, there are different expression patterns in different species. In the present study,traf6 expression was the highest in the blood,elevated in the spleen, and the lowest in the liver. In a previous study, Wang,et al.[25]had reported similar results, in which Nile tilapiatraf6had the highest expression level in the spleen and the lowest expression level in the head kidney. In contrast,traf6was found to be predominantly expressed in the blood of the orange-spotted grouper[22], the head kidney of grass carp[21], and the gills of common carp[20]. These discrepancies among species in expression patterns might be a result of species variation and differences in studied developmental stages. We also measuredtraf6expression across various embryonic developmental stages in Nile tilapia. Our results suggest thattraf6contributes to innate immunity during early developmental stages. The contribution of the innate immune response to protection of the early developmental stages of fish has previously been reported in zebrafish[38,39], carp[40], and rohu (Labeorohita)[41].
In mammals, TRAF6 is involved in various physiological processes, including innate and adaptive immunity[14,42], and it plays playing important roles in the resistance of hosts to pathogen infection[16,17,25,43].In this study,traf6expression was clearly induced in all examined tissue types followingS.agalactiaechallenge (Fig. 1c). Similarly, a noticeable change in orange-spotted groupertraf6expression was reportedly induced in various tissues followingCryptocaryonirritansinfection[22]. Similarly, expression of bothtraf6andtak1 in grass carp was significantly upregulated to varying degrees in the skin, gill, head kidney and spleen at most time points followingIchthyophthiriusmultifiliisinfection[21]. And Wang,et al.[25]additionally assessed Nile tilapiatraf6expression levels in the spleen, head kidney and intestine after infection withS.agalactiae. They found thattraf6was significantly upregulated at 48h, 24h and 3h, respectively, indicating thattraf6is involved in innate immunity of Nile tilapia treated withS.agalactiae; however, the trends in expression level oftraf6were inconsistent with our present study, which is presumably due to differences in tilapia specifications and culture conditions. The present study, along with previous studies, indicates that piscinetraf6might be involved in immune responses to bacterial infection.
3.2 Expression analysis of traf6 in macrophages
We further exploredtraf6expression in head kidney macrophages following treatment with LPS,Poly I: C, Δcpsand WC1535. With the exception of Δcps, these three treatments represent a Gram-negative bacterium, a viral analogue, and a Gram-positive bacterium, respectively[44]. Our results showed that,following treatment with LPS, poly I: C and WC1535,traf6expression in macrophages was significantly induced, indicating thattraf6could be induced by stimulation with Gram-positive bacteria,Gram-negative bacteria and viral analogues.traf6plays crucial roles in Nile tilapia defences against bacterial and viral infections. Moreover, different treatment groups exhibited differential expression profiles, which may be due to the activation of different pathogen-resistant pathways by various infection types. Additionally, Capsule polysaccharide (Cps) is a main virulence factor ofS.agalactiae[45]. Nile tilapiatraf6expression showed no change after treatment with Δcps(capsule polysaccharide gene cluster deleted strains), indicating that it is involved in the signal pathway induced by Cps.
3.3 Signal transduction of TRAF6
TRAF6 is an E3 ubiquitin ligase that plays an essential role in TLR/IL-1R, TNFR and C-type lectin receptor signalling pathways and it has further been shown to be an activator protein for the classical NFκB pathway[12-15]. For this, we tried to perform preliminary functional experiments of tilapiatraf6in the cell lines of Nile tilapia, but all failed. Finally, we conducted it in HEK293T cells with a view to verify some functions of tilapiatraf6. In this study, we found thattraf6overexpression strongly induced NF-кB activity in HEK293T cells, which is consistent with results from Wang,et al.[25]. Similarly, overexpression of orange-spotted groupertraf6in HEK293T cells and zebrafishtraf6in ZFL cells both strongly activate NFкB[19,22], suggesting functional conservation oftraf6activity between fish and mammals. In the Myd88-dependent TLR pathway, IRAK1 dissociates from the Myd88-IRAK4 complex and interacts with TRAF6[11,16,17].In the present study, we also found TRAF6-myc coprecipitated with IRAK1-flag, indicating that TRAF6 interacts with IRAK1 (Fig. 3c). This is consistent with previous studies[11,17], suggesting thattraf6is involved in and plays an important role in the TLR signalling pathway.
InTLR2-,TLR21- andTLR13b- overexpressing fish, the expression level oftraf6was upregulated.This indicates that Nile tilapia TRAF6 is involved in signal transduction for TLR2, TLR21 and TLR13b.Importantly, TRAF6 does not participate in the signal transduction of TLR22 and TLR13a. This is consistent with prior observations that certain TLR signalling transduction is dependent on TRAF6, but not all[17].
In conclusion, Nile tilapiatraf6has a broad tissue expression and can be detected across various embryonic developmental stages. Moreover,traf6expression was significantly induced byS.agalactiaeWC1535in vivoand LPS, Poly I: C andS.agalactiaeWC1535 in Nile tilapia macrophages. Co-IP assays indicated that Nile tilapia TRAF6 interacts with IRAK1. In addition, TRAF6 was localized mainly in the cytoplasm, and overexpression significantly increased NF-κB activation in HEK293T cells.TLR2,TLR21andTLR13boverexpression could upregulate thetraf6expression levelin vivo, which indicates that TRAF6 is involved in the signalling transduction of TLR2, TLR21 and TLR13b.
