Possible therapeutic targets and promising drugs based on unsymmetrical hetaryl-substituted porphyrins to combat SARS-CoV-2
2022-01-19YuryGubrevNtlyShLebedevElenYurinSergeySyrbuAlekseyKiselevMikhilLebedev
Yury A.Gubrev,Ntly Sh.Lebedev,Elen S.Yurin,Sergey A.Syrbu,Aleksey N.Kiselev,b,Mikhil A.Lebedev,b
aG.A.Krestov Institute of Solution Chemistry of the Russian Academy of Sciences,153045,Ivanovo,Russia
bIvanovo State University of Chemistry and Technology,153000,Ivanovo,Russia
Keywords:
Porphyrins
SARS-CoV-2
Molecular docking
Photoinactivation
Peer review under responsibility of Xi’an Jiaotong University.
A B S T R A C T
Coronavirus disease 2019 is a serious disease that causes acute respiratory syndrome and negatively affects the central nervous system.Severe acute respiratory syndrome coronavirus 2(SARS-CoV-2)crosses the blood-brain barrier due to the spike(S)protein on the surface of the viral particles.Thus,it is important to develop compounds that not only have an inhibitory effect but are also capable of completely deactivating the S protein function.This study describes the purposeful modification of porphyrins and proposes compounds,asymmetrically hetaryl-substituted porphyrins with benzothiazole,benzoxazole,and N-methylbenzimidazole residues,to deactivate the S protein functions.Molecular docking of SARS-CoV-2 proteins with hetaryl-substituted porphyrins showed that the viral S protein,nucleocapsid(N)protein,and non-structural protein 13(nsp13)exhibited the highest binding affinity.Hetaryl-substituted porphyrins form strong complexes(13-14 kcal/mol)with the receptor-binding domain of the S protein,while the distance from the porphyrins to the receptor-binding motif(RBM)does not exceed 20 Å;therefore,RBM can be oxidized by1O2,which is generated by porphyrin.Hetarylsubstituted porphyrins interact with the N protein in the serine/arginine-rich region,and a number of vulnerable amino acid residues are located in the photooxidation zone.This damage complicates the packaging of viral RNA into new virions.High-energy binding of hetaryl-substituted porphyrins with the N-and C-terminal domains of nsp13 was observed.This binding blocks the action of nsp13 as an enzyme of exoribonuclease and methyltransferase,thereby preventing RNA replication and processing.A procedure for the synthesis of hetaryl-substituted porphyrins was developed,new compounds were obtained,their structures were identified,and their photocatalytic properties were studied.
1.Introduction
The coronavirus disease 2019(COVID-19)pandemic caused by severe acute respiratory syndrome coronavirus 2(SARS-CoV-2)has clearly demonstrated the vulnerability and defenselessness of humanity to new viral threats.COVID-19 usually causes severe acute respiratory syndrome[1].In addition,new clinical reports are increasing,indicating the dramatic effects of SARS-CoV-2 on the central nervous system.For example,a previous study[2]reported that SARS-CoV-2 preferentially targets neurons in the brain.Imaging of neurons shows that exposure to SARS-CoV-2 is associated with altered tau protein distribution and hyperphosphorylation,ultimately resulting in neuronal death.
The neurological symptoms of COVID-19 are complex and differ among individuals[3].Considering this symptomatology,two possible mutually non-exclusive forms of virus penetration were proposed:through the olfactory nerves(transneuronal)and the hematogenous pathway with penetration via the blood-brain barrier(BBB).In a cytokine storm,BBB permeability is impaired.Several attempts have been made to determine the exact cause of damage to the central nervous system,such as direct penetration of the virus into the brain by binding to the BBB cells or penetration of the virus due to increased BBB permeability during a cytokine storm.To date,the confirmed cause of central nervous system damage is the direct penetration of virus.As a result,the BBB becomes vulnerable due to the presence of spike(S)protein on the surface of the virus,which is the driving unit.A previous study[4]showed that the S protein of SARS-CoV-2 easily crosses the BBB of mice,penetrates into the parenchymal tissue of the brain,and binds to the glycocalyx of the capillaries of the brain,causing damage and apoptosis of endothelial cells and neurons[5].
Human neurodegenerative diseases are often a gradual process that,in some cases,develop over several decades.Neurological and neurodegenerative disorders occur in approximately 35% of patients[6,7].Given the possible long-term impact of COVID-19,it is necessary to develop drugs that target the S protein not just to inhibit its binding to the angiotensin-converting enzyme 2(ACE2),but to completely disrupt its functionality.
Recent studies showed that porphyrins and their structural analogues can be successfully used to inactivate viruses and drugresistant forms of pathogenic bacteria.Our study aimed to design new chemical compounds with the desired properties,namely,hetaryl-substituted porphyrins,using computer methods(molecular docking).In particular,the desired properties are as follows:1)high-energy binding to the SARS-CoV-2 S protein in the receptorbinding domain(RBD)site or in its immediate vicinity(inhibition of binding to ACE2),2)the ability to absorb light energy in the farwave part of the spectrum(for working in the“therapeutic window”)and generate reactive oxygen species(for irreversible damage to the protein during photoirradiation),3)high quantum yield of reactive oxygen species(for irreversible damage to the protein during photoirradiation),4)solubility in physiological media,and 5)the absence of specific interactions(π-π,H-bond between the substance and amino acids of the S protein)to avoid additional dissipation of light energy and increase of the quantum yield of singlet oxygen when porphyrin binds to the S protein of the virus.
2.Materials and methods
2.1.Molecular docking
The D-I-TASSER-predicted structures of SARS-CoV-2 proteins were downloaded from the Zhang Lab website (https://zhanggroup.org//I-TASSER/)[8,9].The heterotrimeric structure of S protein was downloaded from the Protein Data Bank(ID:7bnm)[10].The structures of the porphyrins(Fig.1)were minimized using the ORCA 4.0 program[11]using the DFT b3lyp method.Molecular docking of proteins with porphyrins was performed using Auto-Dock Vina 1.1.2[12]and visualized using PyMol 2.4.1.The ligand and protein structure files were prepared using AutoDockTools 1.5.6.The size of the grid matrix for blind docking was adjusted such that the protein molecules completely overlapped.Owing to the large size of the grid matrix,the exhaustiveness parameter was increased to512[13].Molecular docking enabled the determination of the 20 most favorable structures of each porphyrin.After analyzing the results,redocking was performed using a grid matrix size of 80 Å × 80 Å × 40 Å and at the center near Arg408 of A,B,and C chains in coordinates x/y/z(217/217/265).The most optimal positions are listed in Table 1.Since the porphyrins are charged,the potential of the protein globule was calculated using the adaptive Poisson-Boltzmann Solver method to analyze the docking sites[14].
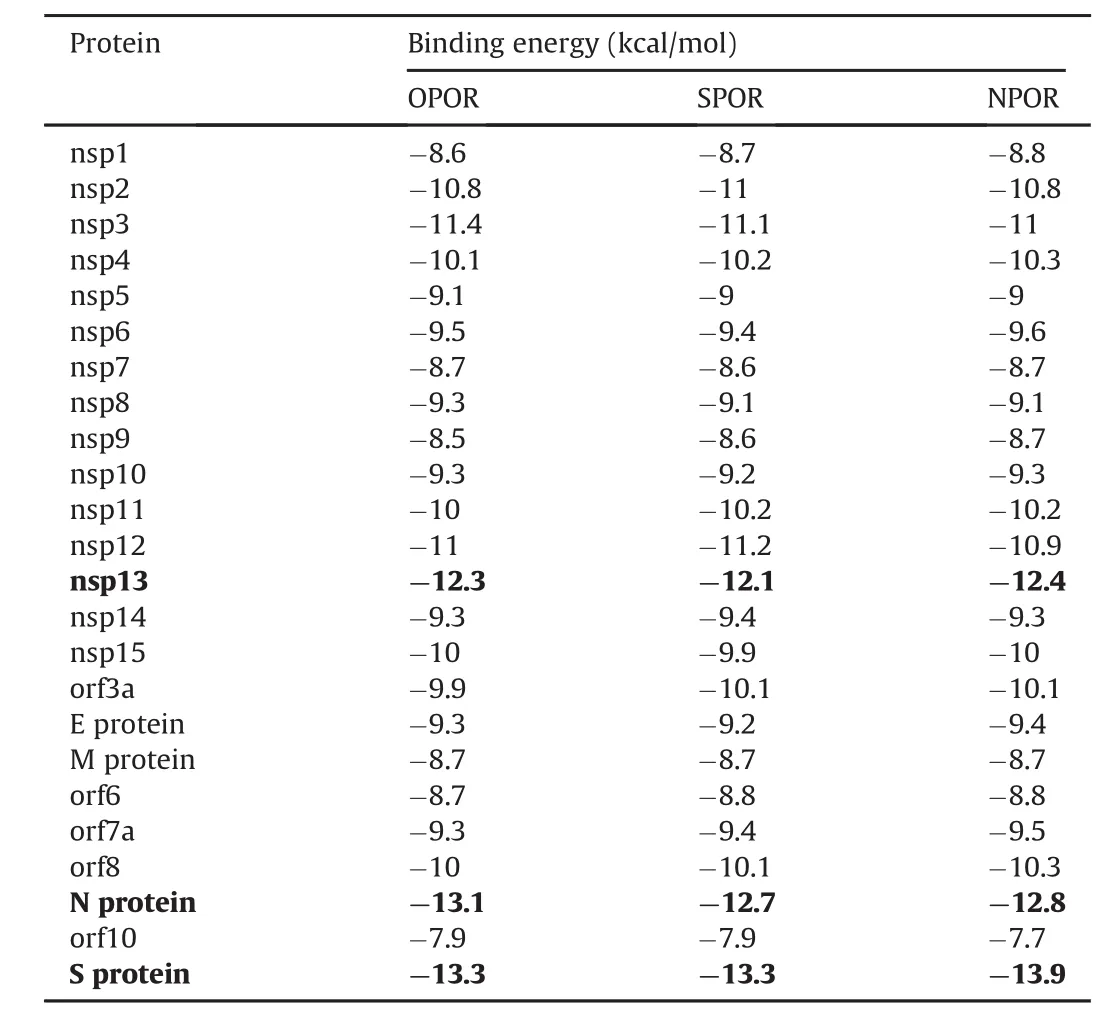
Table 1SARS-CoV-2 proteins binding energy to hetaryl-substituted porphyrins.
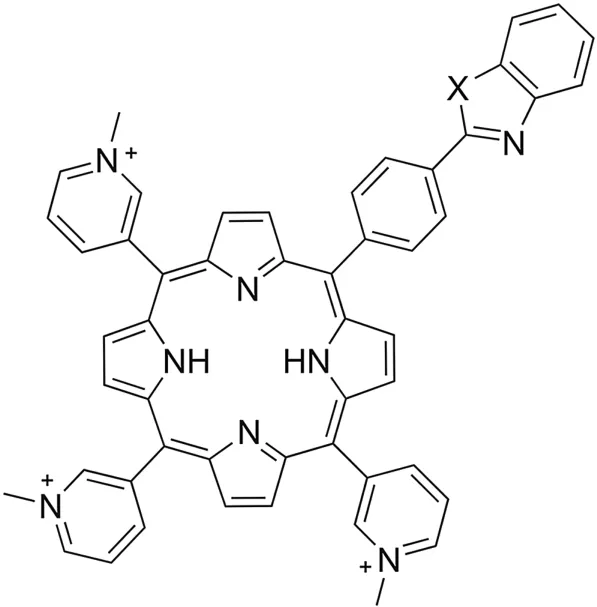
Fig.1.Structural formulas of unsymmetrical hetaryl-substituted porphyrins.5-[4'-(1′,3′-benzothiazol-2′-yl)phenyl]-10,15,20-tri-(N-methyl-3′-pyridyl)-porphyrin triiodide(X=S,SPOR);5-[4'-(1′,3′-benzoxazol-2′′-yl)phenyl]-10,15,20-tri-(N-methyl-3′-pyridyl)-porphyrin triiodide(X=O,OPOR);5-[4’-(N-methyl-1′′,3′′-benzoimidazol-2′′-yl)phenyl]-10,15,20-tri-(N-methyl-3′-pyridyl)-porphyrin triiodide(X=N-CH3,NPOR).
2.2.Porphyrin synthesis
2.2.1.5-(4′-bromophenyl)-10,15,20-tris-(3′-pyridyl)porphyrin
A mixture of 10 mL(0.144 mol)of pyrrole,6.66 g(0.036 mol)of 4-bromobenzaldehyde,and 10.15 mL(0.108 mol)of 3-pyridy lcarboxaldehyde was added to 500 mL of propionic acid and 35 mL(0.27 mol)of propionic anhydride,which were boiled for 20 min while passing gas.The mixture was boiled for another 1.5 h while passing air.Propionic acid was distilled under vacuum.The residue in the flask was diluted with 300 mL of methanol and 30 mL of ammonia solution.The precipitate of the porphyrin mixture was filtered,washed with methanol,and dried at room temperature to achieve a constant weight.The dried precipitate was dissolved in 200 mL of dichloromethane and chromatographed on a column with Al2O3(activity grade III according to Brockmann),eluting with a mixture of ethanol and methylene chloride(10:1,V/V).The third fraction of 5-(4′-bromophenyl)-10,15,20-tris-(3′-pyridyl)porphyrin was collected.The solvent was evaporated,and the porphyrin was rechromatographed on a column with Al2O3(activity grade III according to Brockmann),eluting with a mixture of ethanol and methylene chloride(10:1,V/V).The product was monitored by thinlayer chromatography on Alufol, ultraviolet-visible(UV-Vis)spectrophotometry,1H NMR spectroscopy,and MALDI-TOF-MS.Yield:3.95 g(15.8%).UV-Vis(dichloromethane; λmax,log ε):416(5.91),512(4.60),547(4.58),588(4.48),and 643(4.58)nm.1H NMR(CDCl3; δ,ppm):9.50 s(3H,2′-HPy),9.10 d(3H,6′-HPy,J=5.5),9.02 d(2H,8,12-H,J=4.5),8.89 s(4H,3,7,13,17-H),8.57 d(2H,2,18-H,J=4.5),8.44 d(3H,4′-HPy,J=5.5),8.07 d(2H,2′′,6′′-HPh,J=8.0),7.91 d(4H,5′-HPy,3′′,4′′-HPh,J=8.0),and-2.78 s(2H,NH).MALDI-TOF-MS,m/z calculated for C41H26BrN7:696.60;defined:696.68 [M]+.Retention factor (Rf,silufol):0.72,dichloromethane-ethanol(10:1,V/V).
2.2.2.Zinc 5-(4′-bromophenyl)-10,15,20-tris-(3′-pyridyl)porphyrinate
Furthermore,3g(4.3mmol)of5-(4′-bromophenyl)-10,15,20-tris-(3′-pyridyl)porphyrin and 4.7g(0.021mol)of anhydrous zinc acetate were dissolved in a mixture of 200 mL of methanol and 100 mL of methylene chloride.The mixture was boiled in a flask with a reflux condenser for 1.5 h.Monitoring was carried out using a UV-Vis spectrometer.The mixture was cooled,and the excess solvent was distilled,chromatographed on a column with Al2O3(activity grade III according to Brockmann),and eluted with a mixture of ethanol and methylene chloride(10:1,V/V).The solvent was evaporated under vacuum,and the residue was washed with water,filtered,and dried at room temperature to achieve a constant weight.Yield:3.2 g(98%).UV-Vis(dichloromethane;λmax,logε):416(6.06),549(4.62),and590(4.71)nm.1HNMR(CDCl3;δ,ppm):9.50s(3H,2′-HPy),9.10d(3H,6′-HPy,J=5.5),9.02d(2H,8,12-H,J=4.5),8.89s(4H,3,7,13,17-H),8.57d(2H,2,18-H,J=4.5),8.44d(3H,4′-HPy,J=5.5),8.07d(2H,2′′,6′′-HPh,J=8.0),and7.92d(4H,5′-HPy,3′′,4′′-HPh,J=8.0).MALDI-TOF-MS,m/z calculated for C41H24BrN7Zn:759.97.Defined:760.03[M]+.Rf(silufol):0.67,dichloromethane-ethanol(10:1,V/V).
2.2.3.5-[4’-(1,3-benzothiazol-2-yl)phenyl]-10,15,20-tri-(3′-pyridyl)-porphyrin
In a 100-mL flask equipped with a magnetic stirrer and a reflux condenser,0.03 g of Pd(OAc)2(20%,mol/mol),0.026 g of Cu(OAc)2H2O(20%,mol/mol),0.176 g of triphenylphosphine(1 eq.),0.51 g of 5-(4′-bromophenyl)-10,15,20-tris-(3′-pyridyl) zinc porphinate(0.671 mmol),45 mL of toluene,and 146μL(2 equivalents,1.342 mmol)of benzothiazole were added.The reaction mixture was boiled with stirring for 48 h.Then,the mixture was cooled to room temperature,and 50 mL of methylene chloride was added.The organic solution was filtered,the precipitate was washed with 10 mL of methylene chloride,and the combined organic fractions were evaporated under vacuum.The second fraction was collected.The zinc complex was dissolved in dichloromethane,and 3 mL of concentrated HCl was added to destroy the complex.Then,it was treated with ammonia and washed with water.The solvent was dried by adding anhydrous sodium sulfate and evaporated under vacuum.Chromatography was performed on a column with Al2O3(activity grade III according to Brockmann),eluting first with dichloromenthane and then with a mixture of ethanol and methylene chloride(1:10,V/V).The second fraction was collected.Yield:0.345 g(67%),green-violet crystalline powder.UV-Vis(dichloromethane;λmax,log ε):420(5.96),516(4.66),550(4.57),590(4.60),and 646(4.59)nm.1H NMR(CDCl3; δ,ppm):9.50 s(3H,2′-HPy),9.10 d(3H,6′-HPy,J=5.5),9.02 d(2H,8,12-H,J=4.5),8.89 s(4H,3,7,13,17-H),8.57 d(2H,2,18-H,J=4.5),8.44 d(3H,4′-HPy,J=5.5),8.27 d(2H,2′′,6′′-HPh,J=6.0),7.82-7.79 m(4H,5′-HPy,3′′,4′′-HPh),7.94-8.11 m(2H,from benzothiazole J=8.2),7.46-7.54 m(2H,from benzothiazole),and-2.78 s(2H,NH).MALDI-TOF-MS,m/z calculated for C48H30N8S:750.87;defined:750.89[M]+.Rf(silufol):0.73,dichloromethane-ethanol(10:1,V/V).
2.2.4.5-[4’-(1′′,3′′-benzothiazol-2′′-yl)phenyl]-10,15,20-tri-(N-methyl-3′-pyridyl)-porphyrin triiodide(SPOR)
A mixture of 0.22 g(0.29 mmol)5-[4'-(1,3-benzothiazol-2-yl)phenyl]-10,15,20-tri-(3′-pyridyl)-porphyrin and 0.5 mL(0.46 mmol)of methyl iodide was refluxed in dimethylformamide(30 mL)for 1 h.The solution was cooled and diluted with benzene(1:1,V/V).The precipitate was separated by filtration,washed sequentially with benzene and acetone,and then dried.Yield:0.21 g(98%).UV-Vis(water; λmax,log ε):418(6.09),516(4.69),549(4.63),585(4.65),and 634(4.65)nm.1H NMR(DMSO d6;δ,ppm):9.52 s(3H,2′-HPy),9.11 d(3H,6′-HPy),9.02d(2H,8,12-H),8.89s(4H,3,7,13,17-H),8.57 d(2H,2,18-H),8.44 d(3H,4′-HPy),8.27 d(2H,2′′,6′′-HPh),7.82-7.79 m(4H,5′-HPy,3′′,4′′-HPh),7.94-8,11 m(2H,from benzothiazole),7.46-7.54 m(2H,from benzothiazole),4.72 s(9H,CH3-N),and-2.78bs(2H,NH).MALDI-TOF-MS,m/z calculated for C51H39I3N8S:1176.69;defined:1176.73[M]+.
2.2.5.5-[4’-(1′′,3′′-benzoxazol-2′′-yl)phenyl]-10,15,20-tri-(3′-pyridyl)-porphyrin
Compound 5-[4’-(1′′,3′′-benzothiazol-2′′-yl)phenyl]-10,15,20-tri-(3′-pyridyl)-porphyrin was prepared analogously;instead of benzothiazole,0.159 g(2 equivalents,1.342 mmol)of benzoxazole was used.Yield:0.395 g(79%)and green-violet crystalline powder.UV-Vis(dichloromethane; λmax,logε):420(5.97),516(4.67),551(4.57),590(4.62),and 646(4.62)nm.1H NMR(δ,ppm):9.50 s(3H,2′-HPy),9.10 d(3H,6′-HPy,J=5.5),9.02 d(2H,8,12-H,J=4.5),8.89 s(4H,3,7,13,17-H),8.57 d(2H,2,18-H,J=4.5),8.44 d(3H,4′-HPy,J=5.5),8.26 d(2H,2′′,6′′-HPh,J=6.0),7.82-7.80 m(4H,5′-HPy,3′′,4′′-HPh),7.58-8,02 m(2H,from benzoxazole J=8.3),7.34-7.43 m(2H,from benzoxazole),and-2.79 s(2H,NH)(CDCl3).MALDI-TOF-MS,m/z calculated for C48H30N8O:734.80;defined:734.83[M]+.Rf(silufol):0.73,dichloromethane-ethanol(10:1,V/V).
2.2.6.5-[4’-(1′′,3′′-benzoxazol-2′′-yl)phenyl]-10,15,20-tri-(N-methyl-3′-pyridyl)-porphyrin triiodide(OPOR)
Compound 5-[4’-(1′′,3′′-benzothiazol-2′′-yl)phenyl]-10,15,20-tri-(N-methyl-3′-pyridyl)-porphyrin triiodide(Fig.1)was obtained in the same way;instead of benzothiazole,0.159 g(2 equivalents,1.342 mmol)of 5-[4’-(1′′,3′′-benzoxazol-2′′-yl)phenyl]-10,15,20-tri-(N-methyl-3′-pyridyl)-porphyrin triiodide was used.Yield:0.23 mg(98%)and brown crystalline powder.UV-Vis(water;λmax,logε):418(6.05),516(4.67),550(4.64),585(4.64),and 634(4.66)nm.1H NMR(DMSOd6;δ,ppm):9.51s(3H,2′-HPy),9.10d(3H,6′-HPy),9.02d(2H,8,12-H),8.90s(4H,3,7,13,17-H),8.57d(2H,2,18-H),8.44d(3H,4′-HPy,J=5.5),8.26 d(2H,2′′,6′′-HPh),7.82-7.80 m(4H,5′-HPy,3′′,4′′-HPh),7.58-8.02 m(2H,from benzoxazole),7.34-7.43 m(2H,from benzoxazole),4.71s(9H,CH3-N),and-2.79s(2H,NH).MALDI-TOF-MS,m/z calculated for C51H39I3N8O:1160.62;defined:1160.67[M]+.
2.2.7.5-[4’-(N-methyl-1′′,3′′-benzoimidazol-2′′-yl)phenyl]-10,15,20-tri-(3′-pyridyl)-porphyrin
Compound 5-[4’-(1′′,3′′-benzothiazol-2′′-yl)phenyl]-10,15,20-tri-(N-methyl-3′-pyridyl)-porphyrin was prepared analogously;instead of benzothiazole,0.178 g(2 equivalents,1.342 mmol)of N-methylimidazole was obtained.Yield:0.228 g(45%)and greenviolet crystalline powder.UV-Vis(dichloromethane; λmax,log ε):420(5.97),516(4.67),550(4.62),590(4.59),646(4.62)nm.1H NMR(CDCl3; δ,ppm):9.50 s(3H,2′-HPy),9.10 d(3H,6′-HPy,J=5.5),9.02 d(2H,8,12-H,J=4.5),8.89 s(4H,3,7,13,17-H),8.57 d(2H,2,18-H,J=4.5),8.44 d(3H,4′-HPy,J=5.5),8.26 d(2H,2′′,6′′-HPh,J=6.0),7.82-7.80 m(4H,5′-HPy,3′′,4′′-HPh),7.59 d(2H,from imidazole J=8.4),7.42-7.47 m(2H,from imidazole),3.88 s(1H N-Me),and-2.78 s(2H,NH).MALDI-TOF-MS,m/z calculated for C49H33N9:747.29;defined:747.36 [M]+.Rf(silufol):0.70,dichloromethane-ethanol(10:1,V/V).
2.2.8.5-[4’-(N-methyl-1′,3′-benzoimidazol-2′′-yl)phenyl]-
10,15,20-tri-(N-methyl-3′-pyridyl)-porphyrin triiodide(NPOR)
Compound 5-[4’-(1′,3′-benzothiazol-2′-yl)phenyl]-10,15,20-tri(N-methyl-3′-pyridyl)-porphyrin triiodide(Fig.1)was prepared in the same way;instead of 5-[4’-(1′,3′-benzothiazol-2′-yl)phenyl]-10,15,20-tri-(3′-pyridyl)-porphyrin, 5-[4’-(N-methyl-1′,3′-benzoimidazol-2′-yl)phenyl-10,15,20-tri-(3′-pyridyl)-porphyrin was used.Yield:0.19 g(99%)and brown crystalline powder.UV-Vis(water;λmax,logε):418(6.07);516(4.69),550(4.65),585(4.63),634(4.64)nm.1H NMR(DMSO d6;δ,ppm):9.50 s(3H,2′-HPy),9.10 d(3H,6′-HPy),9.02 d(2H,8,12-H),8.89 s(4H,3,7,13,17-H),8.57 d(2H,2,18-H),8.44 d(3H,4′-HPy),8.26 d(2H,2′,6′-HPh,J=6.0),7.83-7.80 m(4H,5′-HPy,3′,4′-HPh),7.59 d(2H,from imidazole),7.42-7.47 m(2H,from imidazole),4.72 s(9H,CH3-N),3.89 s(1H N-Me),and-2.78 s(2H,NH).MALDI-TOF-MS,m/z calculated for C52H42I3N9:1173.66;defined:1173.74[M]+.
3.Results and discussion
Research on the SARS-CoV-2 has been ongoing since 2019. Thegenome structure, epidemiological and clinical characteristics, andpathogenetic mechanisms of SARS-CoV-2 are known. Information onthe structure of the virus and the role of the structural units of SARSCoV-2 in the life cycle of the virus is constantly increasing. SARS-CoV-2 belongs to β-coronavirus genera and contains a positive-sensesingle-stranded RNA that is enclosed in a nucleocapsid (N) protein.This protein is covered by an envelope containing a S protein and amembrane protein. In addition to the listed structural proteins, SARSCoV-2 contains non-structural and accessory proteins. Structuralproteins ensure virus safety. They are responsible for binding to hostcell receptors and membrane fusion. Non-structural proteins (NSPs)are mainly involved in various stages of the viral life cycle.
Screening of porphyrin,chlorine,phthalocyanine,and porphyrazine compounds,based on our own and literature data on the empirical“structure-property”relationship[15-20],allowed us to propose structures of porphyrins(Fig.1)that are believed to have the above specified properties.
To test our hypothesis using bioinformatics methods,hetarylsubstituted porphyrins were docked with all SARS-CoV-2 proteins.The results are shown in Table 1.Based on the data obtained,hetaryl-substituted porphyrins exhibited the highest affinity for the S protein,followed by the N protein and the nsp13.The rest of the SARS-CoV-2 proteins were also able to bind to hetarylsubstituted porphyrins,but with much less energy gain(Table 1).
Let us consider the results obtained for the docking of hetarylsubstituted porphyrins with S protein,N protein,and nsp13.The S protein protruding on the surface of the viral particle consists of three identical S protein chains.Each chain consists of two subunits,S1 and S2.The S1 subunit is responsible for binding to receptors[21].The S2 subunit is responsible for the fusion of the virus and host cell membranes.The S1 subunit includes an RBD site that binds complementarily to the ACE2 receptor of the host cell.This interaction causes a strong conformational change in the S2 subunit.Then,with the help of the host enzymes(furin and transmembrane serine protease),the S protein is cleaved into the S1 and S2 subunits[22,23].Ultimately,it leads to the fusion of the viral envelope and the host cell membrane,followed by the release of the N into the cytoplasm[24,25].Thus,the S protein is an obvious target.Targeting the S protein,especially in the RBD region,can inhibit viral entry into the cells.As an example,Fig.2 shows the result of hetaryl-substituted porphyrin docking containing a benzoxazole residue with the S protein.The binding of all studied arylporphyrins occurs at the same sites of the S protein,namely,on the RBD surface of each chain.More precisely,the porphyrin compound is located between the RBD regions of two adjacent chains,simultaneously interacting with both RBD amino acid sequences(Table 2).
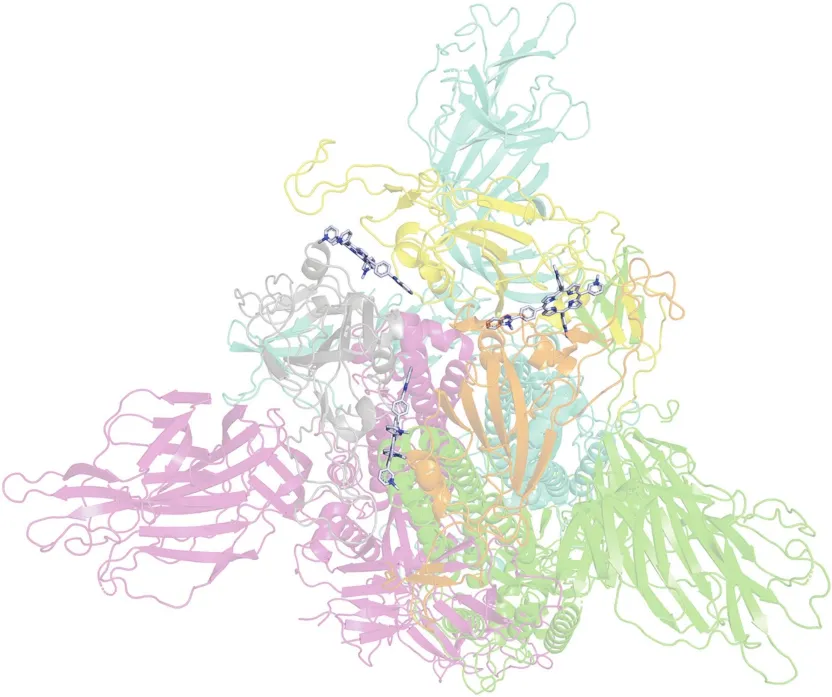
Fig.2.Molecular docking of OPOR with S protein.Green,magenta,and blue indicate the three chains of S protein;gray,orange and yellow indicate receptor-binding domain.
As noted above,the first step of the entry of the virus into the host cell is RBD binding to the ACE2 receptor.In RBD,two parts are distinguished:the RBD nucleus and the receptor-binding motif(RBM).It is a claw-like RBM that interacts with host ACE2.According to the literature[26],the overall amino acid sequence similarities between the S proteins of SARS-CoV-2 and those of SARS-CoV(isolated from humans,civets,or bats)range 76%-78% for the total protein,73%-76% for RBD,and 50%-53% for RBM.Based on this information,it can be concluded that the RBM is a less conserved part of the S protein.Mutations in the RBM region provide the virus with the ability to evolve and adapt to a larger number of cell receptors in different hosts.Owing to their clawlike shape,two critical amino acids(Gln493 and Asn501)are distinguished in the RBM.These acids,which bind to the amino acid residues of ACE2(Lys31 and Lys353),provide recognition and allow the binding of RBM to ACE2[26,27].In addition to the indicated amino acid residues of RBM,Leu455,Phe486,and Ser494 bind to ACE2.We determined the distance between the amino acid residues of RBM and the center of the hetaryl-substituted porphyrins(Table 3).
We believe that this is a very important characteristic,since the removal of the photosensitizer from the indicated amino acid residues determines the possibility of complete blocking of the binding of the virus to ACE2.The reactive oxygen species generated by photoirradiation of porphyrin are very reactive.They have a very short lifetime and a short diffusion path.For singlet oxygen,the estimated lifetime in water is 4μs[28],and the maximum possible path of1O2in the absence of quenchers is no more than 100-150 nm[29-31].This distance is much greater than the distance of the RBM amino acid residues from the hetaryl-substituted porphyrins(Table 3).Therefore,upon photoirradiation of the complex,the probability of oxidation by the generated porphyrins1O2is very high.
The geometry of the complex and the nature of the interaction of S protein with the heteroatom had no influence on the composition of the peripheral porphyrin substituent(Table 2).The nature of the interaction of hetarylporphyrins with S protein is universal and specific.For all studied porphyrins,π-π bound to the Tyr369 of each S protein.The arrangement of the aromatic systems within the complexes was not strictly parallel(Fig.3).The average interplanar distance was approximately 4 Å,which is typical for the π-π complexes of porphyrins.
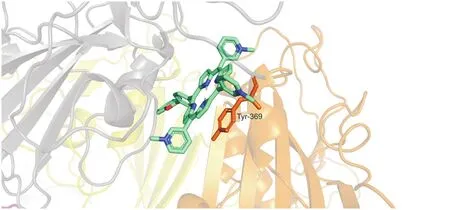
Fig.3.π-π-interaction of OPOR with Tyr369 of S protein.
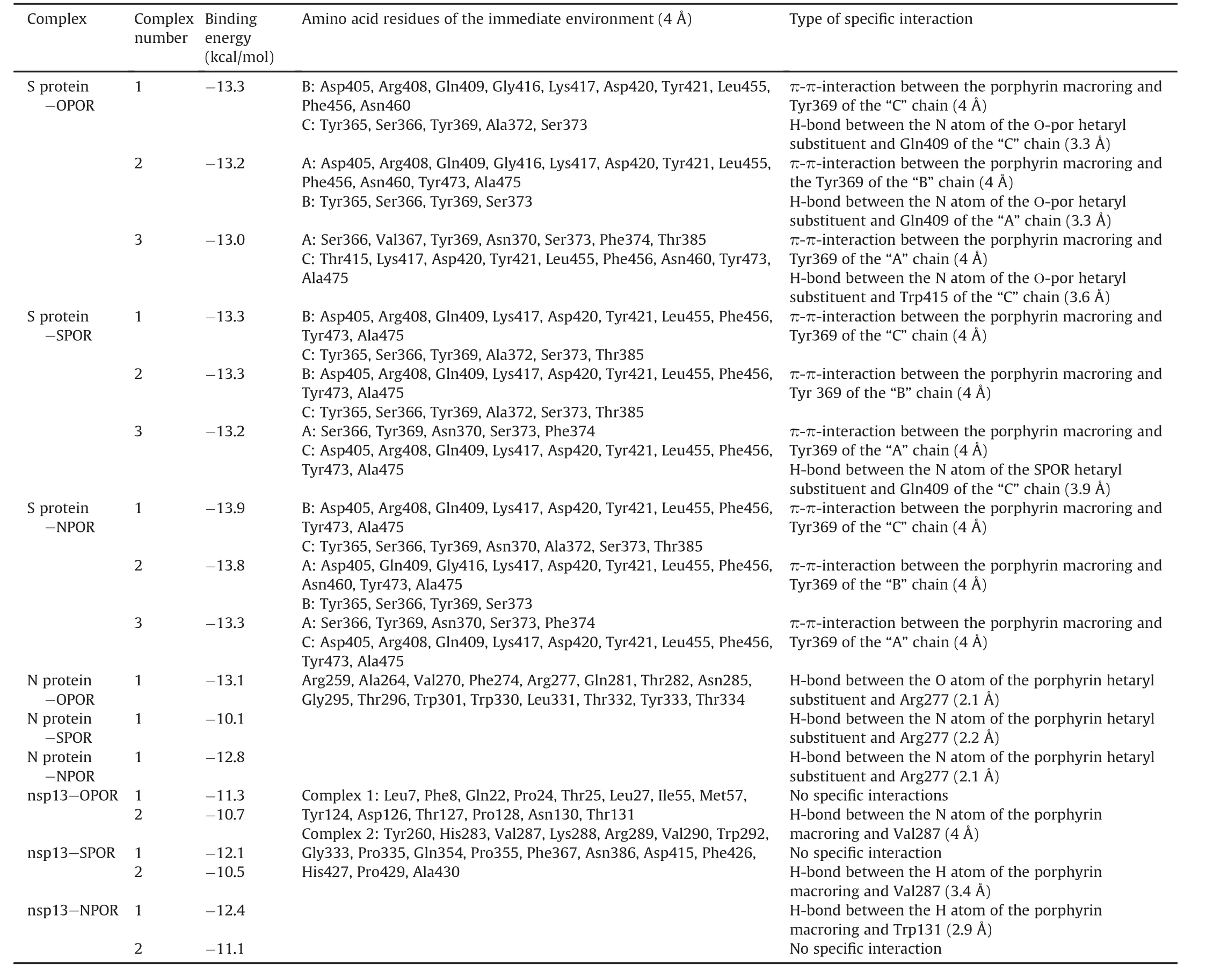
Table 2Molecular docking results of hetaryl-substituted porphyrins with SARS-CoV-2 proteins.

Table 3Distance between amino acid residues of receptor-binding motif(RBM)and the center of hetaryl-substituted porphyrins.
This interaction makes a significant contribution to the stabilization and strengthening of the complex of hetarylporphyrins with the S protein.On the contrary,it is undesirable because it possibly facilitates the scattering of light energy and quenching of the excited triplet state of porphyrin,reducing its photoactivity.The S protein affinity for hetarylporphyrins increased in the following order:SPOR≤OPOR<NPOR.The position of OPOR in this row is unexpected; it is the only hetaryl-substituted porphyrin that forms a H-bond between the oxygen atom of the peripheral porphyrin substituent and Gln409 of each S protein chain.The length of the formed H-bond is approximately 3.3 Å.This value is too high for the H-bond;therefore,it can be considered to be weak.This does not significantly affect the energy of the complex as a whole.In addition to the S protein the studied hetaryl-substituted porphyrins demonstrated high affinity for binding to the N protein and helicase(Table 1).
The N protein is an important structural protein in coronaviruses.After infection,the N protein enters the host cell along with the viral RNA to facilitate its replication in order to ensure the packaging of the viral RNA into new virions[32].The N-protein of SARS-CoV-2 consists of three functional parts:RNA-binding N-tail domain(NTD)(N-arm 1-44 and NTD 44-180);C-tail domain(247-367,C-tail 364-419),which provides oligomerization of the N protein;and a poorly structured central region enriched in Ser and Arg(SR region)(180-247)[32,33].In the aggregation of the N protein,the C-tail domain and a poorly structured central region enriched in Ser and Arg(SR region)are involved.
Judging by the docking results,hetaryl-substituted porphyrins bind to the SR region(Fig.4)due to the universal interactions and the formation of a H-bond.In the case of SPOR and NPOR,the nitrogen atom of the peripheral porphyrin substituent is involved in the formation of the H-bond with the protein.In the case of OPOR,the H-bond with the protein forms an oxygen atom of the peripheral porphyrin substituent(Table 1).The SR region is important for condensing the N protein with RNA through liquid-phase separation[34].In the immediate amino acid environment of hetarylporphyrins,the complex with the N protein contains amino acid residues that are easily photooxidized.These include Tyr333,Trp301,Arg259,and Arg277.The rate constant of their reaction with1O2is higher than those of other amino acids and is(1-7)×107M-1s-1[35,36].We expect that photodamage to this area will have significant inhibitory and virucidal effects.
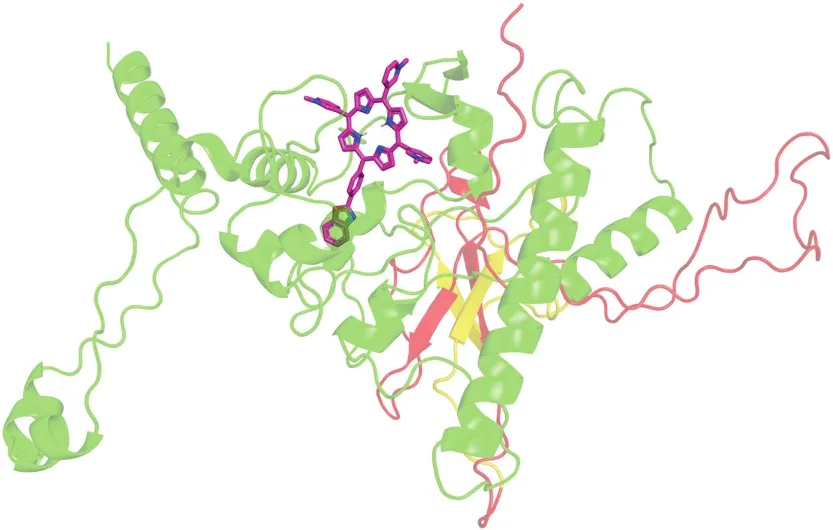
Fig.4.Molecular docking of OPOR with N protein.
The SARS-CoV replication/transcription complex consists of at least 16 NSPs.This complex includes replication enzymes and proteins that carry out post-transcriptional RNA modification(processing).The nsp13 of the coronavirus is unique because it performs the functions of both exoribonuclease(ExoN)and(guanine-N7)-methyltransferase (N7-MTase).The N-terminal domain of nsp13(ExoN)plays a role inproofreading during genome replication:ExoN is thought to mediate proofreading during genome replication.The activity of nsp13 as an exoribonuclease is regulated to avoid the nonspecific degradation of RNA.This process is also regulated by another NSP,nsp10[37,38].The C-terminal domain of nsp13 acts as a N7-MTase for RNA capping;it plays a major role in blocking the 5′-end of viral genomic RNA and avoids innate host immunity[39,40].
Let us consider the results obtained for complex 1 of SPOR with the N-tail domain of nsp13(ExoN)(Fig.5).Of all the hetarylsubstituted porphyrins studied,the H-bonds between the NH groups of the reaction center of the porphyrin and TRP131 form porphyrins, which contain benzothiazole and N-methylbenzimidazole residues.Moreover,the lengths of the H-bonds(Table 1)indicate the low energy of the complexes formed.This fact probably does not significantly affect the decrease in the photocatalytic activity of the considered porphyrins.Analysis of the amino acid environment closest to the porphyrins in a complex with the ExoN domain revealed that only one amino acid residue(Met57)was highly susceptible to photooxidation by singlet oxygen[35].
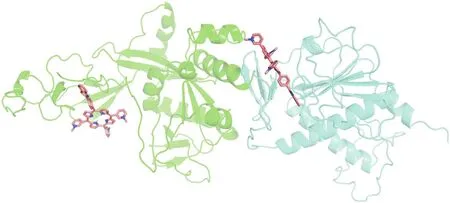
Fig.5.Molecular docking of SPOR with nsp13(exoribonuclease).
During molecular docking,two type of complexes with different binding energies were found(Table 2).The complexes of hetarylsubstituted porphyrins with the C-tail domain of nsp13 are formed due to universal interactions;in the case of porphyrins containing benzothiazole and benzoxazole residues,these complexes are additionally stabilized by the H-bond between the NH groups of the porphyrin macroring and Val287.However,as in the case of complex 1,the energy is very low(Table 1).The nearest amino acid environment of hetaryl-substituted porphyrins in complex 2 in the C-terminal domain of nsp13 is more sensitive to photooxidation.The vulnerable amino acids are Tyr260,Lys288,Arg289,Trp292,and His427[35].
These results demonstrate that hetaryl-substituted porphyrins are able to efficiently bind to parts of the nsp13 that performs an enzymatic function,especially in the C-tail domain.The latter is responsible for the mechanism of RNA capping,which plays a vital role in the escape of viral RNA from immune cells.Therefore,the breaking of binding to porphyrin and even its photoinduced oxidation of nsp13 lead to the degradation of viral RNA.Ultimately,this makes the replication cycle impossible.
We developed a procedure for the synthesis of hetarylsubstituted porphyrins and examined their photochemical properties.The general scheme for the synthesis of water-soluble hetaryl-substituted porphyrins is shown in Fig.6.
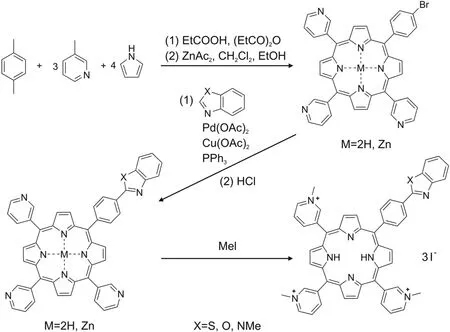
Fig.6.Scheme of synthesis of hetaryl-substituted porphyrins.
The obtained asymmetrically substituted porphyrins have a sufficiently high quantum yield of singlet oxygen:OPOR,0.61;NPOR,0.83;and SPOR,0.86.Based on the obtained quantum yield values,the most promising compounds are NPOR and SPOR,but their ability to oxidize RBM requires experimental verification.We plan to conduct in vitro studies to investigate the different processes involved in the interaction between SARS-CoV-2 proteins and NPOR and SPOR compounds,and to evaluate the possibility of RBM binding to ACE2 in the presence of porphyrins in the dark and under photoirradiation.This will allow the assessment of the inhibitory and photovirucidal activity of NPOR and SPOR.
4.Conclusion
Purposeful synthesis was carried out,and compounds SPOR,OPOR,and NPOR were obtained for the first time.
The photochemical properties of unsymmetrically substituted hetarylporphyrins were studied.They efficiently absorb light energy in the phototherapeutic window[41]and are capable of generating singlet oxygen with a quantum yield of 0.61-0.86.The molecular docking of SARS-CoV-2 proteins with hetarylsubstituted porphyrins made it possible to reveal three promising targets for inhibition and complete deactivation.These include the S protein,N protein,and nsp13.We believe this is a good strategy for targeting a potential drug to multiple targets,especially given the frequent mutations of the virus and the three-tiered defense(at different stages of the virus life cycle).The results obtained require in vitro experimental verification of the selected targets.In case of a positive result,it is necessary to further evaluate the inhibitory and virucidal capacities of hetaryl-substituted porphyrins.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
The reported study was funded by RFBR(Project No.:20-04-60067).
杂志排行
Journal of Pharmaceutical Analysis的其它文章
- Effect of Shengmai Yin on the DNA methylation status of nasopharyngeal carcinoma cell and its radioresistant strains
- Spectroscopic studies of the interaction between phosphorus heterocycles and cytochrome P450
- Impaired tricarboxylic acid cycle flux and mitochondrial aerobic respiration during isoproterenol induced myocardial ischemia is rescued by bilobalide
- Evaluation of the gastrointestinal anti-motility effect of Anacardium occidentale stem bark extract:A mechanistic study of antidiarrheal activity
- Synergistic effects of methyl 2-cyano-3,11-dioxo-18beta-olean-1,-12-dien-30-oate and erlotinib on erlotinib-resistant non-small cell lung cancer cells
- A living cell-based fluorescent reporter for high-throughput screening of anti-tumor drugs
