In-vitro assessment for the control of Fusarium species using a lactic acid bacterium isolated from yellow pitahaya (Selenicereus megalanthus (K.Schum.Ex Vaupel Moran))
2021-12-14LeidyVALENCIAHERNNDEZKarinaPEZPEZEyderMEZPEZLilianaSERNACOCKCristobalAGUILAR
Leidy J.VALENCIA-HERNÁNDEZ,Karina LÓPEZ-LÓPEZ,Eyder D.GÓMEZ-LÓPEZ,Liliana SERNACOCK,Cristobal N.AGUILAR
1 Universidad Nacional de Colombia,Palmira Campus,Valle 763533,Colombia
2 Food Research Department,School of Chemistry,Autonomous University of Coahuila,Saltillo Campus,Coahuila 25280,Mexico
Abstract The fungistatic activity of a lactic acid bacterium,which had been isolated from yellow pitahaya cultures,against fungi associated with basal rot (Fusarium oxysporum and Fusarium fujikuroi) was measured in the present study. Its activity was assessed in three fractions:fermented (S1),metabolic products (S2),and biomass (S3),using two fermentation substrates:Man Rogosa Sharpe agar (MRS) and potato dextrose agar (PDA). The bacterium was molecularly identified as Lactobacillus plantarum. S3 reduced F.fujikuroi growth by 100% over 48 h of fermentation,which occurred during the stationary phase of bacterial growth. The three fractions’ fungistatic activity against F.fujikuroi depended on the substrate employed. The fermentation kinetic parameters for L.plantarum indicated that its specific growth rate was 0.46 h-1,with 93.63% substrate consumption,0.045 kg kg-1 cell yield,and 0.54 kg kg-1 product yield. The kinetic parameters calculated will allow for bacteria production scaling. These in-vitro trials reveal L.plantarum’s possible application as a biocontrol agent for diseases associated with Fusarium. However,further ex-vivo and in-vivo researches are required to demonstrate its behavior in crops.
Keywords:lactic acid bacteria,Fusarium,fungistatic activity,biomass,yellow pitahaya
1.Introduction
One biological control alternative to agrochemical use against fungi and bacteria is the use of antagonistic microorganisms,such as lactic acid bacteria (LAB). LAB use has garnered the attention of researchers,given their potential as biocontrol agents against pathogenic bacteria and fungi,which cause animal and plant diseases. LAB have been widely studied in plants,and are considered to be biocontrol agents (Barrios-Robleroet al.2019).The antifungal compounds produced by LAB include organic acids,short chain fatty acids,hydrogen peroxide,reuterin,diacetyl and bacteriocins and bacteriocin-like inhibitory substances (Shehataet al.2019). The biocontrol potential of LAB is demonstrated in the prevention of fungal infections in both apples and grapes (Gajbhiye and Kapadnis 2016). Daranaset al.(2018) isolated and selected lactic acid strains to controlPseudomonas syringaepv.actinidiaein kiwi fruit,Xantomonas arboricolapv.pruniin prunes andXantomonas fragariaein strawberries. They found thatL.plantarumPM411 and TC92 prevented all three pathogens from infecting their corresponding plant hosts. Tomato seeds treated with LAB were infected withRalstonia solanacearum,in order to promote the production of defense-related enzymes and thus impart resistance to tomato crops (Konappaet al.2016). Shehataet al.(2019) demonstrated the biocontrol potential ofLactobacillussp.RM1 on wheat grains.Fusariumis considered to be among the major plant pathogens that cause significant diseases. Species in this genus are responsible for various plant alterations,including wilt and root and stem rot (Hernández-Cruzet al.2020).Fusariumhas resistance structures which enable it remain latent in soil for up to 10 years. Fungal growth is favored by warm temperatures,which then permit them to colonize the soil and invade the plantviathe xylem (Xuet al.2020). Yellow pitahaya (Selenicereus megalanthus(K.Schum.Ex Vaupel) Moran)) crops are affected byFusariumspecies,includingFusarium oxysporumsp.melonis,Fusarium monoliforme,andFusarium fujikuroi,which cause basal rot in the fruit (Salazar-Gonzálezet al.2016).
Biological control is an alternative approach to agrochemical use for fungal infection treatment. However,the scientific literature does not report on LAB antifungal activity against theFusariumspecies associated with basal rot in yellow pitahaya fruit. The objective of this investigation was to evaluate the fungistatic activity of a LAB (isolated from cultures of yellow pitahaya) against fungi associated with basal rot in said fruitviain-vitrotesting.
2.Materials and methods
2.1.LAB strain isolation
A group of 36 samples of yellow pitahaya cultures that had been infected by basal rot (including fruit) were taken with sterile swabs. Said samples were transported in refrigeration using stuart transport culture medium. Serial dilutions from 10-4to 10-7were made,using 0.1% peptone water. Exactly 0.1 mL of each dilution was seeded,in duplicate,in boxes with Man Rogosa Sharpe agar (MRS) (De Manet al.1960).Aniline blue was added,and these were incubated at 33°C and 37°C respectively for 48 h. Colonies that assimilated aniline blue were successively replicated in MRS agar and incubated at 33°C and 37°C respectively for 24 h,until pure cultures could be obtained. Once the pure cultures were obtained,Gram staining was performed,and morphology was determined. Strains presumed to be LAB were cryopreserved with 45% (w/v) glycerol.
2.2.Measurement of LAB fungistatic activity
Fusarium oxysporumandF.fujikuroihave been isolated from yellow pitahaya crops affected with basal rot disease conducted by Salazar-Gonzálezet al.(2016). These fungi were used to measure the fungistatic activity of cryopreserved LAB isolated from yellow pitahaya crops.
Fusarium oxysporumandF.fujikuroihave been cultured on potato dextrose agar (PDA) at 26°C for 8 d,until sporulation occurred. Spores were subsequently collected in test tubes and 0.002 kg L-1peptone water was added,then tubes were vigorously shaken (Magnussonet al.2003). Suspensions were adjusted into 105spores mL-1and cryopreserved at -4°C,with the addition of 0.004 L L-1glycerol,pending further use. The spore count (so as to adjust to 105spores mL-1) was performed through the microscopic determination of the cell number,using a Neubauer Chamber(Brand,Germany).
Fungistatic activity tests were carried out with two growth media:MRS agar (favorable for LAB growth),or PDA (favorable forF.oxysporumandF.fujikuroigrowth)(Valencia-Hernándezet al.2016). The 10 mL spore suspensions (105spores mL-1) ofF.oxysporumandF.fujikuroi(separately) were placed in the center of Petri dishes (with 20 mL of PDA and MRS agar,each fungus was tested on both media). Paper disks (Whatman No.1)with 0.8 cm diameters,containing 10 μL of isolated bacteria(24 h of incubation,at a concentration of 109CFU mL-1),were placed 0.016 m from the fungus. Petri dishes were incubated at 37°C for two days (boxes with MRS medium),and at 26°C for six days (boxes with PDA medium). For each test,filter paper discs,without bacteria,were used as controls.
Fungus growth was monitored by test photographs,using the photograph technique described by Leónet al.(2006) with several modifications. Fungal growth area measurement was calculated using ImageJ Evaluation Software,version 1.40 (Wayne Rasband,National Institutes of Health,USA).
The fungistatic activity was measured as the fungal growth inhibition percentage (eq.1).
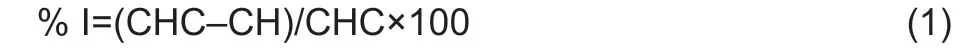
where % I is the inhibition percentage for theFusariumspecies,CHC is the growth area of theFusariumspecies in the control test (m2),CH is the growth area of theFusariumspecies (m2) when inoculated with isolated bacteria.
The bacteria that had the highest fungal growth inhibition percentage was selected. Identification and measurement of kinetic fermentation parameters of fermentation were performed on said bacterium.
2.3.ldentification of the selected bacterium
The selected bacterium was identified morphologically,biochemically,and molecularly. In order to obtain LAB genomic DNA,the procedure described by Dimkićet al.(2013) was employed with several modifications. Exactly 5 mL of the selected bacterium was centrifuged at 8 680×g for 2 min at 4°C. Once the process was finished,the supernatant was discarded and the pellet obtained was resuspended in 50 mmol L-1of 2-amino-2-hidroximetilpropano-1.3-diol-hydrogen chloride (Tris-HCL,pH 8),1 mmol L-12.2´,2´´,2´´´-(ethane-1,2-diyldinitrilo) tetra acetic acid (EDTA),and 25% sucrose and lysozyme (8 g L-1)for 30 min at 37°C. Thereafter,proteinase K (protease K,enzyme commission number E.C.3.4.21.64) and 10%sodium dodecyl sulfate (SDS) were added. Resuspended pellets were incubated for 10 min at 65°C. Subsequently,a volume of phenol-chloroform and a buffer volume (50 mmol L-1of Tris-HCl (pH 8),1 mmol L-1EDTA and 25%sucrose),were added and centrifuged at 17 013×g for 5 min at 4°C,and the supernatant was recovered. A volume of trichloromethane (chloroform) was added,and the supernatant was again recovered. Later,two volumes of cold ethanol and one-tenth of 3 mol L-1sodium ethanoate(sodium acetate) were added,centrifuged at 8 680×g for 2 min at 4°C and supernatant removal was performed. The DNA was obtained from the bottom of the tube,washed with 70% ethanol,and resuspended in 50 mL of RNase and TE buffer (50 mmol L-1Tris,10 mmol L-1EDTA). A 16S ribosomal gene fragment was amplified,using primers W01 and WO12 (Soleret al.2008).
Amplification was performed with PCR,with a total volume of 50 mL with 10×Taqbuffer ((NO4)2SO4),2 mmol L-1of MgCl2,0.4 mmol L-1of each primer WO1 (5´-AGA GTT TGA TC(AC) TGG CTC-3´) and WO12 (5´-TAC GCA TTT CAC C(GT) C TAC A-3´),0.025 U mL-1ofTaqDNA polymerase (2´-deoxyribonucleoside-5´-triphosphate:DNA deoxynucleotidyltransferase,enzyme commission number 2.7.7.7),1 mL of chromosomal DNA,and 5 mmol L-1dNTPs.The amplification was carried out in a thermocycler (Bio-Rad®,USA),with denaturation cycle which runs for 4 min at 94°C and 30 denaturation cycles at 94°C,for 30 s,with primer binding at 50°C for 30 s and elongation at 72°C for 60 s,and a final extension of 5 min at 72°C (Soleret al.2008).
The 16S RNA amplification products were analyzed with agarose gel electrophoresis 0.008 kg L-1in 1× TAE buffer at 70 V for 1 h,with 0.005 g L-13,8-diamino-5-ethyl-6-phenylphenanthridinium bromide (ethidium bromide).The 1-kb molecular weight marker (Fermentas®,USA)was utilized. Bands were visualized using an ultraviolet light transilluminator,and photographs were taken with the Bio-Rad gel imaging and documentation (Bio-Rad®,USA).
The amplified PCR fragment was purified and sequenced in Macrogen-Korea. Sequencing reactions were assembled in the CLC Main Workbench 7.0 Software (Qiagen,Germany). In order to learn the sequence’s identity,a bioinformatic analysis was performed in the NCBI Blastn Software,using the TL/16S_ribosomal_RNA_Bacteria_and_Archaea database. The highest score value was considered for the case in which several strains had the same molecular identity percentage. The sequence data were checked by BLAST analysis (Zhanget al.2000). The phylogenetic analysis of the 16S rRNA sequences of the isolates obtained in the study was conducted with MEGA X using the neighbor-joining method with 1 000 bootstrap replicates (Kumaret al.2018).
2.4.The fermentation kinetics of the selected bacterium
The selected bacterium was grown,in triplicate,in fedbatch fermentations using MRS broth supplemented with 40% glucose (MRS+GLU). Fermentations were performed at 37°C for 48 h with continuous agitation at 100 r min-1,without aeration (shaker orbital model 5000I VWR®,USA),and a workload of 0.5 L. Fermentations were adjusted to pH 6.0,using 1 mol L-1NaOH. The volume of the inoculum was 10% of the substrate.
From the fermentations described above,samples were taken at 0,1,2,4,6,8,12,24 and 48 h of fermentation,and lactic acid,acetic acid,biomass concentration,cell viability,and fungistatic activity,were measured.
In order to measure lactic acid,concentration test strips(Reflectoquant®-lactic acid test,Merck) were used. The strips were read using a reflectoquant equipment (RQflex®plus 10,Germany).
The acetic acid concentration was determined using HPLC (Agilent 1100,USA),through a payment for external services provision.
In order to determine biomass concentration,0.01 L of fermentation substrate was taken for each time period,then centrifuged at 10 000×g (Eppendorf Centrifuge-5804R®,Germany) for 10 min,and the supernatant was removed from each sample. The biomass was washed with brine and oven-dried at 100°C (AOAC 925.10 1990).
Cell viability was measured at each fermentation time.Dilutions from 10-1to 10-10were made with peptone water(0.001 kg L-1). Subsequently,each dilution was seeded in duplicate,in plates containing MRS agar added with aniline blue. Plates were inoculated with 1 mL of each dilution,and were incubated at 37°C for 48 h. The viable cell count was expressed as CFU mL-1.
Total sugar content was determinedviathe DNS method(Miller 1959),and the percentage of substrate consumption was calculated.
From fermentation kinetics,specific growth rate (μ),biomass yield (Yx/s),and product yield (Yp/s),were calculated by eqs.(2) and (3). To measureYp/s,lactic acid production was used as a reference:
whereS0is the initial concentration of total sugar (kg L-1),Sis final concentration of total sugar (kg L-1) until a time at whichP(lactic acid concentration in kg L-1) is the maximum,Xis the final biomass concentration (kg L-1),andX0is the initial biomass concentration (kg L-1).
2.5.The fungistatic activity kinetics of the selected bacterium
The fungistatic activity of fermented (S1),metabolic products(S2),and biomass (S3) was measured. For this purpose,fermented samples were collected at 0,1,2,4,6,8,12,24 and 48 h,and the curves corresponding to the kinetics of fungistatic activity were constructed. S1 corresponded to fermented,and S2 and S3 were obtained by centrifuging the fermented for 15 min at 5 000×g (Eppendorf®Centrifuge-5804R,Germany). S2 was filtered twice,first with a 0.45-mm diameter filter,and then with 0.2-mm diameter filter.The filtrate was packaged. The fungistatic activity tests for S1,S2,and S3 againstF.oxysporumandF.fujikuroiwere carried out in the same manner described above. In these tests,10 mL of S1,S2,and S3 were used. S3 had a concentration of 109CFU mL-1. Similarly,eq.(1) was used to calculate the fungus growth inhibition percentage.
2.6.Statistical analysis
The response variable was fungistatic activity,measured as an inhibition percentage,against theFusariumspecies.The results were analyzed using SAS Statistical Software,version 9.1.3 (Cary,NC). The mean comparison was calculated using the Tukey test,with a probability ofP<0.05.
Standard deviations were calculated for the kinetics of lactic acid production,biomass formation,acetic acid production,cell viability,and substrate consumption.
3.Results
3.1.LAB strain isolation
Of the 36 samples collected from the yellow pitahaya culture,eight Gram-positive bacteria were presumed to be LAB. Isolations were designated with capital letters.Table 1 describes the average percentage of fungal growth reduction,isolation sites,and growth temperatures. Fungus growth inhibition was dependent on theFusariumspecies(P<0.0001),antagonistic strain (P<0.0001),and substrate(P<0.0001). In both MRS substrate and PDA substrate,bacteria “E”,“F”,and “G” had the highest fungistatic activity against the twoFusariumspecies. However,bacterium“G” had higher percentages of fungal growth reduction.Therefore,it was selected for molecular identification and fermentation kinetic parameter measurement.
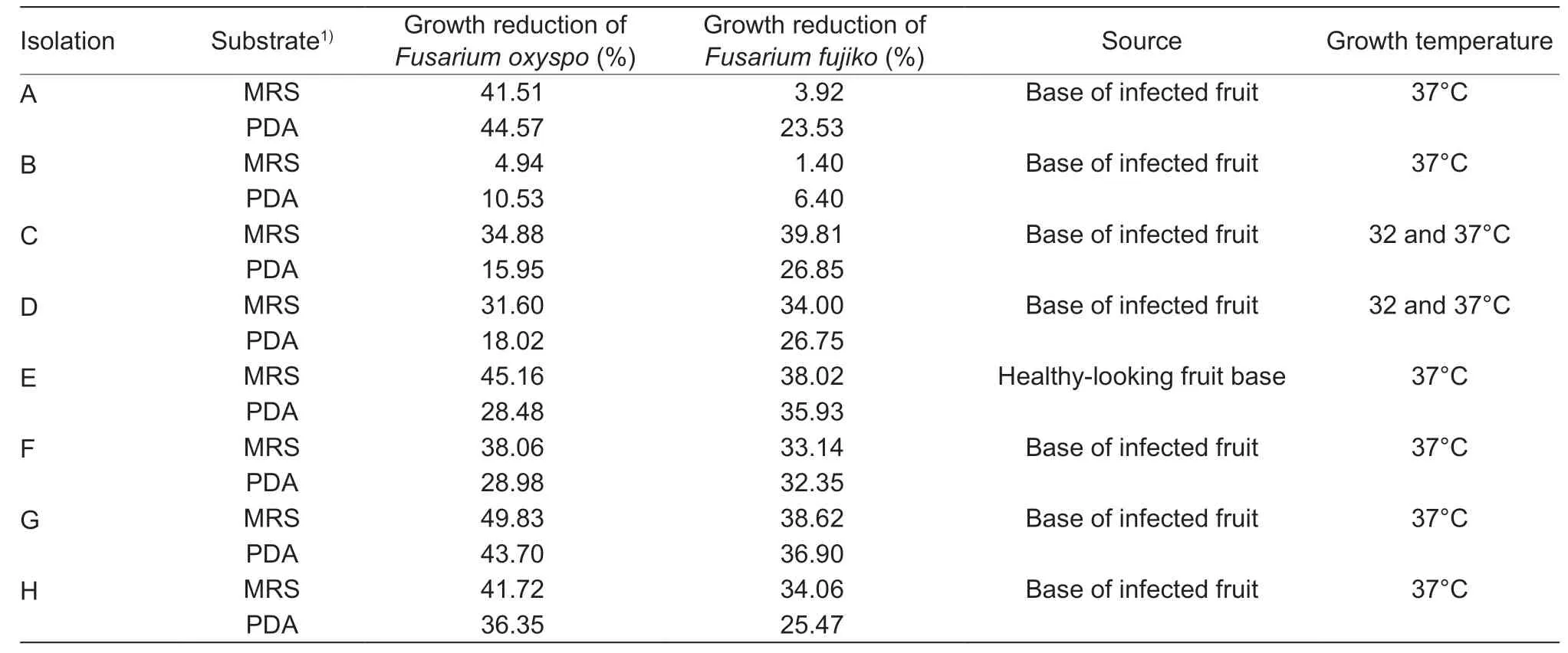
Table 1 Average percentage of fungal growth reduction,isolation site and growth temperature of bacteria isolated from yellow pitahaya crops
3.2.The identification of a lactic acid bacterium isolated from yellow pitahaya cultures
Bacterium “G” presented colonies from 2 to 5 mm,with milky white,smooth,round surfaces,and sizes. Colonies stained blue in the presence of aniline blue. The images generated by optical microscopy showed that the cells were Grampositive,short,rod-shaped,flagella-free,and spore-free.Biochemical tests indicated that this bacterium was glucose and lactose (+),gas production (-),sulfhydric acid (-),TSI(-),catalase (-),gelatin hydrolysis (-),growth at 40°C (+),a producer of lactic acid (1.94 to 33.8 g L-1) between 4 and 24 h,respectively,and had an optimum temperature (32°C).
A fragment of approximately 700 bp of the 16S ribosomal gene for the selected bacterium was amplified with PCR,and subsequently sequenced. The fragment included V1 to V4 variable regions of the 16S rRNA gene. The “G” strain presented a sequence of 683 nt,which showed a 100%identity withL.plantarumstrain WCFS1 16S ribosomal RNA (NR_075041,score 1.384),100%L.pentosus124-26S ribosomal RNA (NR_029133,score 1.368) and 99.85%withL.plantarumstrain NBRC 15891 16S ribosomal RNA(NR_113338,score 1.364).
A phylogenetic tree was performed (Fig.1). Fig.1 shows that BAL G is grouped with the BAL of the plantarum species,which is the further evidence that the isolated BAL in this study belongs to this species. Based on these results,we propose to name the BAL G asLactobacillus plantarumstrain G 4103 (GenBank accession number MT355409).The strain ofL.plantarumidentified in this investigation was deposited into the Bank of Strains and Genes at the Universidad Nacional de Colombia’s Biotechnology Institute(IBUN),with registration number IBUN-090-03774.
3.3.The fermentation kinetics of L.plantarum
When the MRS+glucose substrate was used,L.plantarumproduced between 1.94 and 33.8 g L-1of lactic acid. The highest production of this acid occurred between the 4th and 24th h of fermentation (Fig.2). Štětinaet al.(2010)found that LAB such asLactobacillus casei,Lactobacillus rhamnosus,Lactobacillus plantarum,Lactobacillus paracasei,andLactobacillus curvatus,produced high concentrations of lactic acid (36.03 to 76.65 g L-1) in MRS substrate. These authors related the production of lactic acid to its inhibitory power againstFusariumspecies.Traces of acetic acid were found at the 6th h of fermentation(0.013 g L-1) (Fig.2).
The substrate concentration decreased rapidly in the first 12 h of fermentation (Fig.2),with 23.9% of substrate consumption,corresponding to a rapid increase in the number of viable cells (Fig.2).
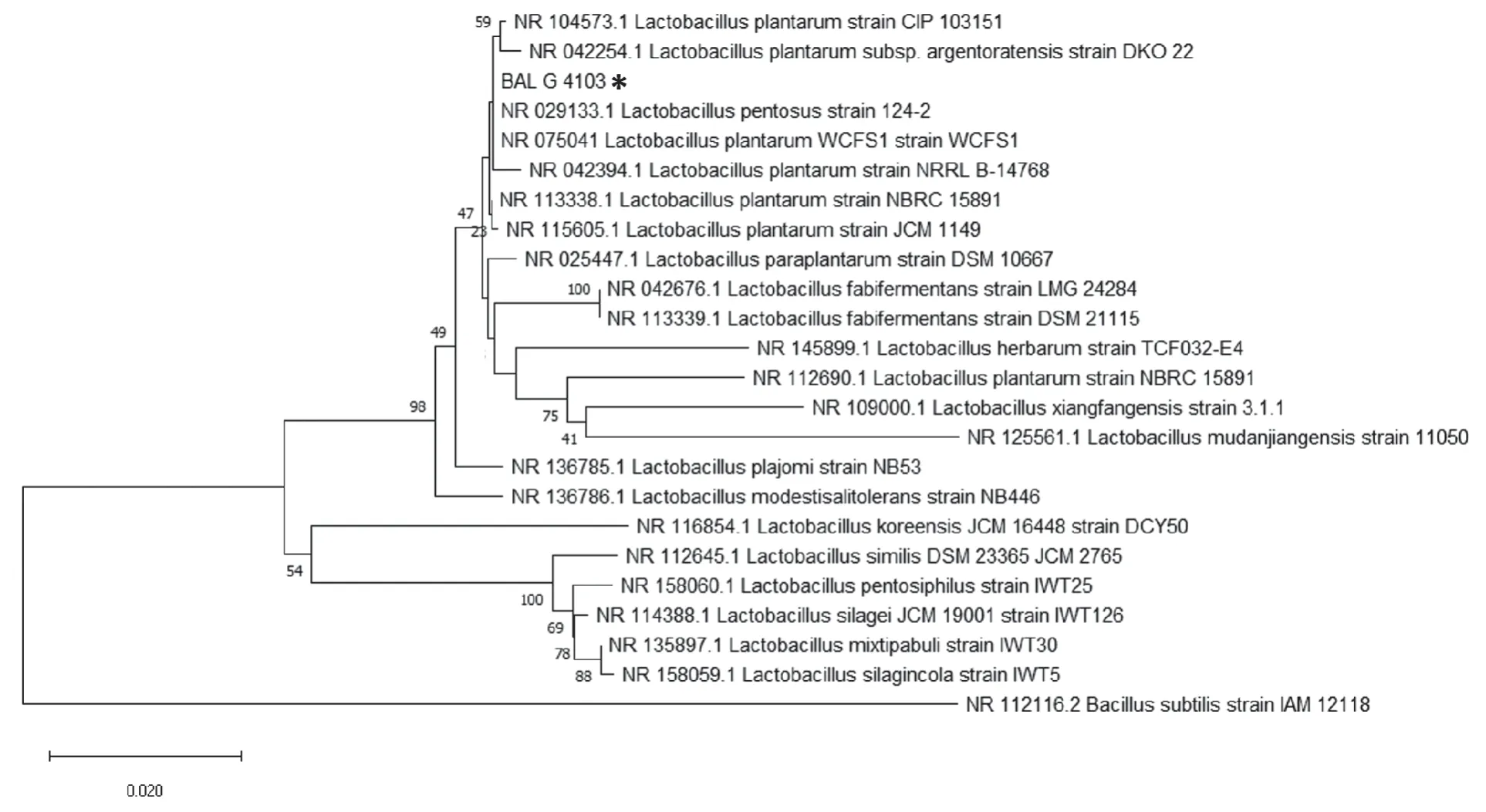
Fig.1 Phylogenetic tree of 16S rRNA sequence of the isolated BAL G 4103 (Lactobacillus plantarum strain G 4103 obtained using Neighbour-Joining (NJ) tree approach included in the MEGA X Software. Bacillus subtilis was used as an outgroup. * indicates the BAL G 4103 characterized in this study. The bar below the tree indicates nucleotide substitutions per site. Only bootstrap values above 40% are shown.
Biomass and viable cells are shown in Fig.3,where a curve with exponential growth is observed,until the 8-h fermentation mark. From the 12-h mark,cell death began,decreasing only one logarithmic unit until 48 h of fermentation.A significant increase for biomass,between the 2nd and 8th h was obtained (2.73 and 6.17 g L-1,respectively). This behavior correlates with the concentration of live cells,where maximum growth was obtained at the 8th h (7.75×109CFU mL-1). The kinetic parameters ofL.plantarumwere also obtained,such asμ,percentage of substrate consumption,Yx/s,andYp/swith values corresponding to 0.46 h-1,93.63%,0.045 kg kg-1,and 0.54 kg kg-1,respectively.
3.4.Fungistatic activity kinetics
S1,S2,and S3 presented fungistatic activity against the two fungi in MRS and PDA. The fungistatic activity of S1,S2,and S3 againstF.fujikuroiandF.oxysporumdepended on the culture medium (Fig.4-A-D). The maximum fungistatic activity of the three substances was obtained at 48 h. At that time,statistically significant differences between S1 and the other two treatments were found. The greatest fungistatic activity was obtained with S1 and S3.
4.Discussion
This investigation found a LAB (L.plantarum),isolated from yellow pitahaya crops,with high fungistatic activity againstFusariumspecies (F.oxysporumandF.fujikuroi),which are plant pathogens and cause basal rot in the yellow pitahaya.
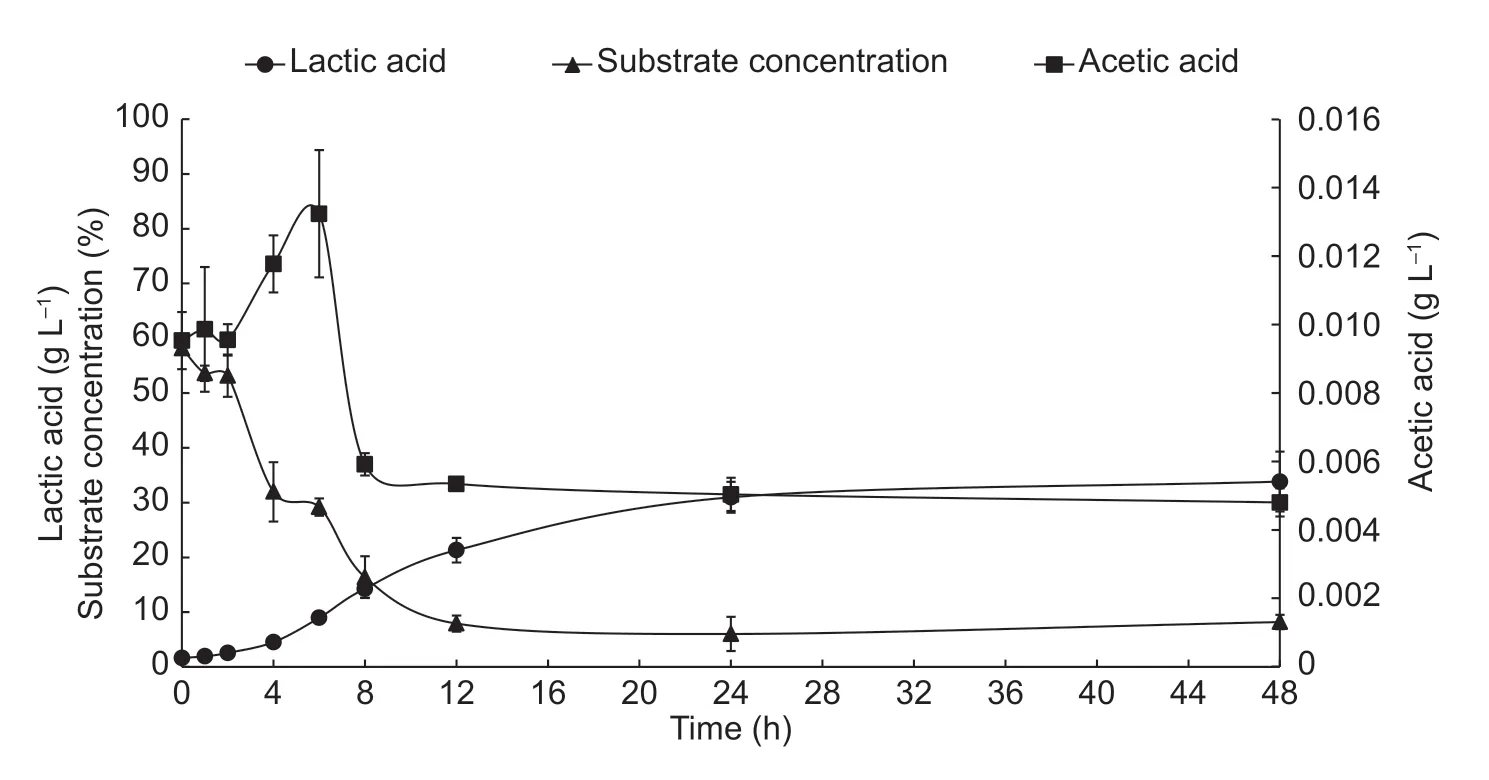
Fig.2 Production kinetics of lactic acid,acetic acid and total sugar concentration (substrate) of Lactobacillus plantarum (S1) during 48 h of fermentation. Error bars represent the standard deviations of the trials with three replicates.
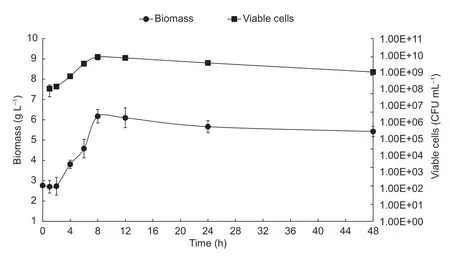
Fig.3 Biomass production kinetics and viable cells of Lactobacillus plantarum during 48 h of fermentation. Error bars represent the standard deviations of the trials with three replicates.
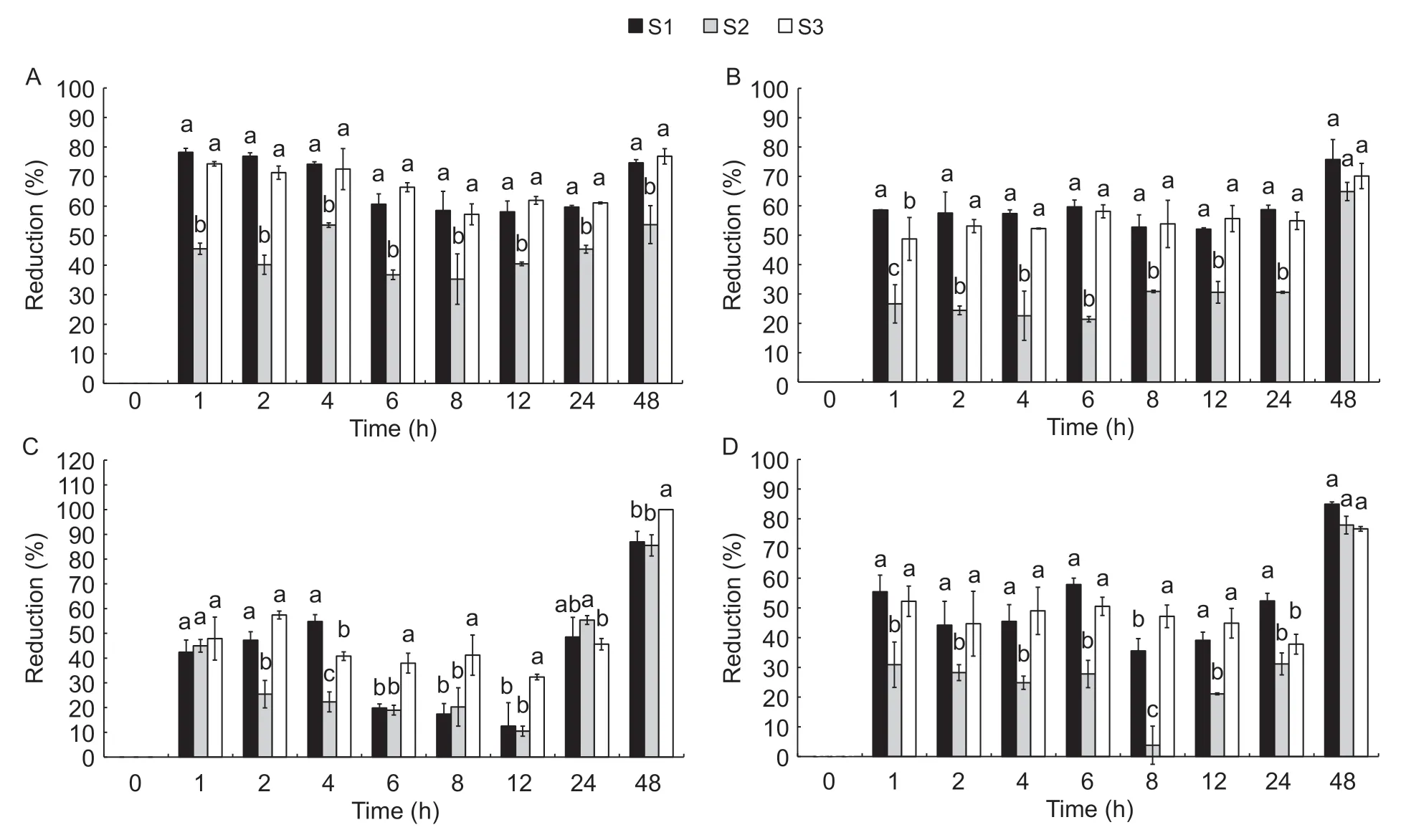
Fig.4 Fungistatic activity kinetics of three substances:ferment (S1),metabolic products (S2) and biomass (S3) of Lactobacillus plantarum. A,against F.oxysporum in MRS substrate. B,against F.oxysporum in PDA substrate. C,against Fusarium fujikuroi in MRS substrate. D,against F.fujikuroi in PDA substrate. Error bars represent the standard deviations of the trials with three replicates.
Lactobacillus plantarumis a heterofermentative microorganism,which is demonstrated by the presence of lactic acid and traces of acetic acid,as metabolic products of fermentation. The metabolites produced by bacteria depend on the oxidation pathway of carbohydrates.Homofermentative LAB uses the energy source and becomes 85% lactic acid,while other heterofermentative LAB convert glucose into different metabolites,such as lactic acid,acetic acid,ethanol,carbon dioxide,acetate,and acetaldehyde (Suskovicet al.2010).
The differences encountered in the fungistatic activity in MRS and PDA agars show the complexity of thein-vitromeasure of fungistatic activity and demonstrates the different nutritional requirements of microbial species.
This research showed thatL.plantarumconsiderably reduces the growth ofFusariumspecies.In-vitroresearch has demonstrated fungistatic activity of this lactic acid bacterium against different fungi (Arasuet al.2014;Shehataet al.2019). However,there are very few reports on the application of antifungal LAB as biocontrol agents in fruit. Wanget al.(2011) foundL.plantarumIMAU100 antifungal activity againstBotrytis cinerea,Alternaria solani,Phytophthora drechsleriTucker,F.oxysporum,andGlomerella cingulate. Barrios-Robleroet al.(2019) found that LAB had activity againstColletotrichum gloeosporioides.Delavenneet al.(2012) demonstrated strongL.harbinensisantifungal activity againstDebaryomyces hansenii,Kluyveromices lactis,Kluyveromyces marxianus,Penicillium brevicompactum,Rhodotorula mucilaginosa,andYarrowia lipolytica.L.plantarumhelps to minimize cucumber rot,caused byRhizopus stoloniferandB.cinerea(Satheet al.2007). Rotting in apples can be controlled by the application of LAB (Triaset al.2008),and grapes can be bioprotected by LAB (Lanet al.2012).
Some researchers associated the fungistatic activity of some bacteria with their exponential growth phase (Wanget al.2011). Herein,S1,S2,and S3 presented fungistatic activity in all stages of growth,and these fractions showed greater activity in the stationary phase ofL.plantarumgrowth. Differentiating the metabolites responsible for fungistatic activity was not the objective of this investigation.However,it may be stated that fungistatic activity could be related to biomass,and primary and secondary metabolites.These metabolites may include lactic acid or other organic acids not identified here,or antibiotics,bacteriocins,or other cellular functions,which would be regulated by the quorum sensing signaling process. This system is used by bacteria to communicate through autoinductors,and is dependent on microbial growth (Papenfort and Bassler 2016).Factors such as temperature,pH,carbon source,and the presence of competing microorganisms affect the microbial population. The above,in turn,facilitates the accumulation of self-inducing molecules that regulate defense responses to adverse environments (Stephenset al.2020). In this investigation,stress levels could be caused by factors such as type of fungus (F.oxysporumandF.fujikuroi),or growth substrate (MRS or PDA).
The antifungal activity ofL.plantarumhas been attributed to many factors. Strömet al.(2002) and Laitilaet al.(2002) attributedL.plantarumfungistatic activity to cyclic dipeptides such as the L-Phe-L-Pro cycle,the L-Phe-trans-4-OH-LPro cycle,and 3-phenylacetic acid. Studies argue that fungal inhibition can occur due to the production of metabolites dependent on specific pH and temperature conditions,as well as the effect of organic acids (Shehataet al.2019). Other authors reported that acetic acid,at low concentrations,is the cause of fungistatic activity,and that this acid is more effective than lactic acid (Corsetti and Settanni 2007). Janisiewiczet al.(2000) proposed that the fungistatic power of LAB against plant pathogens could be due to competition for nutrients and space,considered an antagonism mechanism.
Also,it is well known that the antifungal effect of LAB is attributed to the mixture of more than one compound that may include organic and fatty acids,proteinaceous compounds,cyclic dipeptides,phenolic compounds,and H2O2,propionate,phenyl-lactate,hydroxyphenyl-lactate and 3-hydroxy fatty acids (Voulgariet al.2010; Licandroet al.2020).
All of the kinetic parameters of fermentation presented in this investigation are important to scale the reproduction ofL.plantarum. Scaling is important for future applications of the bacterium as a biocontrol agent.
The information presented in this scientific article is novel. However,ex-vivoandin-vivostudies are required to demonstrate the effect of LAB against pathogens associated with the basal rot of yellow pitahaya fruit.
5.Conclusion
This investigation demonstrated the fungistatic activity of a lactic acid bacterium,which had been isolated from yellow pitahaya cultures (molecularly identified asL.plantarum),againstF.oxysporumandF.fujikuroi,fungi associated with pitahaya fruit basal rot. This study demonstrated that the fermented,metabolic products and biomass ofL.plantarumhave fungistatic activity againstF.oxysporumandF.fujikuroi.Lactobacillus plantarumcan reduce the growth ofF.fujikuroiby up to 100%. Thesein-vitrotrials uncoverL.plantarum’s possible use as a biocontrol agent for diseases associated withFusarium. However,ex-vivoandin-vivoresearches are still needed to demonstrateL.plantarumbehavior in crops. Finally,due to the differences found inin-vitrofungistatic activity,when MRS and PDA culture media were applied,it is concluded that the standardization of fungistatic activity measurement techniques is vital to ensure the comparability of results within the scientific literature.
Acknowledgements
The authors would like to thank the Administrative Department of Science,Technology,and Innovation,Colciencias,Asoppitaya,and the Inter-American Development Bank IDB for funding.
杂志排行
Journal of Integrative Agriculture的其它文章
- lnfectious disease-resistant pigs:Will they fly?
- Crop photosynthetic response to light quality and light intensity
- Alginate oligosaccharides preparation,biological activities and their application in livestock and poultry
- Genome-wide pedigree analysis of elite rice Shuhui 527 reveals key regions for breeding
- TaSnRK2.4 is a vital regulator in control of thousand-kernel weight and response to abiotic stress in wheat
- Development and characterization of new allohexaploid resistant to web blotch in peanut
