lnfectious disease-resistant pigs:Will they fly?
2021-12-14TadSONSTEGARDPerryHACKETT
Tad S.SONSTEGARD,Perry B.HACKETT
1 Acceligen 3388 Mike Collins Drive Suite #1,Eagan,MN 55121,USA
2 Center for Genome Engineering,Deptement of Genetics,Cell Biology and Development,University of Minnesota,Minneapolis,MN 55455,USA
Cloven hoof animals have been a major source of nutritious animal protein for humans for at least 10 000 years. Over this time many were domesticated into livestock and some were put under selection for traits of importance or cultural preference. Accompanying domestication,diseases were transmitted from livestock to humans as a result of living in close quarters. Eight of 15 major chronic,temperate diseases,including diphtheria,influenza A,measles,mumps,pertussis,rotavirus,smallpox,tuberculosis,are thought to have reached humans from domestic animals(Wolfeet al.2012). The list may be longer since only those with pandemic potential become evident. The toll on humans resulting from viruses moving from livestock to people is enormous. For instance,the United States Centers for Disease Control and Prevention (CDC) estimates that influenza,which periodically emerges from swine and birds,annually has resulted in 9-45 million illnesses,140 000-810 000 hospitalizations and 12 000-61 000 deaths,depending on the year (CDC 2020). The estimated cost today of influenza is greater than $25 billion per year(Molinariet al.2007,adjusted for inflation). The costs to farmers of viral infections in livestock can be even larger.For instance,the recent outbreak African swine fever (ASF)in China is estimated to have resulted in a cost of more than$50 billion since 2019 (Carriquiryet al.2020). Vaccines to treat emerging infectious agents that plague livestock take years to develop,are expensive and inefficient to administer,and some fail to provide full immunity over an animal’s lifetime. Vaccines,like those against Foot-andmouth disease virus (FMDV) can be effective at eradicating disease transmission within national borders; but,they may challenge frontline diagnostic testing for large-scale serological surveillance and post-vaccination monitoring(Asforet al.2020). Countries that are free of FMDV without the use of a national vaccination program have an advantageous trade status for movement of live animals and animal products (OIE/FAO 2012).
Genetic selection to improve disease resistance is possible but this approach is rare because animals under intense selection for maximally efficient production are too valuable to be used for rigorous testing of disease-resistance alleles. Identification of important gene variants for resistance is further complicated by number of phenotypes needed for statistical accuracy,pathogen exposure and load,and the polygenic nature of most disease resistant traits (Bishop and Woolliams 2014). Genome-editing technologies have been touted as a potential solution to rapidly introduce alleles into livestock (Carlsonet al.2013)including either inactivating (knocking-out) a known viral receptor or introducing (knocking-in) a validated resistance gene from another species (Lillicoet al.2016). The main impediments to this approach include difficulties to manipulate the genome across a wide variety of animals in a precise and reproducible manner,complex and expensive biosafety containment (BSL-3) facilities,as well as a dearth of genetic targets for manipulation to interfere with infectious life cycles.
With the advent of full genome reference sequences for livestock (Rexroadet al.2019) and gene-editing technologies including CRISPR/Cas9 (Proudfootet al.2019),genetic engineers have the tools to quickly introduce precise changes to key genes and breeders can study host responses to pathogen infection. Many have taken advantage of prior studies of viral entry mechanisms to target “gatekeeper” genes for infection,for instance inactivating CD163,one of the host proteins used by the porcine reproductive and respiratory syndrome (PRRS)virus (e.g.,Whitworthet al.2016; Burkardet al.2017)or CRISPR-targeting the CP204L gene in the ASF virus genome (Hübneret al.2018). However,this approach does not always work; other studies that have relied on changing host immune response to make animals more resilient have not been successful (McClearyet al.2020). All of these studies have focused on single gene changes relative to one infectious agent.
The recent breakthrough reported by Xuet al.(2020)demonstrates the first successful breeding of a food animal withtwogene edits. Together,the two deployed changes to the genome,CD163andpAPN,confer full resistance to two economically important viruses in swine,PRRSV and transmissible gastroenteritis virus (TGEV)respectively (Fig.1). Moreover,reduced susceptibility to a third pathogen,porcine delta coronavirus (PDCoV),was validated. They used the CRISPR/Cas9 system to make double knock-out (DKO) edits in pig embryo fibroblast cells that could be characterized for both intended and unintended (off-targeting) genome alterations before nuclear transplantation into embryos for transplantation. The successful primary cloning efficiency following transplant was less than 0.1% to yield two viable offspring with intended edits and no detected off-targeting events. Cells taken from these two piglets were then used for a second round of cloning that yielded 12 more pigs suitable for phenotypic assessment including resistance to PRRSV,TGEV and PDCoV challenges. Much of the testing was conducted in porcine alveolar macrophages,known targets of PRRSV. One intriguing question is whether the DKOs may have interactive influences; the group observed that in PDCoV-infected DKO pigs that there was a delayed adaptive immune response against the virus. Regardless,the power of multiplex genome editing has now been validated for two important livestock pathogens.
The scientific take-home messages from this work are evident. First,precise and reproducible editing of multiple sites in a large animal genome for a recessive trait or knock-out is feasible using gene-editing tools and cellular screening. Cloning may not be the best reproductive method for deploying in commercial swine; but,clearly the ability to complement the genetic progress made by animal breeders for other traits is tenable for introducing viral resistance to economically important infectious diseases.Second,this work shows the ability to improve animal management and well-being is achievable. This aspect is important for realizing the goals of the international One Health Initiative (https://onehealthinitiative.com/) that couples human and animal health as well as protects our planet by increasing agricultural sustainability through reduced losses from disease. Indeed,the swine acute diarrhea syndrome coronavirus (SADS-CoV),which is related to PDCoV,appears able to replicate in primary human cells thereby suggesting potential zoonotic susceptibility of humans to infection from coronavirus-infected pigs (Edwardset al.2020).
Although the science is secure and sound,the most significant implications of this work may not be forthcoming.Before any genome-edited animal,or crop for that matter,can be deployed for commercial sales,governmental regulatory agencies must assess various consequences to human and animal health as well as address environmental concerns before granting approval. That process in the United States has been far more challenging than the science -only one genome-engineered food animal has been approved by the Food and Drug Administration (FDA)over the past three decades,growth-enhanced salmon(Carrollet al.2016). Indeed,even the far more enlightened United States Department of Agriculture’s Animal and Plant Health Inspection Service allows only a single edit per line in order to best mimic natural processes (USDA 2018).Hence,in an environment in which even single edits in animal genomes are problematic,acceptance of DKO pigs will remain a serious challenge regardless of how much the animals are needed (Hackett and Carroll 2015; Hackett 2020). The worldwide importance of introducing disease resistance genes into animals has never been more evident and it may be that the biggest effect of the paper by Xuet al.(2020) will be to unlock regulatory inertia in order to allow genome-edited animals onto farms throughout the world.
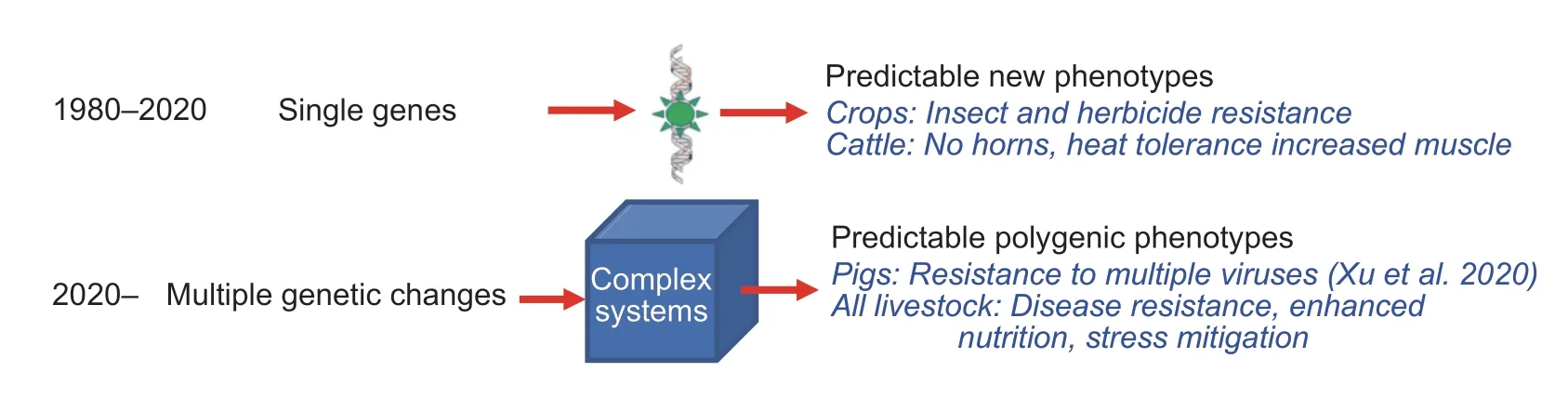
Fig.1 Genome editing allows introduction of multiple genotypic modifications to achieve complex phenotypes in livestock that older transgenic technologies could not support.
杂志排行
Journal of Integrative Agriculture的其它文章
- Crop photosynthetic response to light quality and light intensity
- Alginate oligosaccharides preparation,biological activities and their application in livestock and poultry
- Genome-wide pedigree analysis of elite rice Shuhui 527 reveals key regions for breeding
- TaSnRK2.4 is a vital regulator in control of thousand-kernel weight and response to abiotic stress in wheat
- Development and characterization of new allohexaploid resistant to web blotch in peanut
- Compact plant type rice has higher lodging and N resistance under machine transplanting
