Magnetotactic bacteria from the human gut microbiome associated with orientation and navigation regions of the brain*
2021-12-09RozalynSIMONPurnikaDamindiRANASINGHENawrozBARAZANJIMalinBergmanJUNGESTRJieXUOlgaBEDNARSKALenaSERRANDERMariaENGSTRDennisBAZYLINSKIsaKEITASusannaWALTER
Rozalyn A. SIMON , , Purnika Damindi RANASINGHE , Nawroz BARAZANJI ,Malin Bergman JUNGESTRÖM , Jie XU , Olga BEDNARSKA , Lena SERRANDER ,Maria ENGSTRÖM , Dennis A. BAZYLINSKI , Åsa V. KEITA , Susanna WALTER 1,
1 Center for Medical Image Science and Visualization (CMIV), Linköping University, Linköping 58183, Sweden
2 Department of Health, Medicine, and Caring Sciences, Linköping University, Linköping 58183, Sweden
3 Division of Infection and Inf lammation, Department of Biomedical and Clinical Sciences, Linköping University, Linköping 58183, Sweden
4 Institute of Global Food Security, School of Biological Sciences, Queen’s University Belfast, Belfast BT95DL, United Kingdom
5 School of Life Sciences, University of Nevada at Las Vegas, Las Vegas, Nevada 89154, USA
6 Division of Surgery, Orthopedics and Oncology, Department of Clinical and Experimental Medicine, Linköping University,Linköping 58183, Sweden
Abstract Magnetotactic bacteria (MTB), ubiquitous in soil and fresh and saltwater sources have been identif ied in the microbiome of humans and many animals. MTB endogenously produce magnetic nanocrystals enabling them to orient and navigate along geomagnetic f ields. Similar magnetite deposits have been found throughout the tissues of the human brain, including brain regions associated with orientation such as the cerebellum and hippocampus, the origins of which remain unknown. Speculation over the role and source of MTB in humans, as well as any association with the brain, remain unanswered. We performed a metagenomic analysis of the gut microbiome of 34 healthy females as well as grey matter volume analysis in magnetite-rich brain regions associated with orientation and navigation with the goal of identifying specif ic MTB that could be associated with brain structure in orientation and navigation regions. We identif ied seven MTB in the human gut microbiome: Magnetococcus marinus, Magnetospira sp. QH-2, Magnetospirillum magneticum, Magnetospirillum sp. ME-1, Magnetospirillum sp. XM-1, Magnetospirillum gryphiswaldense,and Desulfovibrio magneticus. Our preliminary results show signif icant negative associations between multiple MTB with bilateral f locculonodular lobes of the cerebellum and hippocampus (adjusted for total intracranial volume, uncorrected P<0.05). These f indings indicate that MTB in the gut are associated with grey matter volume in magnetite-rich brain regions related to orientation and navigation. These preliminary f indings support MTB as a potential biogenic source for brain magnetite in humans. Further studies will be necessary to validate and elucidate the relationship between these bacteria, magnetite concentrations, and brain function.
Keyword: magnetotactic bacteria; human microbiome; microbiota-gut-brain axis; magnetoreception
1 INTRODUCTION
Magnetotactic bacteria (MTB) are phylogenetically diverse, globally ubiquitous, and common in soil and both saline and freshwater sources (Lin et al., 2017).They have the ability to biomineralize intracellular membrane-bounded magnetic nanocrystals of magnetite (Fe3O4) or greigite (Fe3S4) (Bazylinski et al., 2007). These magnetic nanocrystals can act as a compass, allowing the bacteria to align themselves along the Earth’s geomagnetic f ield lines and more easily locate preferable regions of low oxygenation or redox conditions in aquatic environments (Bazylinski et al., 2013). Intriguingly, similar magnetite nanocrystals have also been reported in the human body (Gorobets et al., 2018), including throughout the brain (Kirschvink et al., 1992; Dunn et al., 1995;Schultheiss-Grassi et al., 1999; Brem et al., 2006;Maher et al., 2016; Gilder et al., 2018; Khan and Cohen, 2019), though their origin and role remain a topic of much debate and include navigation as well as the regulation of iron transport within cells(Gorobets and Gorobets, 2012; Gieré, 2016; Maher et al., 2016; Gorobets et al., 2017a; Natan and Vortman,2017; Gilder et al., 2018; Natan et al., 2020). Some have proposed that humans have the inherent ability to biomineralize these magnetic nanocrystals(Kirschvink et al., 1992; Medviediev et al., 2017)while others postulate that they are foreign in origin(Maher et al., 2016), either from air pollution or biogenic sources such as acquisition from magnetiteproducing MTB (Natan et al., 2020). As MTB are found in both saline and freshwater sources throughout the world, it is likely that humans are in regular contact with various MTB species. Recent developments in metagenomic and next-generation sequencing have conf irmed the prevalence of MTB in samples taken from human skin, mouth, and gut(Natan et al., 2020). Moreover, f indings in the brain indicate magnetite to be in higher concentrations in the prefrontal cortex, cerebellum, upper brain stem,hippocampus, and limbic regions (Dunn et al., 1995;Schultheiss-Grassi et al., 1999; Dobson, 2002; Gilder et al., 2018; Khan and Cohen, 2019). Evidence indicates that some forms of iron found in the brain are associated with aging, declining cognitive function, and reduced grey matter volume (Rodrigue et al., 2013; Daugherty et al., 2015; Modica et al.,2015; Daugherty and Raz, 2016; Salami et al., 2021).Accumulating research shows that magnetite in particular is associated with Alzheimer’s disease(Hautot et al., 2003; Collingwood and Dobson, 2006;Pankhurst et al., 2008; Plascencia-Villa et al., 2016).In addition, evidence seems to indicate that magnetite derived from air pollution may represent a more toxic form than that of biogenic sources (Maher, 2019).What potential biological role magnetite might have in brain function has also not been determined (Dunn et al., 1995; Schultheiss-Grassi et al., 1999; Brem et al., 2006), though recent evidence indicates that it may play a similar role in humans as in bacteria and other animals, providing humans the capability to sense magnetic f ields (Wang et al., 2019). As the origin and role of magnetite in the human brain remain elusive, here we aimed to identify specif ic species of MTB in the human gut and explore the potential that these MTB might be associated with magnetite-rich brain regions known to function in human orientation and navigation. Here we identify MTB found in stool samples from healthy individuals and explore potential associations with brain structure in brain regions associated with navigation and orientation, specif ically the cerebellum and hippocampus.
2 MATERIAL AND METHOD
2.1 Subject
Thirty-four healthy females (HC), mean age 33 years(range, 20-55 years), were recruited by advertisement.Exclusion criteria were established via interviews with participants to determine if they suff ered from any organic gastrointestinal disease, metabolic or neurological disorders, or severe psychiatric disease(e.g., schizophrenia, bi-polar disorder, psychosis,etc.), as well as the use of antibiotics, self-reported nicotine intake within the previous two months, and self-reported regular nonsteroidal anti-inf lammatory drugs (NSAID) use. Female study participants were chosen to avoid potential sex-related confounds as previous studies have noted diff erences in the brain associated with sex (Gupta et al., 2014, 2017). The regional ethical review board approved the study(2013/506-32 and 2014/264-32), and all subjects gave their written informed consent.
2.2 Microbiota
2.2.1 Microbiota metagenomic sequencing
Whole-genome next-generation sequencing was done by GATC Biotech (Konstanz, Germany), now Eurof ins Genomics (Ebersberg, Germany). After thawing each stool sample, they underwent cell wall lysis, DNA extraction and purif ication. DNA sequencing was performed using an Illumina Technology HiSeq 4000 (read mode 2×150 bp). The sequence reads were then inspected for base quality from the 3′ and 5′ ends for removal of low-quality reads. Trimmomatic (Bolger et al., 2014) was used to f ilter low-quality reads (leading: 18 trailing: 18 sliding window: 4: 22 minlen: 20). Then, f iltered reads were aligned to the GRCh37/hg19 genome using the BWA sequence aligner (Li and Durbin,2009). Bases with average Phred quality below 15 were considered of low quality, and only mate pairs(forward and reverse read) were used for the next analysis step.
After removing the host sequence reads, taxonomic prof iling of the non-host sequences was performed using the Kraken2 version 2.0.8-beta (Wood and Salzberg, 2014) and Minikraken2 v1 8GB database including RefSeq bacteria, archaea, and viral libraries(available on April 23, 2019). Taxonomic classif ication performed with Kraken2 breaks the sequences into overlappingk-mers and is based on a 31-mer exact alignment. Thek-mers are mapped to their lowest common ancestor of the genomes that contains thatk-mer in a precomputed reference database. For each read, a classif ication tree is found by pruning the taxonomy and only retaining taxa (including ancestors) associated withk-mers in that read. Each node is weighted by the number ofk-mers mapped to the node, and the path from root to leaf with the highest sum of weights is used to classify the read.Kraken2 includes a default library, based on completed microbial genomes in the National Center for Biotechnology Information’s (NCBI) RefSeq database. Abundance measured by the count of taxa assigned reads from various taxonomic levels. Read counts of input samples observed at various taxa levels (phylum, genus, and species) were normalized by using the centered log(2)-ratio (CLR)transformation (Aitchison, 1982) implemented in the R package mixOmics (Lê Cao et al., 2016).Transforming compositional data using log ratios helps to reduce the spurious correlations and allow sub-compositionally coherent comparisons between samples and study groups (Gloor et al., 2016; Calle,2019).
2.2.2 Contig-based taxonomy analysis
Reads were assembled into contigs using the de novo assembler Megahit (version 1.2.9) program (Li et al., 2015). Protein coding sequences were predicted using the eggnog-mapper (Huerta-Cepas et al., 2017)(using the latest version of the PFAM database).Using DIAMOND ‘blastP’ mode (Buchf ink et al.,2015) the protein sequences were compared against the NCBI’s non-redundant (nr) and UniRef90 databases to identify the protein accession, description and taxonomic identif ier of the best matching sequence.
2.3 Magnetic resonance imaging (MRI)
All MR images were acquired using a 32-channel head coil on a 3T Philips Ingenia MRI scanner (Philips Healthcare, Best, the Netherlands) at the Center for Medical Image Science and Visualization at Linköping University, Sweden. T1-weighted 3D FFE images were acquired in all participants using the following parameters: inversion preparation and delay 900 ms,SAG-plane, f ield of view (FOV) 256 mm×240 mm×170 mm, resolution 1 mm×1 mm×1 mm, f lip angle 9°, repetition time (TR)=7 ms, time to echo (TE)=3.2 ms, acquisition time (TA)=5.34 min. The T1-weighted images acquired for all participants were examined to ensure that they were free from any obvious pathologic abnormalities.
2.4 Regional grey matter probability
Grey matter volume (GMV) measurement was performed on the T1 weighted images using CAT12 toolbox (CAT, http://dbm.neuro.uni-jena.de/vbm/) in statistical parametric mapping (SPM) running on MATLAB (R2017a, MathWorks Natick,Massachusetts, USA) (Ashburner and Friston, 2000).Each T1 image was reoriented so that all the images would have the anterior commissure as the point of origin. Prior to segmentation, a non-linear deformation f ield for each image was estimated. Using a tissue probability map, each image was segmented into grey matter, white matter, and cerebrospinal f luid and spatially normalized into MNI space. For betweensubject registration and grey matter modulation, the diff eomorphic anatomical registration using exponentiated lie algebra (DARTEL) toolbox was used (Ashburner, 2007). Final modulation of the voxel values was done according to the Jacobian determinant of the deformation f ield initially estimated. Using the same automated anatomical labeling (aal) atlas ROIs as in the fMRI analysis, grey matter probability was extracted using the MANGO image processing system (Research Imaging Center,UTHSCSA; http://ric.uthscsa.edu/mango). Using the region of interest (ROI) statistics in MANGO we extracted the grey matter probability for each individual in each ROI. ROIs comprised bilateral anterior, posterior, and f locculonodular lobes of the cerebellum, as well as bilateral hippocampi (Fig.1).For negative control regions we chose the bilateral insula and inferior frontal gyrus or Broca’s region, as neither are commonly cited in association with navigation or orientation.
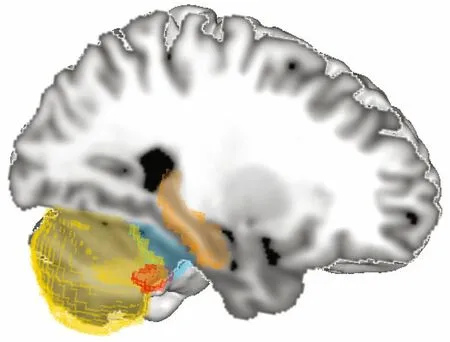
Fig.1 Sagittal view of automated anatomical labeling (aal)atlas regions of interest used in grey matter analysis
2.5 Statistical analysis
As the Shapiro-Wilk test showed non-normality in many of the measures, nonparametric tests were used for all analyses. Nonparametric partial correlation analyses were performed between normalized proportions of MTB species and grey matter volume(GMV) values, while controlling for total intracranial volume (TIV) in IBM SPSS Statistics (Windows v26.0, IBM Corp., Armonk, NY). For each ROI region independently, correction for multiple correlations over the seven tests conducted with MTB species was done using the Holm-Sidak method and reported as adjustedPvalues (P<0.05) in GraphPad Prism (Windows v8.0.0, GraphPad Software, San Diego, California, USA). Additional correlations were conducted to check for interactions between MTB with age and TIV. As this was a preliminary exploratory study, values uncorrected for multiple correlation were additionally reported.
3 RESULT
3.1 Microbiota
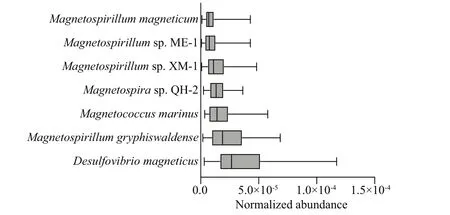
Fig.2 Normalized abundance of MTB species
Using a metagenomic, next-generation sequencing approach we examined the stool samples of 34 healthy females. A total of 3 983 species were detected, of which seven were identif ied as MTB:Magnetococcus marinus,Magnetospirasp.QH-2,Magnetospirillum magneticum,Magnetospirillumsp.ME-1,Magnetospirillumsp.XM-1,Magnetospirillumgryphiswaldense, andDesulfovibrio magneticus. All seven MTB species were found in every participant (Fig.2) and all species were of the phylum Proteobacteria, class Alphaproteobacteria, with the exception ofDesulfovibriomagneticusfrom class Deltaproteobacteria. The majority of these species’typical habitats are shallow freshwater and sediment,whereasMagnetococcus marinusandMagnetospirasp.QH-2 are found in marine waters and sediment.
3.2 Associations with GMV
Partial correlation analysis of each species of MTB with bilateral subregions of the cerebellum and the hippocampus found no signif icant associations with bilateral anterior or posterior regions of the cerebellum but found multiple negative associations with the f locculonodular lobe of the cerebellum (Table 1;P<0.05 uncorrected, adjusted for TIV). Additional associations were found bilaterally in the hippocampus withMagnetospirasp.QH-2 andMagnetospirillumsp.XM-1. After correction for multiple comparisons,Desulfovibrio magneticusshowed signif icant negative associations with bilateral f locculonodular lobes left(ρ=-0.46, adjustedP=0.048) and right lobe (ρ=-0.47,adjustedP=0.047) andMagnetospirasp.QH-2 showed signif icant negative associations with f locculonodular lobe right (ρ=-0.54, adjustedP=0.008) and the left hippocampus (ρ=-0.46, adjustedP=0.048) (Fig.3). No associations were found betweenMagnetococcus marinusand any of the brain regions. Nor were any associations found between MTB with either age or TIV (data not shown). No correlations were found between MTB and negative control regions of the bilateral insula inferior frontalgyrus or Broca’s region—regions not commonly associated with orientation or navigation (data not shown).
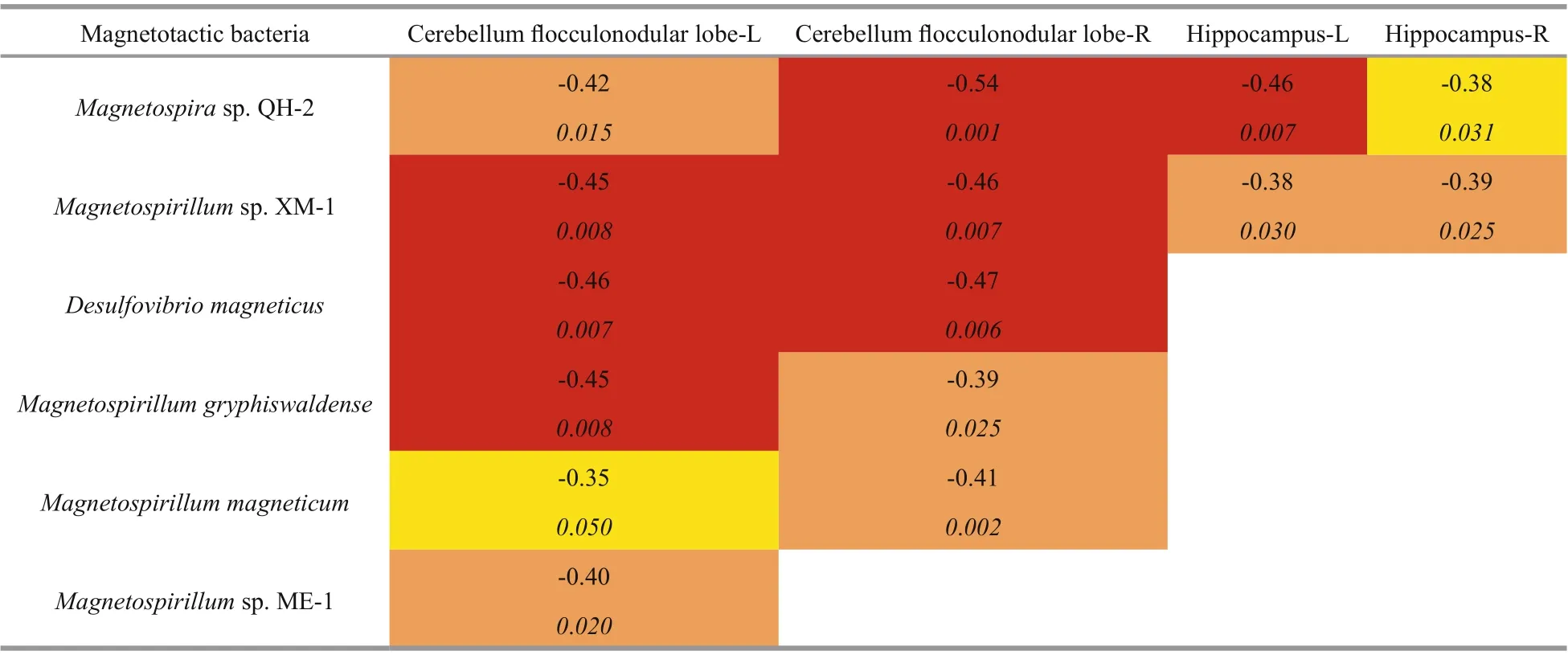
Table 1 Signif icant nonparametric partial correlations between MTB species and regional grey matter brain volume ( P<0.05 uncorrected, adjusted for TIV)
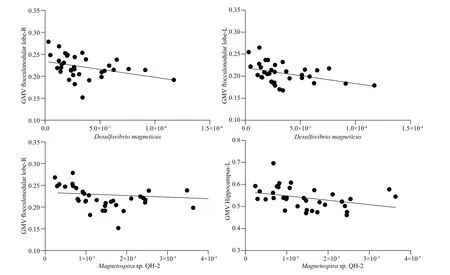
Fig.3 Signif icant nonparametric bivariate correlations between MTB species and regional grey matter brain volumes after correction for multiple comparisons, plotted here unadjusted for TIV (corrected P<0.05)
4 DISCUSSION
As the origin and role of magnetite in the human brain remain elusive and a matter of much debate, we f irst aimed to identify which species of magnetotactic bacteria could be found in the human gut, and then explored the potential that MTB might be associated with magnetite-rich brain regions known to function in human orientation and navigation. From the stool samples of healthy females, we identif ied seven MTB species in low abundance. Further protein blasts conf irmed the presence of MTB by identifying additional associated proteins, but none associated with the magnetosome, likely as a result of the low abundance which greatly limited the representation of the MTB genomes using this method. Our preliminary results indicate that six of the seven MTB found in the human gut have some degree of signif icant association with the regions of the hippocampus and the vestibulof locculonodular lobe of the cerebellum. These associations were strongest amongst MTBMagnetospirasp.QH-2 andDesulfovibrio magneticus. The hippocampus has long been identif ied as a region involved in orientation and navigation, but the specif ic contribution of the vestibulo-f locculonodular lobe of the cerebellum is less commonly cited in association with these tasks(Khan and Chang, 2013; Rochefort et al., 2013;Haines and Mihailoff , 2018). The vestibulof locculonodular lobe is thought to be responsible for computation of the “head-to-world” frame of reference and is involved in the vestibulo-ocular ref lex in orientation of the body (Yakusheva et al.,2013; Laurens and Angelaki, 2016). Notably, in the study conducted by Gilder et al. (2018), the cerebellum was also reported to be one of the regions of highest magnetite concentration. The negative correlation between grey matter volumes in magnetite-rich regions of the brain with MTB proportions in the gut opens the door to further investigation into the direct relationship between these specif ic gut MTB and brain magnetite. If indeed MTB were the source of brain magnetite, then our f indings of a negative association would be in line with the numerous reports of high brain iron associated with reduced brain volumes in disease states (Rodrigue et al., 2013;Daugherty et al., 2015; Daugherty and Raz, 2016),but to our knowledge, the association between grey matter volume and magnetite load specif ically, has not been investigated in healthy individuals. Although brain iron load has been shown to increase with age,in this study we f ind no relationship between age and MTB proportions in healthy females. This f inding aligns with previous assessments of brain magnetite by Gilder et al. (2018) who found no relationship between age and magnetite in the whole brain, and Dobson et al. (2002) who found no age associations in the hippocampus of females, but did in males.
As MTB have been found in many diff ering environments throughout the world, in both freshwater and saline sources, as well as in soil and sediments,human contact with and ingestion of MTB are likely to be quite common; though it is important to note that the metagenomic evidence presented here does not determine viability of the bacteria in the gut. Yet,as MTB are often found in the transition zone between aerobic and anaerobic environments, show varying tolerance to oxygen and pH, with the ability to use their magnetosomes to maintain optimal growth and survival in the oxic-anoxic transition zone, it seems possible that the MTB identif ied, or closely related,could potentially survive in, or adapt to, the widerange of pH and aerobic-anaerobic microenvironments found within the digestive tract (Bazylinski and Lefèvre, 2013; Andrade et al., 2020). If MTB are viable in the gut, potential paths to the brain might include the passage of live bacteria through the gut(Bednarska et al., 2017) or any of the numerous mechanisms of blood brain barrier disruption known by bacteria (Al-Obaidi and Desa, 2018). And although rare, there are indeed reports of Alphaproteobacteria found within the brains of human cadavers (Branton et al., 2013; Roberts et al., 2018). Though metagenomic analysis of the gut microbiome as conducted here,cannot conf irm the viability of MTB, the potential remains that simply the ingestion of nonviable MTB still allows them to be a passive biogenic source of magnetite found throughout the human body, with numerous potential functions beyond the hypothesized navigation and regulation of iron transport (Gorobets et al., 2017b; Wang et al., 2019; Natan et al., 2020).
Further lines of inquiry include the isolation and culture of the specif ic MTB identif ied by this metagenomic sequencing, assessment of iron load and association with MTB proportions, and detailed comparison of magnetite crystals isolated from these MTB with those derived from human brain tissues.Future studies would also benef it from a larger number of participants of both sex, as well as including information concerning female menstrual cycle and menopause, as these could potentially inf luence the gut microbiome (Santos-Marcos et al., 2018; Shin et al., 2019; Zhao et al., 2019).
In conclusion, our f indings conf irm that magnetotactic bacteria are a part of the human gut microbiome. In addition, the preliminary associations detected between gut MTB and brain regions which function in orientation provide motivation for future investigations to conf irm potential relationships between MTB and magnetite found in the human body.
5 DATA AVAILABILITY STATEMENT
The datasets generated during and/or analyzed during this study are available from the corresponding author on reasonable request.
6 ACKNOWLEDGMENT
We wish to thank Viviana Morrillo for her contribution ofMagnetococcusDNA, Mike P. Jones,Mats Fredrikson, and Nikki Hawkins for statistical guidance, and Adriane Icenhour for feedback during early editing.
7 AUTHOR CONTRIBUTION
Conceptualization: RAS, ME, ÅK, and SW.Methodology: RAS, PR, MBJ, JX, OB, NB, ÅK, and SW. Validation: PR, MBJ, JX, and LS. Formal analysis: RAS, PR, NB, and MBJ. Investigation:RAS, PR, NB, MBJ, and JX. Data curation: OB and NB. Writing (original draft preparation): RAS.Writing (review and editing): RAS, ME, DAB, ÅK,and SW. Visualization: RAS. Funding acquisition:DAB, ÅK, and SW.
杂志排行
Journal of Oceanology and Limnology的其它文章
- MamZ protein plays an essential role in magnetosome maturation process of Magnetospirillum gryphiswaldense MSR-1*
- Observations on a magnetotactic bacteria-grazing ciliate in sediment from the intertidal zone of Huiquan Bay, China*
- How light aff ect the magnetotactic behavior and reproduction of ellipsoidal multicellular magnetoglobules?*
- Biocompatibility of marine magnetotactic ovoid strain MO-1 for in vivo application*
- Determination of the heating effi ciency of magnetotactic bacteria in alternating magnetic f ield*
- An approach to determine coeffi cients of logarithmic velocity vertical prof ile in the bottom boundary layer*
