Diabetic kidney disease:Are the reported associations with singlenucleotide polymorphisms disease-specific?
2021-11-25MarekSaracynBartomiejKisielMariaFranaszczykDorotaBrodowskaKaniaWawrzyniecmudzkiRobertMaeckiLonginNiemczykPrzemysawDyrlaGrzegorzKamiskiRafaoskiStanisawNiemczyk
Marek Saracyn, Bartłomiej Kisiel, Maria Franaszczyk, Dorota Brodowska-Kania, Wawrzyniec Żmudzki, Robert Małecki, Longin Niemczyk, Przemysław Dyrla, Grzegorz Kamiński, Rafał Płoski, Stanisław Niemczyk
Marek Saracyn, Dorota Brodowska-Kania, Wawrzyniec Żmudzki, Stanisław Niemczyk, Department of Internal Diseases, Nephrology and Dialysis, Military Institute of Medicine, Warsaw 04-141,Poland
Marek Saracyn, Dorota Brodowska-Kania, Grzegorz Kamiński, Department of Endocrinology and Isotope Therapy, Military Institute of Medicine, Warsaw 04-141, Poland
Bartłomiej Kisiel, Clinical Research Support Center, Military Institute of Medicine, Warsaw 04-141, Poland
Maria Franaszczyk, Department of Medical Biology, Molecular Biology Laboratory, Institute of Cardiology, Warsaw 04-628, Poland
Robert Małecki, Department of Nephrology, Międzyleski Specialist Hospital in Warsaw,Warsaw 04-749, Poland
Longin Niemczyk, Department of Nephrology, Dialysis and Internal Diseases, Warsaw Medical University, Warsaw 02-097, Poland
Przemysław Dyrla, Department of Gastroenterology, Military Institute of Medicine, Warsaw 04-141, Poland
Rafał Płoski, Department of Medical Genetics, Medical University of Warsaw, Warsaw 02-106,Poland
Abstract BACKGROUND The genetic backgrounds of diabetic kidney disease (DKD) and end-stage kidney disease (ESKD) have not been fully elucidated.AIM To examine the individual and cumulative effects of single-nucleotide polymorphisms (SNPs) previously associated with DKD on the risk for ESKD of diabetic etiology and to determine if any associations observed were specific for DKD.METHODS Fourteen SNPs were genotyped in hemodialyzed 136 patients with diabetic ESKD(DKD group) and 121 patients with non-diabetic ESKD (NDKD group). Patients were also re-classified on the basis of the primary cause of chronic kidney disease(CKD). The distribution of alleles was compared between diabetic and nondiabetic groups as well as between different sub-phenotypes. The weighted multilocus genetic risk score (GRS) was calculated to estimate the cumulative risk conferred by all SNPs. The GRS distribution was then compared between the DKD and NDKD groups as well as in the groups according to the primary cause of CKD.RESULTS One SNP (rs841853; SLC2A1) showed a nominal association with DKD (P = 0.048;P > 0.05 after Bonferroni correction). The GRS was higher in the DKD group (0.615± 0.260) than in the NDKD group (0.590 ± 0.253), but the difference was not significant (P = 0.46). The analysis of associations between GRS and individual factors did not show any significant correlation. However, the GRS was significantly higher in patients with glomerular disease than in those with tubulointerstitial disease (P = 0.014) and in those with a combined group (tubulointerstitial,vascular, and cystic and congenital disease) (P = 0.018).CONCLUSION Our results suggest that selected SNPs that were previously associated with DKD may not be specific for DKD and may confer risk for CKD of different etiology,particularly those affecting renal glomeruli.
Key Words: Diabetic kidney disease; Chronic kidney disease; End-stage kidney disease;Single-nucleotide polymorphism; Diabetes mellitus
INTRODUCTION
A diabetes mellitus (DM) epidemic is underway; in 2019, approximately 463 million people worldwide were estimated to have DM, and half of those cases were considered undiagnosed. By 2045, the prevalence is expected to increase to 700 million cases[1-3]. In Poland, around 3.5 million people (9.1% of the total population) suffer from DM[4]. Roughly 30% of patients with type 1 DM (DM1) and 40% with type 2 DM(DM2) develop a serious complication of the small renal vessels:diabetic kidney disease (DKD)[5,6]. The occurrence of DKD considerably worsens the long-term prognosis - the risk of premature death in end-stage kidney disease (ESKD) patients requiring renal replacement therapy (RRT) is 18-fold greater than in the general population[7]. Understandably, DKD is the strongest single predictor of death in a diabetic patient[8]. It is estimated that the number of DKD-associated deaths in DM patients increased by 94% from 1990 to 2012[9]. The recognized risk factors for DKD are hyperglycemia, acute kidney injury, hypertension, and obesity; however, age,gender, race, and family history are also influential factors[10].
Both clinical and epidemiological studies on DKD show a familial association of the disease[11-13], suggesting that heredity plays an important role in its development.Investigators aiming to elucidate the genetic basis of DKD have used two different approaches:candidate gene studies (CGS) and genome-wide association studies(GWAS). During the last two decades, CGS have tested more than 150 loci for their potential relationship with the renal complications of DM[14-16] and have reported an association between several single-nucleotide polymorphisms (SNPs) and albuminuria, glomerular filtration rate (GFR), DKD, and ESKD in both DM1 and DM2 patients.The majority of the proteins encoded by these genes are components of nephron(glomerulus, epithelium, and ion channels), but some are related to the extracellular matrix, immune response, phagocytosis, and cell migration[17]. During the last decade, GWAS have replaced CGS as the study method of choice. The results of GWAS have confirmed some of the CGS findings and have also revealed new associations[18,19]. These initial findings must be corroborated in replication studies in different populations.
With the increasing number of identified genetic risk factors for multifactorial diseases, tools that assess the cumulative impact of these factors on disease risk have been developed. One such tool is the genetic risk score (GRS), which our group has successfully applied in earlier studies[20,21]. GRS is particularly useful in situations wherein the individual effect carried by a single genetic variant is small. Previous studies have shown the capability of GRS to measure the genetic risk of various multifactorial diseases, including myocardial infarction, atrial fibrillation, stroke,rheumatoid arthritis, and psoriasis[22]. Thus, several studies have used this approach to evaluate the risk of renal complications in DM[23-25].
Previous analyses have searched for associations between genetic markers and the risk of diabetic renal complications but may have overlooked a critical issue:whether these genetic markers are specific for DKD or are, rather, associated with the risk for chronic kidney disease (CKD), regardless of its explicit underlying cause.
The aim of our study was to examine the individual and cumulative effects of SNPs previously described in DM2 patients with DKD on the risk for ESKD of diabetic etiology in a population of hemodialyzed (HD) patients and to determine if any associations observed were specific for DKD.
MATERIALS AND METHODS
This study was approved by the Military Institute of Medicine Ethics Committee,Warsaw, Poland. Informed consent was obtained from each patient. All procedures were performed in accordance with the Helsinki Declaration of 1975, revised in 1983.
Patients and controls
From an initial group of 1246 ESKD/HD patients, 136 consecutive patients with DM2 and DKD as the primary cause of ESKD were included in this study; they formed the DKD group. The control group was composed of 121 age-matched ESKD/HD patients with non-DKD (NDKD); this was the NDKD group. The inclusion criteria were as follows:(1) ESKD treated by hemodialysis; and (2) Age ≥ 18 years. The exclusion criterion was the presence of malignancy. The patients were recruited from the Military Institute of Medicine in Warsaw, Poland, and the Mazovian Centers of Dialysis in the following cities in Poland:Radom, Ciechanów, Grodzisk Mazowiecki,Maków Mazowiecki, Sokołów Podlaski, Skierniewice, Warszawa Międzylesie,Wołomin, and Otwock. Patients were classified as DKD or NDKD on the basis of histopathological examinations and diagnoses made by two independent nephrologists in each of the Mazovian Dialysis Centers. Nineteen DKD patients and 11 NDKD patients were excluded from the study because of poor DNA quality or incomplete genotyping data; the final DKD and NDKD groups consisted of 117 and 110 patients, respectively. Nineteen patients in the NDKD group were classified by nephrologists as having kidney disease of complex pathogenesis (CP group). A structured questionnaire was used to collect data regarding age, gender, smoking habits, body mass index (BMI), history of kidney disease in the pre-ESKD/HD period,presence and volume of diuresis, and coexistence of hypertension and current treatment. Then, the patients were re-classified on the basis of the 2012 Kidney Disease Improving Global Outcomes (KDIGO) guidelines and CKD classification system based on the primary cause of their kidney disease. These classifications included (1)glomerular disease (GD); (2) tubulointerstitial disease (TID); (3) vascular disease (VD);and (4) cystic and congenital disease (CCD). Patients with CP were considered separately[26].
SNP selection
SNPs previously associated with DKD in DM2 patients were selected for this study[27-38]. The inclusion criteria for SNP selection were as follows:(1) Association with DKD in the presence of DM2 confirmed in a GWAS, meta-analysis, or large-scale casecontrol study; (2) Odds ratio (OR) ≥ 1.2 (or ≤ 0.83); and (3) Minor allele frequency ≥ 0.1 in a Caucasian population (based on data from the HapMap CEU). The exclusion criteria were as follows:(1) Association limited to non-Caucasian populations; or (2)Lack of studies on Caucasian populations. On the basis of these criteria, 14 SNPs were selected for genotyping. However, five of the genotyped SNPs were excluded from the analysis because of a low genotyping success rate (< 80%) or Hardy-Weinberg equilibrium (HWE) deviation:rs9521445 (MYO16/IRS2), rs1801133 (MTHFR),rs2241766 (ADIPOQ), rs5186 (AGTR1), and rs4880 (SOD2). The complete list of the analyzed SNPs is shown in Supplementary Table 1.
Genotyping
The salting out method was used to isolate DNA form whole blood samples[39]. For SNP genotyping, a custom array was designed (Taqman®OpenArray®Genotyping Plate, Custom Format 16 QuantStudio™ 12 K Flex, Life Technologies, Carlsbad, CA,United States), and genotyping was performed following the manufacturer’s protocols on a QuantStudio™ 12 K Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, United States).
Statistical analysis
The statistical methods used in this study were reviewed by Kisiel B of the Clinical Research Support Center, Military Institute of Medicine, Warsaw, Poland. The differences in allele distribution between cases and controls as well as HWE were evaluated using the PLINK v1.07 statistical software package[40]. The deviation from HWE was considered significant atP< 0.05. The rest of statistical analyses was undertaken with the use of Statistica 12 package (StatSoft Inc). A Bonferroni correction(Pvalue × number of tested SNPs) was used to adjust for multiple comparisons. To evaluate the cumulative risk of multiple loci a GRS (computed by summing the products of the number of risk alleles and the natural logarithm of the OR for each SNP) was calculated. For the calculation of GRS we used ORs from previous studies(Supplementary Table 1). The GRSs of the DKD and NDKD groups were compared using Student’st-test, whereas the GRSs of GD, VD, TID, CCD, and CP groups were compared using ANOVA. Pearson's test was used to assess the correlations between different parameters.
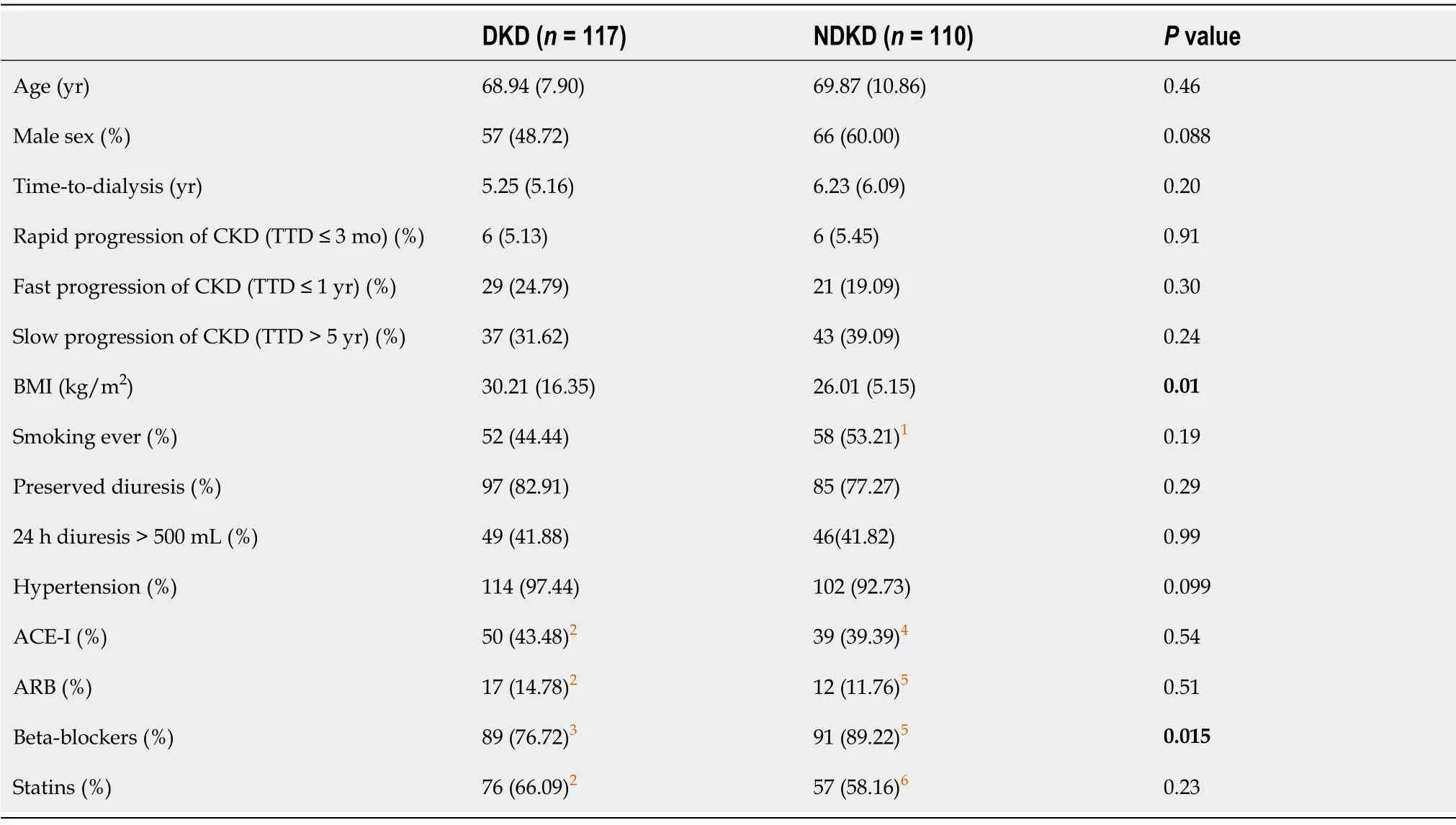
Table 1 Study and control group characteristics
RESULTS
The clinical and demographic data of the subjects are presented in Table 1. The DKD and NDKD groups differed significantly with respect to BMI (P= 0.01) and treatment with beta-blockers (P= 0.015).
SNP associations with DKD are detailed in Table 2. Only one SNP (rs841853)showed a nominal association with DKD (P= 0.048;P> 0.05 after Bonferroni correction).
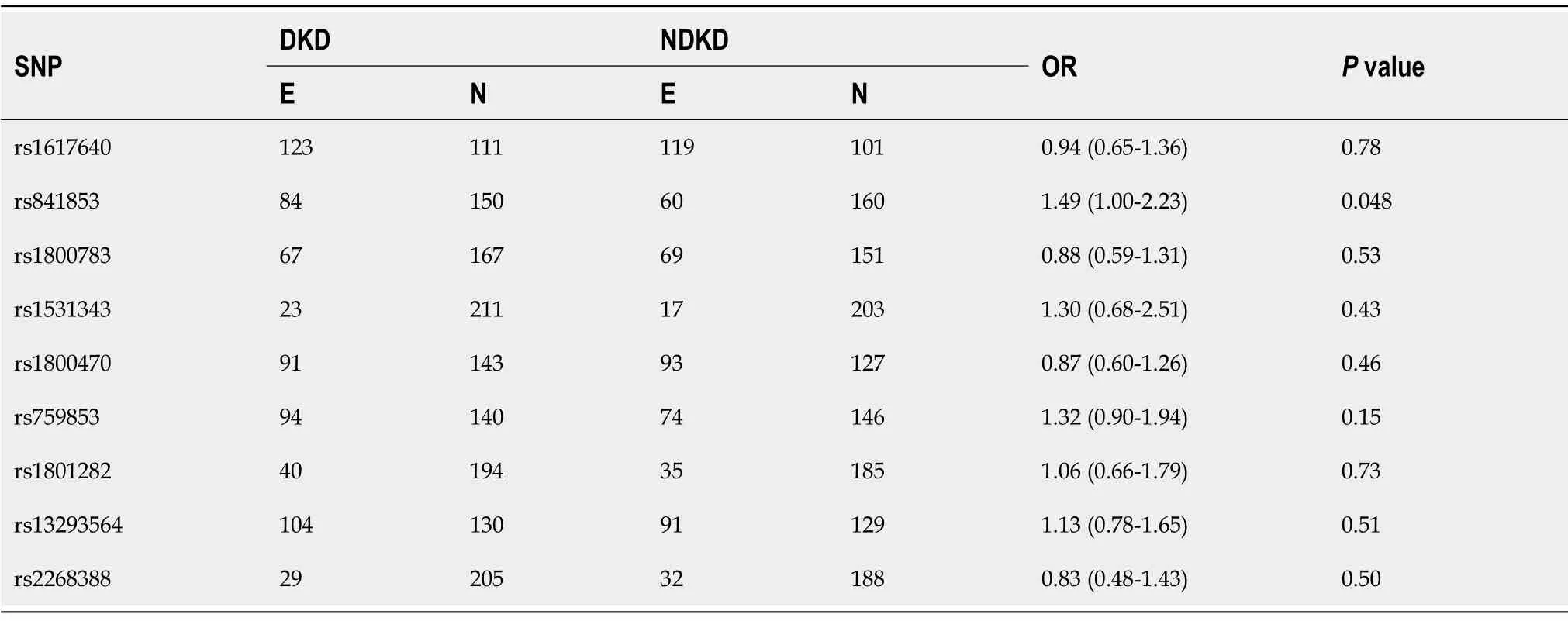
Table 2 Single-nucleotide polymorphisms’ associations with diabetic kidney disease
To evaluate the cumulative risk of multiple loci, we calculated GRS; it was slightly higher in the DKD group (0.615 ± 0.260) than in the NDKD group (0.590 ± 0.253), but the difference was not significant (P= 0.46). The GRS difference remained not significant (P= 0.34) after the exclusion of 19 CP patients from the NDKD group:0.615± 0.260 for the DKD group and 0.582 ± 0.244 for the NKD group. The analysis of associations between GRS and the individual factors of gender, diuresis, and rate of CKD progression in the DKD and NDKD groups showed a significant correlation of GRS with diuresis in the NDKD group (0.643 ± 0.22 for diuresis > 500 mLvs0.535 ± 0.253 for diuresis < 500 mL,P= 0.035, Table 3).
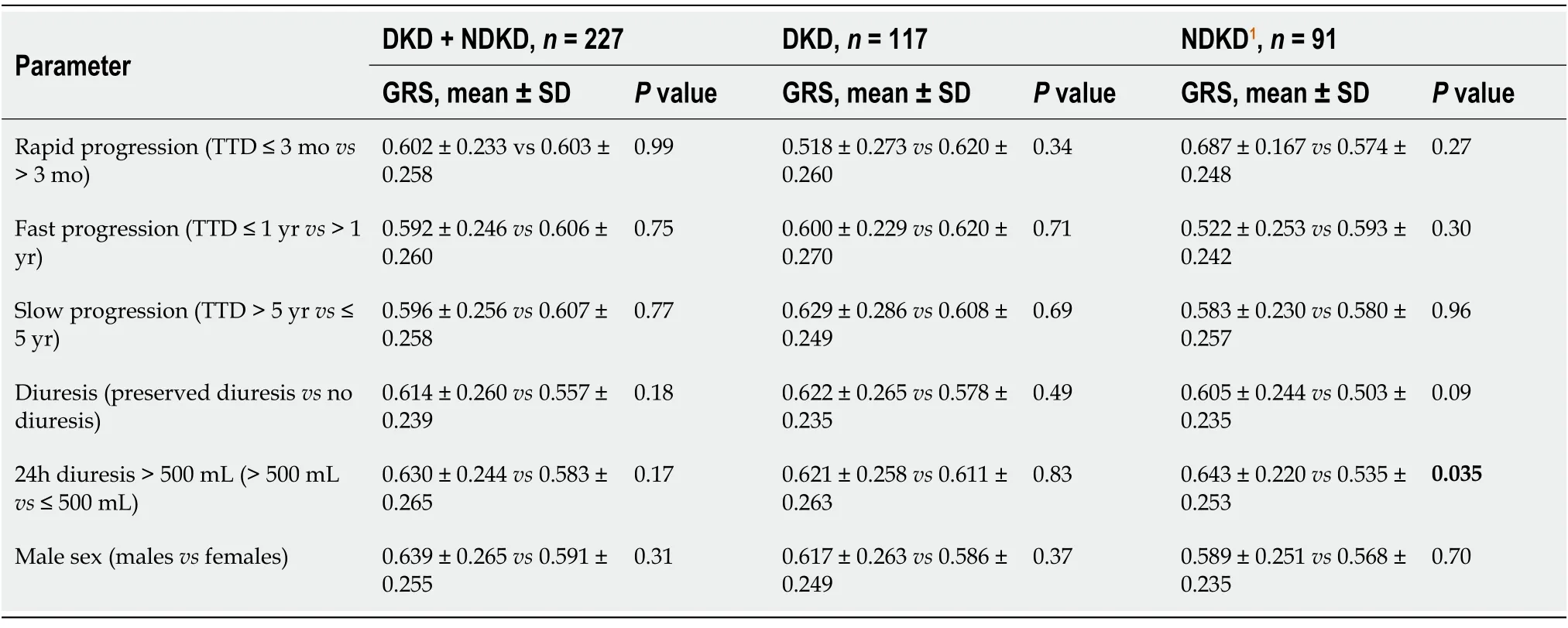
Table 3 Associations between genetic risk score and different parameters in diabetic kidney disease and non-diabetic kidney disease patients
We analyzed the distribution of GRS in the GD, TID, VD, CCD, and CP groups using ANOVA; the differences between groups were not significant (Table 4).However,post hocanalysis showed a significantly higher GRS in the GD group (0.628 ±0.256) than in the TID group (0.461 ± 0.218) (P= 0.014) as well as a higher GRS in the GD group (0.628 ± 0.256) than in the combined TID+VD+CCD group (0.536 ± 0.235) (P= 0.018). The analysis of associations between GRS and the individual factors of gender, diuresis, and rate of CKD progression did not show any significant correlation in the GD group (Supplementary Table 2).
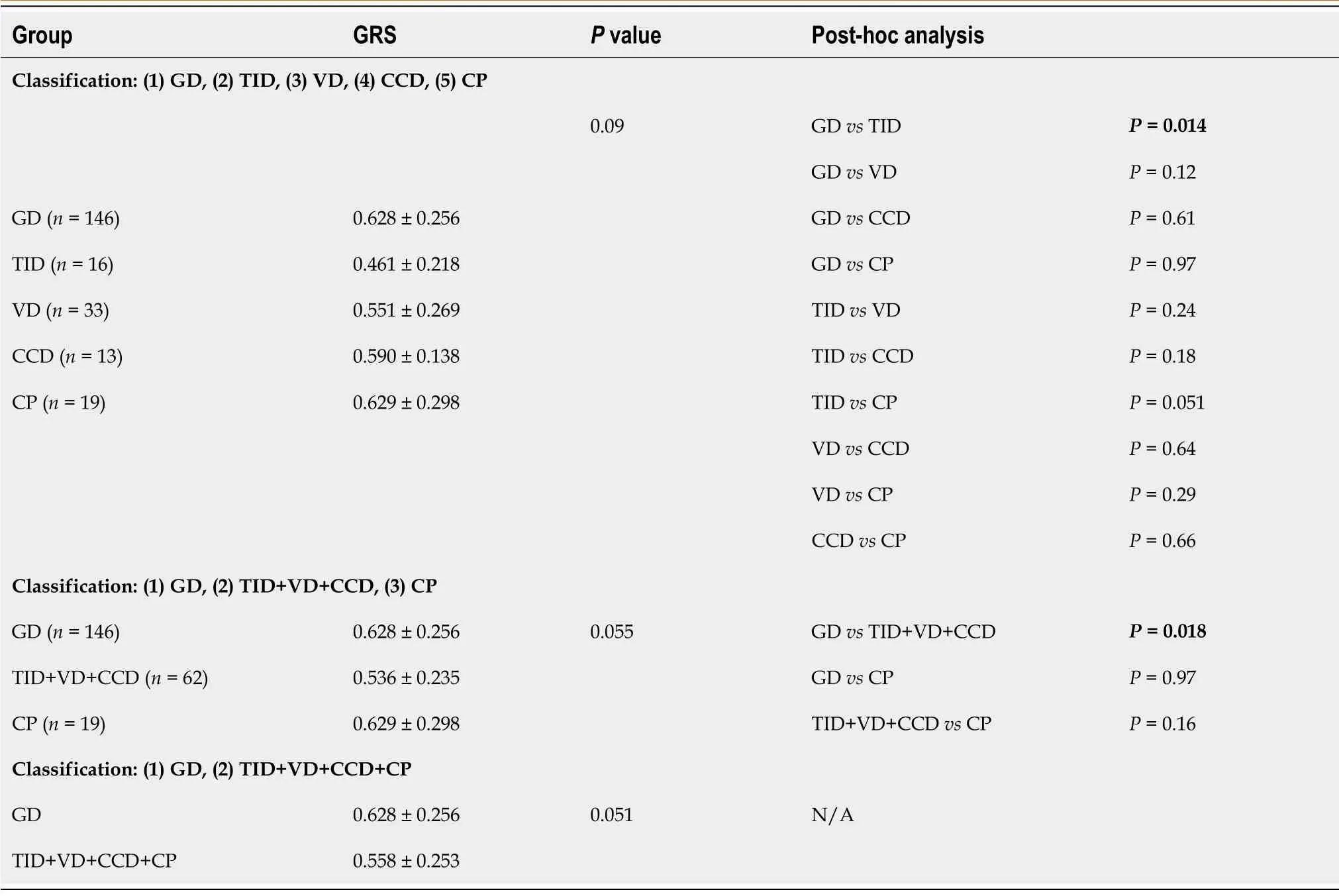
Table 4 The distribution of genetic risk score in particular end-stage kidney disease subgroups
DISCUSSION
To the best of our knowledge, this study may be the first to assess the genetic risk of DKD and other nephropathies leading to ESKD and RRT in a relatively large group of HD patients. Unexpectedly, none of the selected SNPs previously described in DM2 patients and associated with DKD development significantly correlated with DKD in our group of ESKD patients requiring hemodialysis, except one (rs841853) that showed a nominal association with DKD (P= 0.048;P> 0.05 after Bonferroni correction).Moreover, the GRS composed of all genotyped SNPs, carrying the cumulative risk of all tested loci, did not differ significantly between the DKD and NDKD groups and did not associate with any of the analyzed clinical parameters, including the rate of renal failure progression. This observation may have three explanations:(1) The analyzed SNPs are not associated with DKD in a Polish population; (2) The analyzed SNPs are associated with earlier stages of DKD; or (3) The analyzed SNPs are not specific for DKD but are associated with different kidney diseases and their progression.
It should be emphasized that many earlier studies on the genetic background of DKD have used DM patients without DKD as controls. This approach has identified risk factors for DKD development but has been unable to assess their specificities. That is, the genetic factors that have been pinpointed to be associated with DKD are not necessarily specific for DKD. To address this issue, we re-classified our patients on the basis of the KDIGO guidelines and found that GRS was significantly higher in the GD group than in the TID group and the combined TID+VD+CCD group. This observation suggests that these SNPs that were previously described as associated with DKD are in fact associated with the risk of CKD and ESKD of different etiologies,particularly those primarily affecting renal glomeruli.
In recent years, research has focused on the genetic basis of DKD. Nearly 160 genes have been implicated in DKD development as found in CDS and GWAS, and there appears to be a multigenic etiology of disease as gauged by GRS[14-19]. In a study involving 1100 DM2 patients, Wanget al[23] demonstrated that a GRS composed of six SNPs in genes involved in lipid metabolism (PON1, PON2, CETP), hemostasis (ITGA2), and inflammation (LTA1, LTA3) was associated with a significantly higher risk of DKD; risks were lower when SNPs were individually assessed. Toddet al[24]examined a GRS composed of 32 validated BMI loci to determine the relationship between BMI and macroalbuminuria, ESKD, and DKD and found that a 1 kg/m2higher BMI increased the risk of macroalbuminuria by 28%, of ESKD by 43%, and of DKD by 33%. Barbieuxet al[25] explored the association between 18 CKD-related SNPs and CKD G5 in 1,300 DM2 patients and 300 DM1 patients; however, neither a single SNP nor the 18-SNP GRS was linked to the deterioration of renal function, need for RRT, or death. Our study cannot be directly compared with the ones referenced above because it followed a completely different, exclusively DKD-related set of SNPs and a dissimilar study design in which non-diabetic patients were used as controls for diabetic ESKD patients.
The main finding of our study is that SNPs reported to be associated with DKD alone may, in fact, be correlated with CKD of different etiology. In this context, two papers are worth mentioning. A large-scale GWAS on 130000 subjects of European ancestry by Pattaroet al[41] showed that 53 known and novel SNP loci were highly correlated with GFR in individuals with or without diabetes; the effects on both patient groups were of similar magnitude. A study of even larger scale (including over 280000 subjects from the Million Veteran Program, over 765000 subjects from the CKD Gen Consortium, and more than 90000 diabetic patients) published by Hellwegeet al[42] made similar observations:32 SNP loci (17 known and 15 new) were significantly and with similar effect size associated with GFR in both diabetic and non-diabetic groups. Of course, reduced GFR is not synonymous with CKD (or DKD) and ESKD;however, the findings by Pattaroet al[41] and Hellwegeet al[42] raise the possibility that CKDs of different etiologies may, to some extent, share a common genetic basis.
To address this issue, we reviewed recent publications regarding the possible relationship of our SNPs with CKD/ESKD of etiology other than DKD[43-50]. Most reports, whether on small or large populations, testing single or many variants, did not include the exact SNPs selected for our study, although they may have involved the same genes. For example, Hellwegeet al[42] found an association between GFR andNOS3(nitric oxide synthase type 3) (rs3918226), but it was a different variation, which showed only a moderate linkage disequilibrium with ourNOS3SNP (rs1800783) (R2=0.17)[42]. Similarly, our SNPs were not any of those used in a GWAS by Wuttkeet al[43], the largest meta-analysis to date. Essential differences exist between our study and the other studies cited, and simple comparisons are not applicable. First, the aforementioned studies characterized associations between SNPs and GFR in the general population or in specific subpopulations such as diabetic patients, whereas our study aimed to delineate relationships between the selected SNPs and ESKD.Undoubtedly, changes in GFR are not synonymous with ESKD. In fact, a GWAS by Linet al[44] indicated that the majority of loci do not overlap between early stages of CKD and ESKD, suggesting that distinct genetic factors may dominate at different stages of CKD. Second, a GWAS detects only the strongest associations, as corrections for a number of analyses are routinely used, possibly neglecting more nuanced but still meaningful effects on outcome measures. Third, studies on the general population,which include patients with CKDs of different etiologies, may simply not pick up disease-specific associations due to a water-downed subject pool.
The association of the SNPs selected for our study with CKDs of different etiologies,and not only DKD, is supported by the fact that their corresponding loci carry information affecting the structure and function of nephrons or interstitial renal tissue,the disorders of which are not restricted to diabetic etiology. In fact, the most common causes of CKD, such as DKD, hypertensive nephropathy, atherosclerotic nephropathy,renal mass reduction, obstructive nephropathy, and glomerular diseases, share many common pathophysiological pathways[45]. The factor initiating kidney injury is clearly different, but the progression of renal damage may involve mutual mechanisms, such as the renin-angiotensin-aldosterone system; endothelial, podocyte,mesangial, and tubular cell activation; platelet activation; common cytokine activation pathways (transforming growth factor-β1); the spread of inflammation; and repair of damaged tissues (fibrosis). A shared pathophysiology is even more probable in diseases affecting the same region of the kidney - in this case, the renal glomeruli.
This study has several strengths. First, the study group included only patients with ESKD, which addresses the suggestion made by earlier research that different CKD stages may be driven by distinctive genetic factors. Second, non-diabetic ESKD patients comprised our control group; this allowed us to answer the question whether the SNPs previously associated with DKD are specific for that type of CKD; indeed,we demonstrated that the SNPs in question were not specific for ESKD of diabetic etiology.
We must also acknowledge the major limitation of this study:its size. The study group was relatively small for a genetic analysis. However, owing to our strict inclusion criteria, the initial population of approximately 1200 patients with ESKD requiring RRT from dialysis centers in Mazovia, Poland, which has a general population of about 5.5 million, was winnowed down to those patients with documented DKD as the primary cause of ESKD and an appropriate, non-DKD control group with ESKD of etiology other than diabetes. Due to relatively small study group our results need to be confirmed in studies on larger populations.
CONCLUSION
Our study may be the first to demonstrate that selected SNPs that were previously associated with DKD may not be specific for DKD and may confer genetic risk for CKDs of different etiologies, particularly those affecting renal glomeruli. Our results need to be confirmed in studies on larger populations.
ARTICLE HIGHLIGHTS
Research background
A diabetes mellitus (DM) epidemic is underway; 40% patients with type 2 DM (DM2)develop a serious complication, diabetic kidney disease (DKD), and the occurrence of DKD considerably worsens the long-term prognosis. However, the genetic backgrounds of DKD and end-stage kidney disease (ESKD) have not been fully elucidated.
Research motivation
Previous studies have searched for associations between genetic markers and the risk of diabetic renal complications but may have overlooked a critical issue:whether these genetic markers are specific for DKD or are, rather, associated with the risk for chronic kidney disease and ESKD itself, regardless of its explicit underlying cause.
Research objectives
The aim of our study was to examine the individual and cumulative effects (as a genetic risk score, GRS) of SNPs previously described in DM2 patients with DKD on the risk of diabetic ESKD in a population of hemodialyzed patients and to determine if any associations observed were specific for DKD.
Research methods
Fourteen SNPs were genotyped in 136 patients with diabetic ESKD (DKD group) and 121 patients with non-diabetic ESKD (NDKD group). Patients were also classified on the basis of the KDIGO guidelines and CKD classification system based on the primary cause of CKD, such as glomerular disease (GD), tubulointerstitial disease(TID), vascular disease (VD), and cystic and congenital disease (CCD). Patients with complex pathogenesis (CP) were considered separately. The distribution of alleles was compared between diabetic and non-diabetic groups as well as between different subphenotypes. The weighted multilocus GRS was calculated to estimate the cumulative risk conferred by all SNPs. The distribution of GRS was then compared between the DKD and NDKD groups as well as in the groups according to KDIGO guidelines.
Research results
One SNP (rs841853; SLC2A1) showed a nominal association with DKD (P= 0.048;P>0.05 after Bonferroni correction). The GRS was higher in the DKD group (0.615 ± 0.260)than in the NDKD group (0.590 ± 0.253), but the difference was not significant (P=0.46). The analysis of associations between GRS and the individual factors of gender,diuresis, and rate of CKD progression in either study group did not show any significant correlation. However, the GRS was significantly higher in the GD group than in the TID group (P= 0.014) and the combined TID+VD+CCD group (P= 0.018).
Research conclusions
Our study may be the first to demonstrate that selected SNPs that were previously associated with DKD may not be specific for DKD and may confer genetic risk for CKDs of different etiologies, particularly those affecting renal glomeruli.
Research perspectives
Further studies are needed to confirm a similar correlation of other SNPs previously associated with DKD with the development and etiology of ESKD.
ACKNOWLEDGEMENTS
We thank all medical staff members and technicians of dialysis centers who agreed to participate in this study.
杂志排行
World Journal of Diabetes的其它文章
- Non-alcoholic fatty liver disease, diabetes medications and blood pressure
- Diabetes patients with comorbidities had unfavorable outcomes following COVID-19:A retrospective study
- Utility of oral glucose tolerance test in predicting type 2 diabetes following gestational diabetes:Towards personalized care
- Metabolic and inflammatory functions of cannabinoid receptor type 1 are differentially modulated by adiponectin
- Medication adherence and quality of life among type-2 diabetes mellitus patients in India
- Galectin-3 possible involvement in antipsychotic-induced metabolic changes of schizophrenia:A minireview
