Comprehensive genetic screening reveals wide spectrum of genetic variants in monogenic forms of diabetes among Pakistani population
2021-11-23IbrarRafiqueAsifMirShajeeSiddiquiMuhammadArifNadeemSaqibAsherFawwadLucMarchandMuhammadAdnanMuhammadNaeemAbdulBasitConstantinPolychronakos
Ibrar Rafique,Asif Mir,Shajee Siddiqui,Muhammad Arif Nadeem Saqib,Asher Fawwad,Luc Marchand,Muhammad Adnan,Muhammad Naeem,Abdul Basit,Constantin Polychronakos
Ibrar Rafique,Asif Mir,Department of Biological Sciences,International Islamic University,Islamabad 44000,Pakistan
Ibrar Rafique,Luc Marchand,Constantin Polychronakos,Departments of Pediatrics and Human Genetics,McGill University Health Centre Research Institute,Montreal H4A 3J1,Canada
Ibrar Rafique,Research Development and Coordination,Pakistan Health Research Council,Islamabad 44000,Pakistan
Shajee Siddiqui,Department of Medicine,Pakistan Institute of Medical Sciences,Islamabad 44000,Pakistan,Pakistan
Muhammad Arif Nadeem Saqib,Department of Medical Laboratory Technology,National Skills University,Islamabad,44000,Pakistan
Asher Fawwad,Department of Biochemistry,Baqai Institute of Diabetology and Endocrinology,Baqai Medical University,Karachi 74600,Sindh,Pakistan
Muhammad Adnan,PHRC Research Centre,FJMU,Pakistan Health Research Council,Lahore 54000,Pakistan
Muhammad Naeem,Department of Biotechnology,Quaid-I-Azam University,Islamabad 44000,Pakistan
Abdul Basit,Department of Medicine,Baqai Institute of Diabetology and Endocrinology,Baqai Medical University,Karachi 74600,Sindh,Pakistan
Abstract BACKGROUND Monogenic forms of diabetes(MFD)are single gene disorders.Their diagnosis is challenging,and symptoms overlap with type 1 and type 2 diabetes.AIM To identify the genetic variants responsible for MFD in the Pakistani population and their frequencies.METHODS A total of 184 patients suspected of having MFD were enrolled.The inclusion criterion was diabetes with onset below 25 years of age.Brief demographic and clinical information were taken from the participants.The maturity-onset diabetes of the young(MODY)probability score was calculated,and glutamate decarboxylase ELISA was performed.Antibody negative patients and features resembling MODY were selected(n= 28)for exome sequencing to identify the pathogenic variants.RESULTSA total of eight missense novel or very low-frequency variants were identified in 7 patients.Three variants were found in genes for MODY,i.e.HNF1A(c.169C>A,p.Leu57Met),KLF11(c.401G>C,p.Gly134Ala),and HNF1B(c.1058C>T,p.Ser353Leu).Five variants were found in genes other than the 14 known MODY genes,i.e.RFX6(c.919G>A,p.Glu307Lys),WFS1(c.478G>A,p.Glu160Lys)and WFS1(c.517G>A,p.Glu173Lys),RFX6(c.1212T>A,p.His404Gln)and ZBTB20(c.1049G>A,p.Arg350His).CONCLUSION The study showed wide spectrum of genetic variants potentially causing MFD in the Pakistani population.The MODY genes prevalent in European population(GCK,HNF1A,and HNF4a)were not found to be common in our population.Identification of novel variants will further help to understand the role of different genes causing the pathogenicity in MODY patient and their proper management and diagnosis.
Key Words:MODY;Diabetes;Genetics;Monogenic diabetes;Monogenic forms of diabetes
INTRODUCTION
Monogenic forms of diabetes(MFD)result from changes in single gene.Maturityonset diabetes of the young(MODY)is a monogenic form of diabetes.It is inherited in an autosomal dominant pattern[1]and is often misdiagnosed as type 1 or type 2 diabetes[2].It is estimated that MODY accounts for 1%-2% of all the diabetic cases[3].There are fourteen sub-types of MODY listed in OMIM(On-Line Mendelian Inheritance in Man)(#606391).Most common of them areGCK,HNF1A,andHNF4A.The other listed genes arePDX1,HNF1B,NEUROD1,KLF11,CEL,PAX4,INS,BLK,ABCC8,KCNJ11,andAPPL1[1].Mutations in a number of additional genes are also known to cause diabetes.
The subtypes of MODY differ with respect to hyperglycemia,age of disease onset,treatment pattern,and complications reported[4].Therefore timely diagnosis is of vital importance in MFD.Previously,most common genes,i.e.GCK,HNF1A,andHNF4Awere generally sequenced for suspected cases,but n ow with the advent of latest technologies,targeted panel sequencing or exome sequencing are standard[5,6].
Pakistan has a huge burden of diabetes.The recent surveys showed that one out of every four people in the general population is suffering from diabetes in the country[7,8].Being a resource poor country,advanced diagnostic facilities are not available to the public.The literature from Pakistan is scarce on MFD[9,10].Therefore,this study was designed to enroll suspected MFD patients and assess the causal variants involved.To our knowledge,this was the first comprehensive study to be conducted in Pakistan on MFD.
MATERIALS AND METHODS
A total of 184 patients with diabetes onset before 25 years of age were considered for participation in the study.The participants were chosen from the sources indicated below:
National Diabetes Survey(n = 80)
The record of patients who had an onset of disease before 25 years of age was obtained from National Diabetes Survey data.This was a population-based survey carried out all over the four provinces of Pakistan.The detailed methodology is described elsewhere[7].According to World Health Organization,the patients were diagnosed as diabetic if having fasting glucose level > 126 mg/dL(7.0 mmol/L)or HbA1c > 6.5%(48 mmol/mol).Clinical information in 34 cases suggested MFD[young onset,low body mass index(BMI),mild hyperglycemia].The glutamate decarboxylase 65(GAD-65)ELISA testing was negative in 5 cases,which were selected for exome sequencing.
Prospective enrollment from Lahore(n = 15)
A total of 15 patients were enrolled from The Diabetes Clinic at PHRC Research Centre,Fatima Jinnah Medical University,Lahore.The inclusion criteria were onset of diabetes before 25 years of age with preferential first-degree family history of diabetes.The demographic information along with history of disease(onset,complications,treatment details and family history)was collected on proforma.On the basis of BMI and preserved fasting C-peptide,GAD-65 ELISA was performed on three samples where serum was available.The two that were found negative for GAD-65 and,along with another three patients having clinical features like low BMI but for whom serum was not available,were finally selected for exome sequencing.All the selected subjects had normal c-peptide values(range:0.8-3.8 ng/mL)
Prospective enrollment from Karachi(n = 89)
The patients coming to the diabetes clinic for type-1 an d type 2 diabetes treatments were enrolled if onset of the disease was below 25 years of age,with family history of diabetes as preference.Information on demography,treatment,and diagnosis was taken on pre-designed proforma.Height and weight were recorded for BMI.The GAD-65 ELISA was performed on 59 patients.Patients that were GAD-65 negative and additional patients with low BMI but no available serum for testing were selected for exome sequencing(n= 18)(Figure 1).
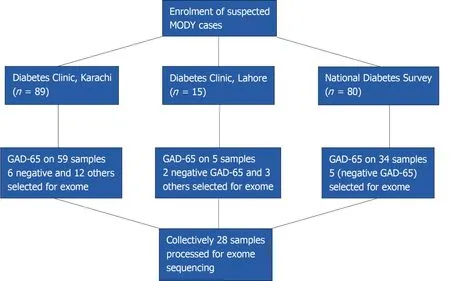
Figure 1 Flow chart for enrollment of patients.GAD-65:Glutamate decarboxylase.
A total of 28 patients were selected for exome sequencing.Their median age at diagnosis was 18 years and median BMI was 22.Among them,17 were taking treatment(14 insulin and 3 were taking OHA agent)and 5 were not taking any treatment.
The DNA was extracted using a QIAamp DNA mini kit(QIAGEN).The GAD-65 autoantibody test was done by using a KRONUS Elisa kit.C-peptide test was commercially tested in a diagnostic laboratory.Exome sequencing was performed by Genome Quebec,Canada.Exome sequencing was carried out with 50 Mb Agilent Sure select array and sequencing on Illumina Hi-seq at 50 × depth.
Written informed consent was taken from participants/parents/guardians prior to enrollment.The Ethical Clearance was taken from Institutional Bioethics Committee of Pakistan Health Research Council and Ethical Review Board of Pakistan Institute of Medical Sciences,Shaheed Zulfiqar Ali Bhutto Medical University,Islamabad.
Data analysis
For bioinformatics analysis,the FastQ files were processed using the best practices recommendations of the genome analysis tool kit(GATK).The reads were mapped to human reference genome GRCh37 using Burrow-Wheeler alignment(BWA-MEM).The Picard tool was used to mark and remove duplicate alignments and then indel realignment and base quality score recalibration was done.The gVCFs were generated with GATK HaplotypeCaller and joint variant calling was done.The variants with low map reads(DP < 20)and low genotype quality(GQ < 20)were discarded.Annovar was used to annotate the variants.UTR,synonymous,and intronic(unless splicing)variants were discarded by standard procedure to focus on protein altering ones.These variants were filtered for frequency in three public databases(Exome Aggregation Consortium,1000 genomes,and gnomAD version v2.1.1).For dominant genes,maximal allele frequency cutoff was 0.0001 in any population while it was 0.005 for recessive genes(ACMG PM2 criterion).The missense variants were selected only if predicted disease-causing by the majority of 10 algorithms used(see legend to Table 1)which satisfies PP3 by ACMG/AMP criteria).The computational evidence supports deleterious effects on the gene[11].Analysis of the exome focused on the genes listed in the University of Chicago monogenic diabetes panel(https://dnatesting.uchicago.edu/tests/monogenic-diabetes-panel).Variant coordinates were searched for functional domains in uniport.org(results shown in Table 1).
RESULTS
A total of 80 cases selected from the National Diabetes Survey with the criteria of having diabetes and onset of disease before 25 years of age.The median age was 24 years.Most of them(62%)were females and average BMI was 28 kg/m2(Table 2).Of them,based on clinical criteria and negativity for antibody,five were selected for exome sequencing.The results revealed three novel missense variants identified in three patients.Two of the variants belong to the OMIM-listed MODY genes,i.e.HNF1A(c.169C>A,pLeu57Met)andKLF11(c.401G>C,p.Gly134Ala)and one inRFX6(c.919G>A,p.Glu307Lys),a gene recently described as mutated in dominant,MODYlike diabetes.
In the prospective enrollment from Lahore,15 patients were enrolled.The mean age for onset of disease in patients was 23 years and mean BMI was 21.5 kg/m2.The fasting blood glucose on average was 239.26 mg/dL.The HbA1c ranged from 6% to 11%(Supplementary Table 1).The exome analysis revealed a variant inHNF1B(c.1058C>T,p.Ser353Leu)for one patient.
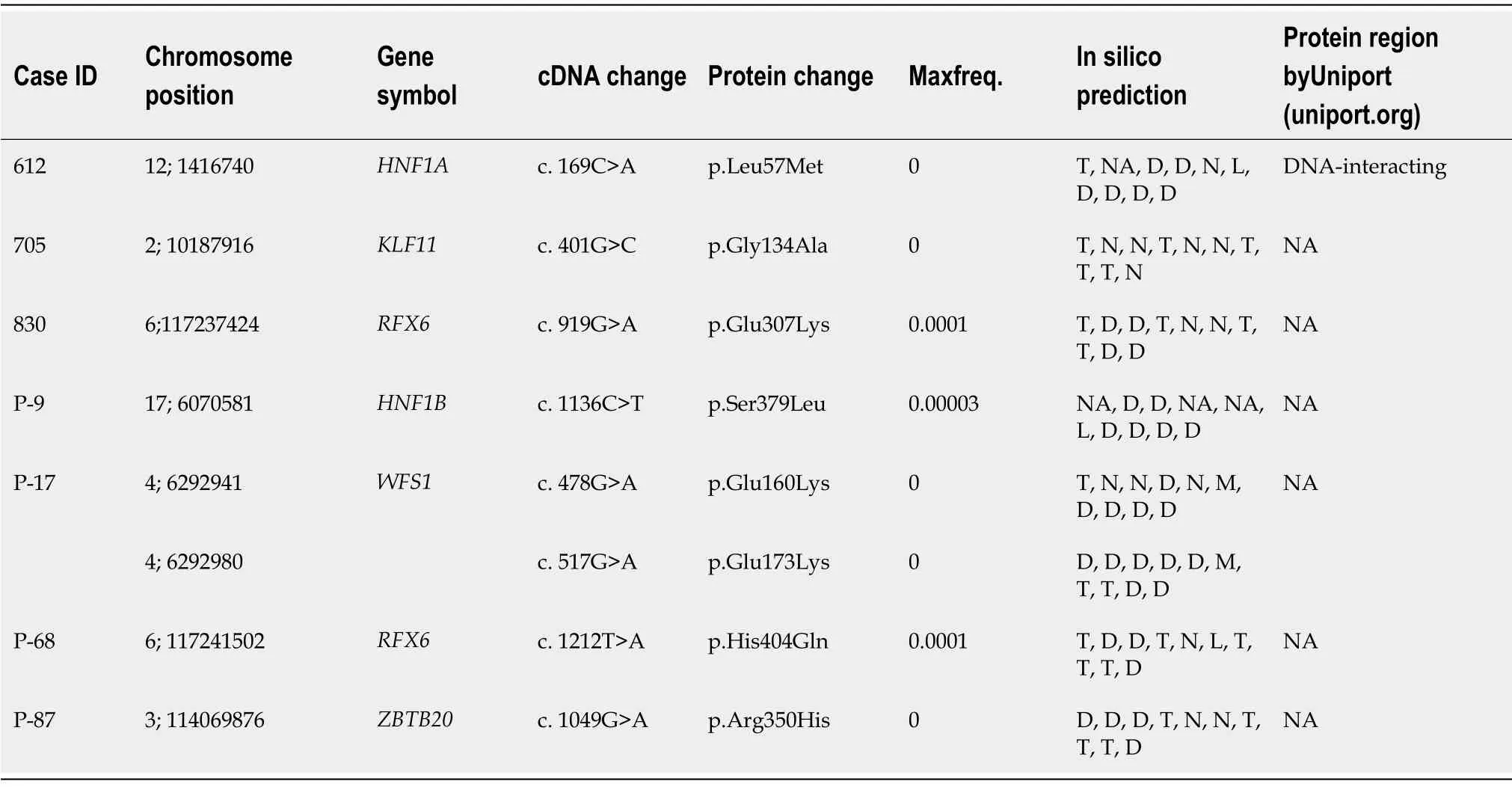
Table 1 Rare variants in cases of monogenic forms of diabetes
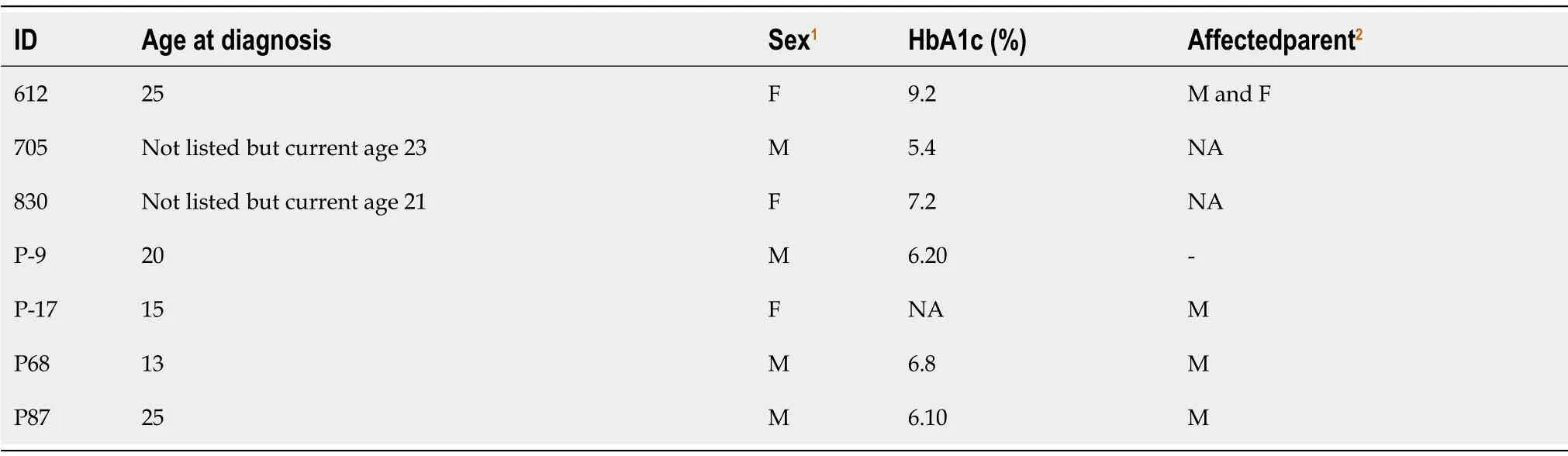
Table 2 Demographic features of patients having causal variants
In the prospective enrollment from Karachi,89 diabetic patients were enrolled.The mean age of onset of disease was 16.5 years and the current age was 26.2 years.Mean BMI was 24.3 kg/m2.Most of them(95%)had family history of diabetes.Among them,93% were taking treatment for diabetes and 68% were taking insulin.About 25% of thepatients had a MODY probability score more than 75%.The serum was available for 59 patients.The GAD-65 autoantibody test was conducted on all patients and 15 of them were negative.The patients whose serum was not available were shortlisted on the basis of low BMI,family history,and HbA1c levels.
Among them,18 were selected for exome sequencing,which revealed potentially causal variants in three patients(Table 1).One had a variant inRFX6(c.1212T>A,p.His404Gln)and a second one was a compound heterozygote(Supplementary Figure 1)forWFS1(c.478G>A,p.Glu160Lys and c.517G>A,p.Glu173Lys).RecessiveWFS1mutations cause Wolfram syndrome,but non-syndromic diabetes alone is also seen[12].A third patient had a variant inZBTB20(c.1049G>A,p.Arg350His).
The mutation in this gene is responsible for causing primrose syndrome[13].The patient did not have the other manifestations of this syndrome.
All variants reported here were missense,all satisfied the PP3 and PM2 criteria but,being novel and not having previously been reported,all classified as VUS.Nevertheless,our extremely low cut-off for allele frequency of 0.0001(more than two orders of magnitude lower than the ACMG/AMP cut-off for a VUS),minimizes the probability of spurious variants.The MODY probability score was calculated for all the participants.More than half of the patients in National Diabetes Survey,all patients from Lahore and one fourth of patients from Karachi had probability scores more than 75%.The probability score was calculated by MODY probability calculator developed for Caucasian population.
DISCUSSION
Exome sequencing of 28 suspected patients for MFD identified missense novel variants in 7 patients(with the caveat thatKLF11,BLK,andPAX4are not universally accepted as genes whose mutation causes diabetes).Previous studies from Pakistan have discussed the importance of diagnosing MFD in Pakistan[9,10].However,to the best of our knowledge,this is the first comprehensive study from the country to enroll suspected MFD patients for exome sequencing.
We enrolled diabetic patients with early onset of disease below 25 years of age with clinical features suggestive of MFD and negative(or unknown)for GAD65 auto antibodies.The probability score was calculated by using the MODY probability calculator developed for the Caucasian population[14].However there is a need to validate this with South Asian populations[15].
In determining the type of MFD,ethnic differences play an important role.There is wide variation of prevalence of different MFD types in different areas.TheHNF1AMODY type was reported to be more prevalent in northern Europe whileGCKMODY types in southern Europe[16].There were similar findings from United States regarding the three major prevalent types of MODY in their population[17].The study on Russian children with non-type 1 diabetes showed that the most prevalent MODY type wasGCK,with only 18% of variants in other than known MODY genes.The studies from China reported thatHNF4AMODY types were relatively less common as compared to Europeans[18,19].A study from Korea also has a similar finding that common MODY types prevalent in Europe were not common there,but instead they found variants in three new genes,including theWFS1gene[20].The MODY landscape in India is also complex,reporting MODY types other than common genes known in European population[21,22].A study from Oman reported that variants were not found in three common MODY genes[23].Similar findings were also reported from Tunisia that common MODY types were not found there and concluded that other genes might be responsible for young onset diabetes in their population[24,25].These discrepant results may be only partially explained by different methodologies and different selection criteria for testing.
We found in our study in a total 28 screened patients that 3 of the patients tested had variants in OMIM-listed MODY genes while 4 had variants were in other genes also known to be mutated in MFD.Similar findings were also reported from Norway[26],France[27]and Sweden[28].Two novel missense variants were found inRFX6(c.G919A,p.E307K).In addition to Europeans[29],variants from this gene was also reported in studies from India[22]and Japan[30].One patient was compound heterozygous atWFS1,the gene mutated in Wolfram syndrome and also responsible for nonsyndromic diabetes.WFS1variants have been reported from India[22],China[12,31],Korea[20,32,33],Russia[6],and European ancestry[34].Finally,one variant was identified inZBTB20,a transcription factor that regulates the function of beta cells and glucose homeostasis[35-37].In humans,dominantZBTB20mutations cause Primrose syndrome.
The three variants in OMIM-listed MODY genes were found inHNF1A,KLF11,andHNF1B.TheHNF1A(MODY 3)is most common type prevalent in some European and Asian countries[38]and variants in this gene have been reported from various countries all around the world[5,39,40].The patients withHNF1Avariants respond well to sulfonylurea therapy[41].The other variant was found in theKLF11(MODY 7)[42];variants from this gene have been reported from France[43]and Japan[44],although recent literature disputes this gene as true MODY gene.One variant was found inHNF1B(MODY 5)as reported to account for 2%-6% of all the diagnosed MODY cases[45].This type was generally found to be associated with kidney dysfunction[46].Variants in this gene have been reported from different countries[47-49].Although it has been considered thatKLF11,BLK,andPAX4are not MODY causing genes,the OMIM and recent literature reported it as involved in MODY[3,6].
Pakistan,being a developing country,is facing a huge burden of diabetes as evident from the recent survey findings that 26% of adults in the general population were suffering from diabetes[7].There is a need to identify the genetic basis of the diabetes in Pakistan,with large-scale efforts of screening.As Pakistan is a limited-resource society,it is important to develop sensitive and population-specific criteria.We propose our paper contributes as the first step in this direction.
A study reported that 56% of the marriages were consanguineous,and among them,over 49% were first cousin marriages[50,51],suggesting unknown recessive genes,such as seen in non-syndromicWFS1cases.Studies from Pakistan on MFD were very scarce.According to American Diabetes Association,the diagnosis of MFD should be considered when there is a family history of diabetes with atypical features of diabetes,such as lacking obesity[52].There is a need to conduct large scale genetic studies on young onset diabetes to understand the genetic aspects from our country.
CONCLUSION
A wide spectrum of genetic variants involved in MFD was identified from this study.The common genes prevalent in European countries were not found common in this study.The genes other than commonly known MODY genes were identified.There is need for large scale genetic studies on early onset of diabetes in the country.
ARTICLE HIGHLIGHTS
Research background
The data on monogenic forms of diabetes(MFD)was lacking from Pakistan.
Research motivation
The identification of MFD from Pakistan will paved the way for better diagnostics and treatment for patients
Research objectives
To identify the genetic variants for MFD.
Research methods
Exome sequencing was used.
Research results
The wide spectrum of genetic variants was identified.
Research conclusions
The MODY genes prevalent in other countries,like those in Europe,were not found common in our population
Research perspectives
More studies are required,keeping in view the consanguinity rate in Pakistan
ACKNOWLEDGEMENTS
The authors thank all the participants and their guardians for participating in the study.Thanks to Ms.Raheela and Bilal Tahir from Baqai Institute of Diabetology and Endocrinology for assistance in patient enrollment and all National Diabetes Survey of Pakistan members.
杂志排行
World Journal of Diabetes的其它文章
- Letter to editor ‘Gastroenteropathy in gastric cancer patients concurrent with diabetes mellitus’
- Adherence to Mediterranean diet and advanced glycation endproducts in patients with diabetes
- Association between admission hemoglobin level and prognosis in patients with type 2 diabetes mellitus
- Factors influencing the effectiveness of using flash glucose monitoring on glycemic control for type 1 diabetes in Saudi Arabia
- p66Shc-mediated oxidative stress is involved in gestational diabetes mellitus
- Profilin-1 is involved in macroangiopathy induced by advanced glycation end products via vascular remodeling and inflammation
